Abstract
HIV-1 protease has been an important drug target for the antiretroviral treatment of HIV infection. The efficacy of protease drugs is impaired by the rapid emergence of resistant virus strains. Understanding the molecular basis and evaluating the potency of an inhibitor to combat resistance are no doubt important in AIDS therapy. In this study, we first identified residues that have significant contributions to binding with six substrates using molecular dynamics simulations and Molecular Mechanics Generalized Born Surface Area calculations. Among the critical residues, Asp25, Gly27, Ala28, Asp29, and Gly49 are well conserved, with which the potent drugs should form strong interactions. We then calculated the contribution of each residue to binding with eight FDA approved drugs. We analyzed the conservation of each protease residue and also compared the interaction between the HIV protease and individual residues of the drugs and substrates. Our analyses showed that resistant mutations usually occur at less conserved residues forming more favorable interactions with drugs than with substrates. To quantitatively integrate the binding free energy and conservation information, we defined an empirical parameter called free energy/variability (FV) value, which is the product of the contribution of a single residue to the binding free energy and the sequence variability at that position. As a validation, the FV value was shown to identify single resistant mutations with an accuracy of 88%. Finally, we evaluated the potency of a newly approved drug, darunavir, to combat resistance and predicted that darunavir is more potent than amprenavir but may be susceptible to mutations on Val32 and Ile84.
Keywords: HIV-1 protease, drug resistance, MM/GBSA, molecular dynamics simulations, darunavir
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) protease is essential for cleavage of the viral gag and pol polyproteins, releasing both structural and enzymatic proteins necessary for viral maturation.1,2 Since inhibition of the HIV protease function will prevent the maturation of these viral proteins and thus the replication of the virus, HIV-1 protease has been an important target of AIDS therapy. HIV-1 protease is a homodimeric aspartic protease and its substrate binding pocket includes the Asp25(25′)-Thr26(26′)-Gly27(27′) catalytic triad and flap regions, which presumably open and close to allow entry and binding of substrates or inhibitors.3,4 The active site of HIV-1 protease can be divided into eight subsites S4-S3-S2-S1-S1′-S2′-S3′-S4′ and the eight corresponding substrate residues are denoted as P4-P3-P2-P1-P1′-P2′-P3′-P4′, where the scissile bond is between P1 and P1′. Currently, there are nine Food and Drug Administration (FDA)-approved protease drugs: saquinavir (SQV), ritonavir (RTV), indinavir (IDV), nelfinavir (NFV), amprenavir (APV), lopinavir (LPV), atazanavir (AZV), tipranavir (TPV), and darunavir (TMC114). These inhibitors are peptidomimetics designed based on the natural substrates, in which the scissile peptide bond was replaced by an uncleavable isostere mimicking the transition state. In the presence of inhibitors, HIV-1 protease variants are under selection pressure and those not binding to the inhibitors but still maintaining catalytic activity would become dominant in the virus population because of the high replication rate and low fidelity of the viral replication.5-7 Resistant mutations include residues directly interacting with inhibitor and those far away from the inhibitor binding site.5,8
Computational methods have been developed to investigate the molecular mechanism of drug resistance and evaluate the potency of an inhibitor to combat resistance before it goes through the long and costly procedure of drug development.9 The methods based on protein structure are particularly useful for mechanistic study and de novo prediction of resistant mutants with little or no prior knowledge. The performance of these methods depends on the availability and quality of the structural template as well as the scoring functions for binding affinity estimation. The recent advancements of force fields and free energy calculation have made structure-based methods more feasible in the studies of HIV-1 protease drug resistance.10
HIV-1 protease drug resistance is caused by mutations that significantly impair the protease’s interaction with drugs but not with substrates. While most structure-based approaches only consider the effect of resistant mutations on inhibitor binding,11-15 Wang and Kollman developed a procedure in 2001 to study the HIV-1 protease’s drug resistance using free energy decomposition and sequence conservation analysis. They compared the contribution of each protease residue to binding with five drugs approved by FDA then and with one substrate, and proposed the following mechanism for drug resistance: if a residue is not conserved, presumably it is not important for viral function, but interacts more favorably with a drug than with the substrate, mutations of this residue may not affect the function of the protease but may impair the binding of the drug and thus cause resistance to the drug.16 Because no crystal structure of any protease-substrate was available at that time, they modeled the substrate-protease complex based on the crystal structure of a peptide inhibitor JG365 bound to the HIV PR. In 2002, crystal structures of six natural peptide substrates complexed with an inactive form of HIV-1 protease were solved,17 which laid the ground for further investigation of the molecular basis for drug resistance. In the current study, we analyzed the competitive binding between inhibitors and substrates of the HIV protease using a protocol similar to that proposed by Wang and Kollman. Compared with the previous work, we used crystal complex structures for six substrates and the accuracy of predictions has been improved. Given the significantly more computation involved in the current study, Molecular Mechanics Generalized Born Solvent Area (MM/GBSA) was used for free energy calculation. MM/GBSA is an approximation that is more efficient but may be slightly less accurate than MM/PBSA. Our calculations showed that MM/GBSA was sufficiently accurate to achieve comparable results with those obtained by MM/PBSA in the previous work. We showed the success of MM/GBSA on predicting single resistant mutations, which suggests the potential of applying the protocol to predict multiple resistant mutations and/or evaluate the potency of a large number of inhibitors during drug development. When this study started, darunavir (TMC114) was being tested in Phase-III clinical trials. Darunavir was approved in June 2006 but not much resistant data has been accumulated. We thus also predicted its potency to combat resistance caused by single mutations of HIV-1 protease.
RESULTS AND DISCUSSION
Identification of critical residues for substrate binding
We first calculated binding free energies for six substrates and eight drugs (Table I, Fig. 1). Since using separate molecular dynamics (MD) trajectories for complex, protein and ligand in binding free energy calculation often introduced significant fluctuation,15,18 we chose to run a single trajectory on the complex such that the simulation errors and noise could be partially cancelled. It is not surprising that on average inhibitors bind more tightly to the protease than substrates: the average binding affinities of the eight inhibitors and six substrates were -36.7 kcal/mol and -31.4 kcal/mol, respectively. To identify critical residues for substrate binding, we calculated the contribution of each individual protease residue to binding with each substrate (Fig. 2 and Fig. S2 in the supplemental data). Since there were a large number of energy decomposition calculations involved in our analysis, the PB model used in our previous study16 was too computationally expensive and was replaced by a Generalized Born (GB) model to compute solvation free energy more efficiently. The contribution of individual residue to binding varies in the range of +1 to -10 kcal/mol. The significant residues to binding are mainly located in three regions, the catalytic site (Asp25, Gly27, Ala28, Asp29, and Asp30), flap (Ile47, Gly48, Gly49, and Ile50) and the C-terminus region (Pro81 and Ile84). Among these residues, Asp25, Gly27, Ala28, Asp29, and Gly49 are well conserved and presumably functionally important, with which potent drugs should form strong interactions. The interactions between individual protease residues and six drugs were also calculated (Fig. S3 in the supplemental data). The overall interaction spectrums of substrates and inhibitors are similar but the subtle differences cause drug resistance. It should be noted that in these five conserved residues, four of them are located in the core region of the active site, including Asp25, Gly27, Ala28, and Gly49. All studied inhibitors have very strong interactions with these four residues. Moreover, no space exists between inhibitors and these four residues to grow new functional groups from the existing inhibitors. So, we think that Asp29 is more important than the other four residues when we want to design inhibitors that are less prone to resistance.
Table I.
Binding Free Energies of the Substrates and Inhibitors (kcal/mol)a
| Name | ΔGbind (exp.) | ΔEele | ΔEvdw | ΔGSA | ΔGGB | TΔS | ΔGtotal |
|---|---|---|---|---|---|---|---|
| MC-CAb | N/A | -101.6 ± 8.5 | -95.5 ± 2.2 | -13.8 ± 0.1 | 137.1 ± 6.5 | -40.4 ± 1.2 | -33.4 ± 4.3 |
| CA-P2b | N/A | -39.7 ± 4.0 | -84.9 ± 0.5 | -12.5 ± 0.0 | 82.2 ± 2.9 | -34.2 ± 0.6 | -20.8 ± 2.6 |
| P2-NCb | N/A | -39.7 ± 12.7 | -95.5 ± 0.4 | -12.6 ± 0.1 | 78.3 ± 15.0 | -41.3 ± 1.4 | -28.3 ± 2.9 |
| P1-P6b | N/A | 82.3 ± 5.8 | -93.6 ± 0.1 | -13.4 ± 2.9 | -41.1 ± 6.0 | -37.6 ± 0.1 | -28.3 ± 0.0 |
| RT-RHb | N/A | -323.0 ± 8.7 | -87.5 ± 0.8 | -12.1 ± 0.3 | 350.4 ± 8.0 | -32.5 ± 0.7 | -39.8 ± 1.0 |
| RH-INb | N/A | -49.4 ± 14.9 | -110.3 ± 2.7 | -14.7 ± 0.1 | 102.6 ± 16.0 | -33.9 ± 0.9 | -37.8 ± 1.7 |
| SQV | -13.0 | -46.3 ± 0.5 | -73.5 ± 1.9 | -10.3 ± 0.0 | 66.5 ± 2.9 | -32.0 ± 0.8 | -31.6 ± 0.4 |
| IDV | -12.4 | -63.3 ± 2.0 | -74.3 ± 0.4 | -9.9 ± 0.0 | 75.1 ± 0.7 | -27.0 ± 0.3 | -45.5 ± 0.9 |
| RTV | -13.7 | -47.8 ± 4.9 | -77.5 ± 0.5 | -10.5 ± 0.2 | 64.3 ± 3.2 | -31.8 ± 0.8 | -39.6 ± 1.0 |
| NFV | -12.8 | -54.9 ± 3.1 | -66.2 ± 1.2 | -10.0 ± 0.0 | 70.2 ± 1.9 | -29.7 ± 1.1 | -31.2 ± 0.0 |
| APV | -13.2 | -50.8 ± 2.2 | -64.5 ± 0.5 | -9.2 ± 0.0 | 61.2 ± 2.0 | -27.0 ± 0.4 | -36.2 ± 0.6 |
| LPV | -15.1 | -27.9 ± 0.3 | -75.7 ± 0.8 | -10.0 ± 0.0 | 50.1 ± 0.3 | -33.2 ± 0.7 | -30.4 ± 0.8 |
| TPV | N/A | -49.4 ± 1.0 | -65.0 ± 0.5 | -8.4 ± 0.0 | 57.0 ± 0.4 | -24.8 ± 1.2 | -40.9 ± 2.0 |
| TMC114 | -15.2 | -41.7 ± 1.0 | -66.8 ± 0.5 | -9.4 ± 0.0 | 53.5 ± 1.4 | -26.0 ± 0.1 | -38.4 ± 0.6 |
The experimental binding free energies for SQV, IDV, RTV, NFV, APV, and LPV were reported by Ohtaka et al.,21 and that for TMC114 was reported by King et al.19
The sequences of the six natural substrates are VSQNYPIVQN (MC-CA), KARVLAEAMS (CA-P2), PATIMMQRGN (P2-NC), RPGQFLQSRP (P1-P6), GAETFYVDGA (RT-RH), and IRKILFLDGI (RT-IN).
Figure 1.
The 2D chemical structures of the eight FDA-approved drugs.
Figure 2.
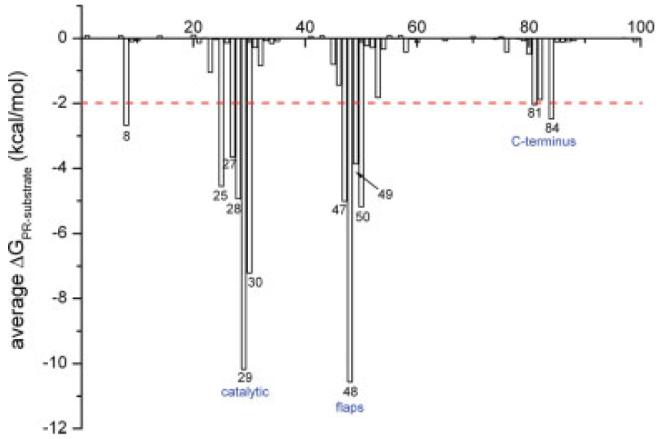
The average free energy interaction between the six substrates and each residue of the HIV-1 protease.
De novo identification of resistant mutations
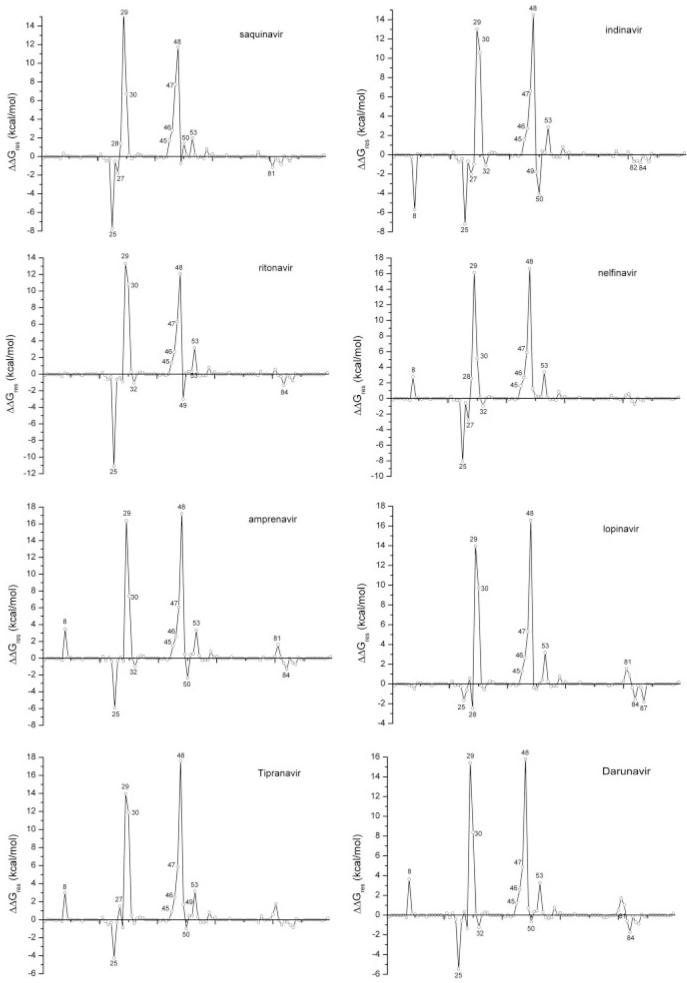
To identify and quantitatively evaluate resistant mutations, we first calculated ΔΔGres, the difference between the protease residues’ contributions to the binding free energy of drugs and substrates, using MM/GBSA (see Fig. 3). The HIV-1 protease is a dimer and a single mutation of its gene is a double mutation of the protein. The ΔΔGres values shown in Figure 3 were averaged over two monomers. Seven residues, including two in the catalytic region (Asp29 and Asp30) and five in the flap region (Lys45, Met46, Ile47, Gly48, and Phe53), interact at least 2.0 kcal/mol more favorably with substrates than with inhibitors. The structural alignment between seven drugs and one substrate MA-CA (Fig. S4 in the supplemental data) showed that these residues can form effective contacts with the P3 and P4 groups in the substrates but not with drugs.
Figure 3.
Free energy difference between each residue’s contribution to the binding with drugs and substrates (ΔΔGres = ΔGresdrug - ΔGressubstrate). The average interaction spectrum for the six substrate-PR complexes was used in the comparison with that for each drug.
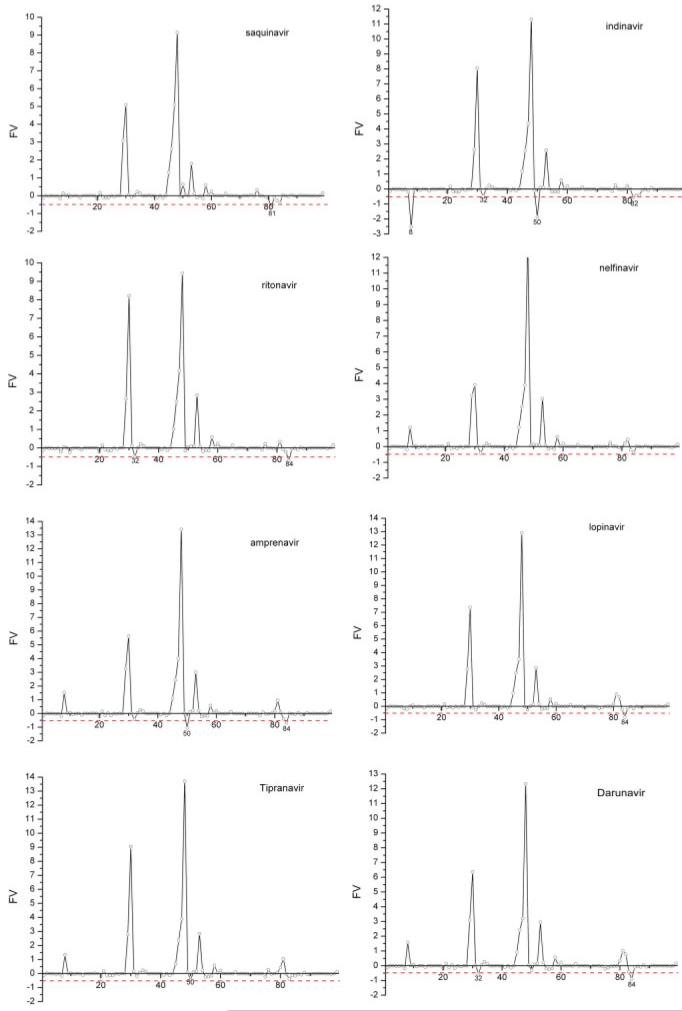
We then used a heuristic FV value to quantitatively combine free energy and conservation information. A large negative FV value often suggests that a residue is variable (larger variability, less conserved) and it interacts more favorably with drugs than with substrates. Mutation on this residue may severely impair the binding of drugs but not that of substrates, and the enzymatic function of the protease is presumably not affected because this residue is presumably functionally unimportant. We have calculated the FV values for all single mutations and predicted whether such a mutation would cause resistance to drugs (Fig. 4 and Table II). The residues with FV values smaller than -0.5 are listed in Table S1 in the supplemental data. In Table II, only five FDA-approved drugs were included for comparison because affinities for lopinavir and tipranavir binding to single mutants of HIV protease had not been reported when we conducted the study. In the present work, a single protease mutation causing a decrease of binding affinity for a drug by at least 10 folds, that is, 1.4 kcal/mol of binding free energy, is considered resistant to the drug. A residue with variability higher than 0.35 is considered not conserved. Using a value of -0.5 (= 1.4 × 0.35) as the threshold for the FV value, the average accuracy (the ratio of correctly predicted mutations to the total mutations) of our predictions for five drugs is 88%, which is a significant improvement over the 76% accuracy of the previous study.16 This improvement of performance is likely to be the result of comparing the interacting profiles of inhibitors to those of six substrates as well as starting MD simulations from the crystal structures of the inactive substrates. The improved accuracy of prediction also shows that the efficient GB model can be used to identify resistant mutations of HIV protease, which is critical for evaluating the resistance effects of multiple mutations and screening large chemical compound databases to search for new potent inhibitors.
Figure 4.
The distributions of the FV values for eight HIV protease drugs (the residues with FV values larger than 2.0 are not shown). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Table II.
The Predictions of Drug Resistance Based on the Predicted FV Values
| Resistance |
||||
|---|---|---|---|---|
| Position | Experimental Ki (mut)/Ki (wt) | FV | Experiment (prediction) | Accuracy |
| Saquinavir | ||||
| V32 | 1.6-7.3 (V32I)50 | -0.1 ± 0.0 | N (N) | 75% |
| M46 | 1.0 (M46I)51 | +2.6 ± 0.4 | N (N) | |
| I47 | 1.0 (I47V)51 | +5.1 ± 0.3 | N (N) | |
| G48 | 13.5 (G48V)52 163.6 (G48V)53 | +8.0 ± 1.3 | Y (N) | |
| I50 | 21.0 (I50V)51 | +0.6 ± 0.1 | Y (N) | |
| V82 | 0.7-3.7 (V82A/F/I)50 3.3-7.3 (V82F/A/I)54 | -0.1 ± 0.3 | N (N) | |
| I84 | 5.8 (I84V)50 10.7 (I84V)54 12.0 (I84V)51 | -0.4 ± 0.0 | N (N) | |
| L90 | 3.0 (L90M)52, 55 20.7 (L90M)53 | -0.0 ± 0.1 | N (N) | |
| Amprenavir | ||||
| M46 | 1.0 (M46I)51 | +2.5 ± 0.3 | N (N) | 100% |
| I47 | 1.0 (I47V)51 | +4.0 ± 0.2 | N (N) | |
| G48 | 3.5 (G48V)53 | +13.4 ± 1.3 | N (N) | |
| I50 | 83.0 (I50V)51 | -1.1 ± 0.6 | Y (Y) | |
| V82 | 0.4-3.3 (V82A/F/I)54 | +0.0 ± 0.1 | N (N) | |
| I84 | 23.0 (I84V)51 2.7 (I84V)54 | -0.7 ± 0.1 | Y (Y) | |
| L90 | 2.7 (L90M)53 | -0.1 ± 0.2 | N (N) | |
| Ritonavir | ||||
| M46 | 4.0 (M46I)51 | +2.5 ± 0.2 | N (N) | 86% |
| I47 | 3.0 (I47V)51 | +4.2 ± 0.5 | N (N) | |
| G48 | 66.7 (G48V)53 | +9.4 ± 1.9 | Y (N) | |
| I50 | 10.0 (I50V)51 | -0.0 ± 0.4 | N (N) | |
| V82 | 0.8-14.7 (V82A/F/I)54 | -0.1 ± 0.3 | N (N) | |
| I84 | 11.2 (I84V)54 20.0 (I84V)51 | -0.7 ± 0.2 | Y (Y) | |
| L90 | 6.7 (L90M)53 | -0.0 ± 0.0 | N (N) | |
| Indinavir | ||||
| V32 | 8.0 (V32I)50 | -0.5 ± 0.2 | N (Y) | 86% |
| M46 | 4.3 (M46I)50 | +2.6 ± 0.6 | N (N) | |
| I47 | 3.0 (I47V)51 | +4.4 ± 2.3 | N (N) | |
| G48 | 6.3 (G48V)53 | +11.3 ± 1.8 | N (N) | |
| V82 | 0.6-6.4 (V82A/F/I)54 6.9-84.7 (V82A/F/I)50 | -0.6 ± 0.2 | Y (Y) | |
| I84 | 2.6 (I84V)54 10.0 (I84V)50 | -0.4 ± 0.3 | N (N) | |
| L90 | 5.8 (L90M)53 | -0.0 ± 0.3 | N (N) | |
| Nelfinavir | ||||
| G48 | 1.0 (G48V)53 | +13.0 ± 0.1 | N (N) | 100% |
| V82 | 0.8-17.5 (V82F/A/I)54 | +0.5 ± 0.5 | N (N) | |
| I84 | 3.5 (I84V)54 | -0.4 ± 0.2 | N (N) | |
| L90 | 3.5 (L90M)53 | -0.0 ± 0.5 | N (N) |
Among the resistant mutations, mutations on Val32, Ile50, and Ile84 are resistant to most FDA-approved drugs. Correspondingly, most multiple-residue resistant mutations often include mutations on these three positions (Stanford HIV database8), which is consistent with the large negative FV values of these residues for most drugs in Figure 4. As these three residues are hydrophobic and close to the binding site, resistant mutations of these positions cause the inhibitors lose more van der Waals interaction with the protease than with the substrates. The qualitative study by King et al. also supports our quantitative results.19 King et al. compared the overlapping van der Waals volume of four substrates (substrate envelop) and that of eight inhibitors (inhibitor envelop). They showed that primary active site mutations form extensive contacts with inhibitors but not with substrates.
We observed several residues, most prominently Asp29, Asp30, Ile47, and Gly48 that form much more favorable interactions with substrates than with drugs and also have large positive FV values. Among these residues, Asp29 is well conserved (variability smaller than 0.25),16 which suggests that the potency of the current drugs to combat resistance can be improved if their interactions with Asp29 are enhanced. A feasible modification is to add polar groups to inhibitors at the P3/P3′ positions such that electrostatic interactions between Asp29/Asp29′ and inhibitors become more favorable. Our analysis suggests that this may be the reason for darunavir being more potent than amprenavir (see below). All drugs interact less favorably with Ile47 and Gly48, two residues in the flap region that are not conserved (variability >0.65) and presumably not important for viral function, than with substrates. In Table II, in vitro binding affinity assay showed that saquinavir and ritonavir but not amprenavir, indinavir and nelfinavir were sensitive to mutations on Gly48 and none of the above drugs were sensitive to Ile47 mutations. Our predictions agreed well with the experimental measurements except the resistant mutations of Gly48 to saquinavir and ritonavir. Wittayanarakul et al. studied the saquinavir resistance caused by Gly48 mutation using MD simulations and free energy calculations.20 They showed that the protein flaps underwent significant conformational changes after mutating Gly48 to Val48. Gly48 was in close contact with the Phe53 side chain of the protease and the P2 and P3 groups of saquinavir. The substituted valine at this position could not fit to the original hydrophobic pocket because the dimethyl groups had steric clash with the Phe53 and P3 side chains. Gly48Val mutation thus caused conformational change of the binding cavity and deteriorated the inhibitor’s binding. Therefore, it is possible to improve the potency of saquinavir by reducing the size of its P3 group.
In our analysis of resistant mutations, the conformational entropy of the protease and drugs/substrates were not included. The premise was that the main contribution to the change of the binding free energy caused by single mutations was from enthalpy, which was partially supported by the experimental measurements by Freire and coworkers.21 According to the thermodynamic dissection of the effects of V82F/I84V on the binding affinities of five HIV-1 protease drugs (indinavir, nelfinavir, saquinavir, ritonavir, and amprenavir) and two secondgeneration inhibitors (KNI764 and TMC126), for all inhibitors but TMC126, the entropy changes were negligible while the enthalpy changes were dominant.
Prediction of the potency of darunavir to combat resistance
Since drug development is expensive and time consuming, it is important to evaluate the potency of a drug lead at the early stage of drug development. The analysis described in the present study to identify resistant mutations for protease inhibitors can serve this purpose well and we demonstrated its usefulness on darunavir. Darunavir is a newly approved drug and has shown excellent promise as a treatment for HIV-1 infection in treatmentexperienced patients.22 The chemical structure of darunavir is quite similar to that of amprenavir with inhibition of several mutant viral strains resistant to other drugs. Darunavir was also shown to have a low liability for development of resistance compared with amprenavir and lopinavir by in vitro mutation experiments.23 We analyzed the potency of darunavir to combat resistance (Figs. 3 and 4). Our analysis showed that two residues (Val32 and Ile84) have FV values smaller than -0.5, which suggests that mutations on Val32 and Ile84 may cause resistance to darunavir. Therefore, we believe darunavir may be therapeutically effective when combined with other protease drugs insensitive to mutations on Val32 and Ile84 such as saquinavir and tipranavir.
The chemical structures of darunavir and amprenavir only differ by a second tetrahydrofuran ring, part of which is referred as bis-THF moiety. It is interesting to investigate whether and how this small difference can improve the potency of darunavir. We examined and compared the contribution of each residue to binding with darunavir and amprenavir (see Fig. 5). Eight residues with a difference larger than 1.0 kcal/mol are labeled in Figure 5. The distribution of these eight residues in the protease is shown in Figure S5 in the supplemental data. Among the eight residues, Ala28, Asp29, Ile47, and Gly48 have more favorable interactions with darunavir than with amprenavir and Ala28 and Asp29 are conserved. Only Gly27 is conserved among the four residues Gly27, Asp30, Ile50, and Val82 that have more favorable interactions with amprenavir than with darunavir.
Figure 5.
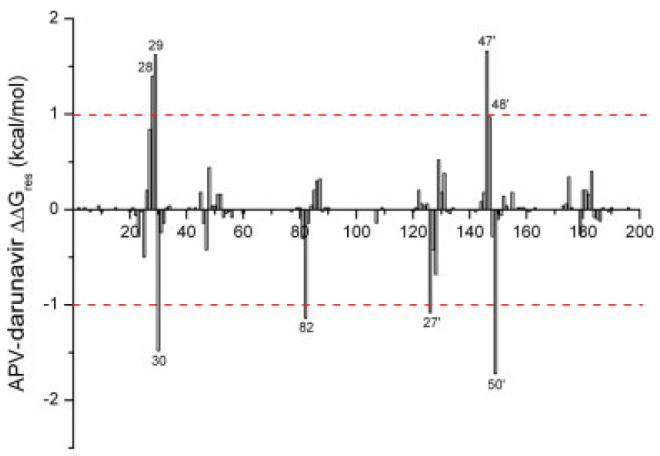
Free energy difference between each residue’s contribution to the binding with amprenavir and with darunavir. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The four residues with more favorable interaction with darunavir, Ala28, Asp29, Ile47, and Gly48, are located near bis-THF and the benzenesulfonamide moieties. As shown in Figure S5, a stable hydrogen bond is formed between the bis-THF and the backbone nitrogen atom of Asp29. Moreover, the electrostatic interaction between Ala28 and darunavir was stronger than that between Ala28 and amprenavir. Because both Ala28 and Asp29 are highly conserved, more favorable interactions with these residues make darunavir more potent than amprenavir. As we discussed in the previous and the present study,24 the potency of the currently FDA-approved drugs to combat resistant mutations might be enhanced if their interaction with Asp29 can be improved. The chemical modification of amprenavir to generate darunavir is along the line we have suggested. On the other hand, the hydrophobic benzene group at the P2′ site of darunavir can also form more favorable van der Waals interaction with the surrounding residues, especially with Ile47′, than amprenavir. The strong van der Waals interaction between Ile47′ and darunavir drags Gly48 closer to darunavir to form more favorable interactions, mainly electrostatic interactions, than that between Gly48 and amprenavir. As Ile47 and Gly48 are not conserved, darunavir may be more vulnerable to mutations on these two residues than amprenavir, which also suggests that the further improvement of darunavir is to reduce its interactions with these residues. Four residues, Asp30, Val82, Gly27′, and Ile50′, have less favorable interactions with darunavir than with amprenavir, among which Asp30, Ile50, and Val82 are highly variable and in fact mutations on Val82 and Ile50 cause resistance to multiple drugs. The weaker interaction of darunavir with Val82 and Ile50′ than amprenavir suggests that darunavir may be more potent to combat resistant mutations on Val82/Val82′ and Ile50/Ile50′ than amprenavir.
METHODS
Molecular dynamics simulations
The crystal structures of the wild-type HIV-1 protease complexed with eight inhibitors and six substrates were obtained from the Protein Data Dank (PDB).25 The PDB entries are: 1HXB (saquinavir),26 1HSG (indinavir),27 1HXW (ritonavir),28 1OHR (nelfinavir),29 1HPV (amprenavir),30 1MUI (lopinavir),31 1D4S (tipranavir),32 1SG6 (darunavir),33 1KJ4 (MA-CA),17 1F7A (CA-p2),17 1KJ7 (p2-NC),17 1KJF (p1-p6),17 1KJG (RT-RH),17 and 1KJH (RH-IN).17 No complex structure for atazanavir was available in PDB. The 2D structures for all inhibitors are shown in Figure 1. The Asn25 residues in the inactive HIV-1 protease complexed with six substrates were mutated back to Asp25 using SYBYL.34 The terminal Arg residue of substrate p1-p6 (1KJF) was not complete in the crystal structure and was removed in our simulation. Phe82 and Val84 in the crystal structure of tipranavir (1D4S) were mutated back to the wild type residues Val82 and Ile84, respectively. Hydrogen atoms were added using the leap module in AMBER 8.0 software package.35 The protonation states of the ionizable residues, except for Asp25/Asp25′, were assigned based on their pKa values at pH = 7. Since kinetic studies of HIV-1 protease have shown that only one of the catalytic Asp residues is protonated and the other is not, only the mono-protonated protease was considered36 and a proton was added to oxygen OD1 of Asp25, the oxygen closest to the inhibitor or substrate. The AMBER03 force field was used for standard amino acids37 and the general AMBER force field (gaff) was used for inhibitors.38 The force field parameters for the inhibitors were generated using the Antechamber program in AMBER8.0.35 Hydrogen atoms were first added to the crystal structure coordinates of inhibitors with hybridization of covalent bonds being considered. Next, after AM1 geometry optimization of the inhibitor structure, the electrostatic potential was calculated using Gaussian9839 with single-point Hartree-Fock (HF)/6-31G*, to which the atomic partial charges were fitted using RESP.40 Each protein-substrate/inhibitor complex was immersed in a rectangular box of TIP3P water molecules,41 and all water molecules in the crystal structures were kept. The water box extended at least 10 Å away from any solute atom. Particle mesh Ewald (PME) was employed for an adequate treatment of long-range electrostatic interactions.42 Counter ions of Cl- were placed to grids of the largest positive electrostatic potentials around protease to neutralize the entire system.
Prior to MD simulations, the whole system was relaxed using 500 steps of steepest descent followed by 2500 steps of conjugate gradient optimization. The MD simulations were performed in the NPT ensemble with a target temperature of 300 K and target pressure of 1 atm. A Berendsen coupling time of 0.2 ps was used to maintain the temperature and pressure of the system.43 SHAKE was used to constrain all bonds involving hydrogen atoms44 and the time step of MD was 2.0 fs. The system was first gradually heated from 10 to 300 K over 20 ps. Initial velocities were assigned from a Maxwellian distribution at the starting temperature. The system was equilibrated at 300 K for 100 ps followed by a production phase between 100 and 500 ps.
MM/GBSA free energy calculations
The free energy calculations were conducted using MM/GBSA45,46 in AMBER8.0.35 The binding free energy ΔGbinding was calculated as:
| (1) |
where (Gcomplex), (Gprotein), and (Ginhibitor) are free energies of protein, and inhibitor, respectively, and each of them was estimated by summing the contributions of gas phase energy Egas, solvation free energy Gsolvation, and conformational entropy—TS. Egas was obtained by summing molecular mechanical (MM) energies of internal, electrostatic, and van der Waals. Gsolvation equals the sum of polar (Gpolar) and nonpolar contribution (Gnonpolar). Gpolar was calculated by the GB model using the parameters developed by Onufriev et al.47 and the values of the interior and the exterior dielectric constants were set to 1 and 80, respectively. Gnonpolar was calculated from solvent accessible surface area determined by the LCPO method48: Gnonpolar = 0.0072 × SASA. Egas, Gpolar, and Gnonpolar were computed from 100 snapshots taken every 4 ps from 100 ps to 500 ps of the MD simulation on the complex. Normal mode analysis was performed to estimate the conformational entropy change upon ligand binding by the nmode program in AMBER8.0.35 Because of the high computational demand, only 20 snapshots were used in normal mode analysis and each snapshot was optimized for 100,000 steps using a distance-dependent dielectric of 4rij (rij is the distance between atoms i and j) until the root-mean-square of the gradient vector was less than 5 × 10-5 kcal mol-1 Å-1.
Evaluation of drug resistance based on energy decomposition and sequence analysis
The empirical parameter free energy/variability (FV) value was defined by Wang and Kollman,16 which is the product of , the difference of residues’ contributions to the binding free energy (ΔGres) between drugs and substrates, and the variability of each position. The variability Vi is calculated as:
| (2) |
where Wj is the weight of the jth sequence. Wj is calculated for each sequence in the alignment of 80 different sequences on the basis of sequence identity, and Pij represents how likely the amino acid aj in the jth sequence can be mutated to the amino acid ai in the ith sequence. The variability at each position of the HIV-1 protease is shown in Figure S1 in the supplemental data. ΔGres was computed using the MM/GBSA decomposition analysis in the mm_pbsa module of AMBER for each HIV protease residue in all protease-substrate/inhibitor complexes. ΔGres is the sum of van der Waals (ΔGvdw), electrostatic (ΔGele), and solvation (ΔGsolvation) energies.
| (3) |
All energy components in Eq. (2) were calculated using the 100 snapshots taken between 100 and 500 ps MD simulations. The framework of GB allows the decomposition of electrostatics on per-residue (atom) basis. In MM/GBSA, the contribution of residue j to the electrostatic free energy is calculated as49:
| (4) |
In Eq. (4), the first term determines the electrostatic contribution to solvation free energy calculated by GB for residue j in snapshot i of molecule x, and the second term is the gas-phase Coulombic interaction energy. ∈W is the dielectric constant of solvent; qk and ql are atom partial charges; rkl is the distance between atom k and l; is a smooth function that depends on atom radii and the distance between two atoms. The detailed description of MM/GBSA decomposition procedure used in AMBER was reported by Gohlke et al.49 The free energy decomposition based on MM/PBSA or MM/GBSA is only a pragmatic way to estimate the contribution of each residue to protein—ligand interactions.
CONCLUSIONS
Using MD simulation and free energy calculation, we have identified critical residues for substrate binding, among which Asp25, Gly27, Ala28, Asp29, and Gly49 are well conserved, with which the potent drugs should form strong interactions. We then compared the single residue interaction profiles of seven FDA-approved drugs with six natural substrates of HIV protease based on the recently solved complex structures of inactive protease and substrates. Our analysis further supports the molecular basis of drug resistance proposed in our previous study: when a mutation of a not-well conserved residue, presumably unimportant for viral function, impairs the binding of inhibitors more than substrates, drug resistance may occur. We thus suggest that an ideal drug should only form strong interactions with the most conserved residues such as Asp25, Gly27, Ala28, Asp29, and Gly49. Since mutations on these residues would harm the enzymatic function of the HIV protease, drug resistance mutations are less likely to occur. The potency of current drugs to combat resistance can be improved if their interactions with conserved residues particularly Asp29 are enhanced.
On the basis of the proposed resistance mechanism, a heuristic parameter FV value was used to successfully identify most single drug resistance mutations. The average accuracy of our prediction is 88%, which is significantly improved from 76% in our previous study.16 The improvement is mainly due to the use of more substrates (six compared with one) and the use of crystal structures of the protease/substrates complexes rather than a modeled complex. More encouragingly, we used a GB model in calculating single residue contribution to binding, which is more efficient than the PB model used in the previous study. This more efficient protocol makes it possible to evaluate resistance properties of multiple residue mutations as the combination of mutations is a large number. It also makes it possible to evaluate the potency of a large number of drug leads before they go through the expensive and time consuming drug development process.
The results from our protocol are good at predicting single mutations that strongly convey resistance according to large changes in experimental binding affinity. It is no doubt that the resistance mechanism is more complicated particularly when mutations causing significant conformational change of the HIV protease are involved. Nevertheless, the proposed protocol provides a pragmatic way to evaluate the potency of drugs/inhibitors and how their potency can be improved. We showed one such application of our protocol on a newly approved drug, darunavir. Our analysis suggested that darunavir is more potent than amprenavir particularly to mutations of Ile50 and Val82 mainly because it significantly improves the interaction with the conserved residue Asp29. The possible resistant mutation to darunavir may likely to occur at Val32 and Ile84. These predictions are waiting for verification.
Supplementary Material
ACKNOWLEDGMENTS
The MD simulations were performed on the Linux cluster in the Center for Theoretical Biological Physics (CTBP) at UCSD. T. H. is supported by a CTBP postdoctoral scholarship. We thank Prof. J. Andrew McCammon for providing access to SYBYL and InsightII molecular simulation packages. WAM is supported by an NIH training grant (5 T32DK07233).
Grant sponsor: NSF PFC-sponsored Center for Theoretical Biological Physics; Grant numbers: PHY-0216576, PHY-0225630; Grant sponsor: NIH (PI Elizabeth Komives); Grant number: 5 T32DK07233; Grant sponsor: Center for AIDS Research at UCSD.
This project is partially supported by a developmental grant in AIDS research from USCD Center for AIDS Research (to W.W.).
REFERENCES
- 1.Darke PL, Nutt RF, Brady SF, Garsky VM, Ciccarone TM, Leu CT, Lumma PK, Freidinger RM, Veber DF, Sigal IS. HIV-1 protease specificity of peptide cleavage is sufficient for processing of gag and pol polyproteins. Biochem Bioph Res Co. 1988;156:297–303. doi: 10.1016/s0006-291x(88)80839-8. [DOI] [PubMed] [Google Scholar]
- 2.Kohl NE, Emini EA, Schleif WA, Davis LJ, Heimbach JC, Dixon RAF, Scolnick EM, Sigal IS. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller M, Jaskolski M, Rao JKM, Leis J, Wlodawer A. Crystal-structure of a retroviral protease proves relationship to aspartic protease family. Nature. 1989;337:576–579. doi: 10.1038/337576a0. [DOI] [PubMed] [Google Scholar]
- 4.Swain AL, Miller MM, Green J, Rich DH, Schneider J, Kent SBH, Wlodawer A. X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus-1 and a substrate-based hydroxyethylamine inhibitor. Proc Natl Acad Sci USA. 1990;87:8805–8809. doi: 10.1073/pnas.87.22.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clavel F, Hance AJ. Medical progress: HIV drug resistance. New Engl J Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 6.Condra JH, Schleif WA, Blahy OM, Gabryelski LJ, Graham DJ, Quintero JC, Rhodes A, Robbins HL, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires KE, Deutsch PJ, Emini EA. Invivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan AH, Michael SF, Wehbie RS, Knigge MF, Paul DA, Everitt L, Kempf DJ, Norbeck DW, Erickson JW, Swanstrom R. Selection of multiple human-immunodeficiency-virus type-1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc Natl Acad Sci USA. 1994;91:5597–5601. doi: 10.1073/pnas.91.12.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shafer RW, Stevenson D, Chan B. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 1999;27:348–352. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao ZW, Han LY, Zheng CJ, Ji ZL, Chen X, Lin HH, Chen YZ. Computer prediction of drug resistance mutations in proteins. Drug Discov Today. 2005;10:521–529. doi: 10.1016/S1359-6446(05)03377-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Donini O, Reyes CM, Kollman PA. Biomolecular simulations: recent developments in force fields, simulations of enzyme catalysis, protein—ligand, protein—protein, and protein—nucleic acid noncovalent interactions. Annu Rev Biophys Biomol. 2001;30:211–243. doi: 10.1146/annurev.biophys.30.1.211. [DOI] [PubMed] [Google Scholar]
- 11.Boutton CW, De Bondt HL, De Jonge MR. Genotype dependent QSAR for HIV-1 protease inhibition. J Med Chem. 2005;48:2115–2120. doi: 10.1021/jm049596h. [DOI] [PubMed] [Google Scholar]
- 12.Shenderovich MD, Kagan RM, Heseltine PNR, Ramnarayan K. Structure-based phenotyping predicts HIV-1 protease inhibitor resistance. Protein Sci. 2003;12:1706–1718. doi: 10.1110/ps.0301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YZ, Gu XL, Cao ZW. Can an optimization/scoring procedure in ligand—protein docking be employed to probe drug-resistant mutations in proteins? J Mol Graph Model. 2001;19:560–570. doi: 10.1016/s1093-3263(01)00091-2. [DOI] [PubMed] [Google Scholar]
- 14.Nair AC, Bonin I, Tossi A, Welsh WJ, Miertus S. Computational studies of the resistance patterns of mutant HIV-1 aspartic proteases towards ABT-538 (ritonavir) and design of new derivatives. J Mol Graph Model. 2002;21:171–179. doi: 10.1016/s1093-3263(02)00149-3. [DOI] [PubMed] [Google Scholar]
- 15.Hou TJ, Yu R. Molecular dynamics and free energy studies on the wild-type and double mutant HIV-1 protease complexed with amprenavir and two amprenavir-related inhibitors: mechanism for binding and drug resistance. J Med Chem. 2007;50:1177–1188. doi: 10.1021/jm0609162. [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Kollman PA. Computational study of protein specificity: the molecular basis of HIV-1 protease drug resistance. Proc Natl Acad Sci USA. 2001;98:14937–14942. doi: 10.1073/pnas.251265598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure. 2002;10:369–381. doi: 10.1016/s0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- 18.Lee MS, Olson MA. Calculation of absolute protein—ligand binding affinity using path and endpoint approaches. Biophys J. 2006;90:864–877. doi: 10.1529/biophysj.105.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem Biol. 2004;11:1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Wittayanarakul K, Aruksakunwong O, Saen-oon S, Chantratita W, Parasuk V, Sompornpisut P, Hannongbua S. Insights into saquinavir resistance in the G48V HIV-1 protease: quantum calculations and molecular dynamic simulations. Biophys J. 2005;88:867–879. doi: 10.1529/biophysj.104.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtaka H, Velazquez-Campoy A, Xie D, Freire E. Overcoming drug resistance in HIV-1 chemotherapy: the binding thermodynamics of amprenavir and TMC-126 to wild-type and drug-resistant mutants of the HIV-1 protease. Protein Sci. 2002;11:1908–1916. doi: 10.1110/ps.0206402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorbera LA, Castaner J, Bayes M. Darunavir-anti-HIV agent HIV protease inhibitor. Drug Future. 2005;30:441–449. [Google Scholar]
- 23.Surleraux DLNG, Tahri A, Verschueren WG, Pille GME, de Kock HA, Jonckers THM, Peeters A, De Meyer S, Azijn H, Pauwels R, de Bethune MP, King NM, Prabu-Jeyabalan M, Schiffer CA, Wigerinck PBTP. Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor. J Med Chem. 2005;48:1813–1822. doi: 10.1021/jm049560p. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Kollman PA. Computational study of protein specificity: the molecular basis of HIV-1 protease drug resistance. Proc Natl Acad Sci USA. 2001;98:14937–14942. doi: 10.1073/pnas.251265598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krohn A, Redshaw S, Ritchie JC, Graves BJ, Hatada MH. Novel binding mode of highly potent HIV-proteinase inhibitors incorporating the (R)-hydroxyethylamine isostere. J Med Chem. 1991;34:3340–3342. doi: 10.1021/jm00115a028. [DOI] [PubMed] [Google Scholar]
- 27.Chen ZG, Li Y, Chen E, Hall DL, Darke PL, Culberson C, Shafer JA, Kuo LC. Crystal-structure at 1.9-angstrom resolution of human-immunodeficiency-virus (HIV)-Ii protease complexed with L-735,524, an orally bioavailable inhibitor of the HIV proteases. J Biol Chem. 1994;269:26344–26348. [PubMed] [Google Scholar]
- 28.Kempf DJ, Marsh KC, Denissen JF, Mcdonald E, Vasavanonda S, Flentge CA, Green BE, Fino L, Park CH, Kong XP, Wideburg NE, Saldivar A, Ruiz L, Kati WM, Sham HL, Robins T, Stewart KD, Hsu A, Plattner JJ, Leonard JM, Norbeck DW. Abt-538 is a potent inhibitor of human-immunodeficiency-virus protease and has high oral bioavailability in humans. Proc Natl Acad Sci USA. 1995;92:2484–2488. doi: 10.1073/pnas.92.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaldor SW, Kalish VJ, Davies JF, Shetty BV, Fritz JE, Appelt K, Burgess JA, Campanale KM, Chirgadze NY, Clawson DK, Dressman BA, Hatch SD, Khalil DA, Kosa MB, Lubbehusen PP, Muesing MA, Patick AK, Reich SH, Su KS, Tatlock JH. Viracept (nelfinavir mesylate. AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J Med Chem. 1997;40:3979–3985. doi: 10.1021/jm9704098. [DOI] [PubMed] [Google Scholar]
- 30.Kim EE, Baker CT, Dwyer MD, Murcko MA, Rao BG, Tung RD, Navia MA. Crystal-structure of HIV-1 protease in complex with Vx-478, a potent and orally bioavailable inhibitor of the enzyme. J Am Chem Soc. 1995;117:1181–1182. [Google Scholar]
- 31.Stoll V, Qin WY, Stewart KD, Jakob C, Park C, Walter K, Simmer RL, Helfrich R, Bussiere D, Kao J, Kempf D, Sham HL, Norbeck DW. X-ray crystallographic structure of ABT-378 (lopinavir) bound to HIV-1 protease. Bioorgan Med Chem. 2002;10:2803–2806. doi: 10.1016/s0968-0896(02)00051-2. [DOI] [PubMed] [Google Scholar]
- 32.Thaisrivongs S, Skulnick HI, Turner SR, Strohbach JW, Tommasi RA, Johnson PD, Aristoff PA, Judge TM, Gammill RB, Morris JK, Romines KR, Chrusciel RA, Hinshaw RR, Chong KT, Tarpley WG, Poppe SM, Slade DE, Lynn JC, Horng MM, Tomich PK, Seest EP, Dolak LA, Howe WJ, Howard GM, Schwende FJ, Toth LN, Padbury GE, Wilson GJ, Shiou LH, Zipp GL, Wilkinson KF, Rush BD, Ruwart MJ, Koeplinger KA, Zhao ZY, Cole S, Zaya RM, Kakuk TJ, Janakiraman MN, Watenpaugh KD. Structure-based design of HIV protease inhibitors: Sulfonamide-containing 5,6-dihydro-4-hydroxy-2-pyrones as non-peptidic inhibitors. J Med Chem. 1996;39:4349–4353. doi: 10.1021/jm960541s. [DOI] [PubMed] [Google Scholar]
- 33.Nichols CE, Hawkins AR, Stammers DK. Structure of the ‘open’ form of Aspergillus nidulans 3-dehydroquinate synthase at 1.7 angstrom resolution from crystals grown following enzyme turnover. Acta Crystallogr D. 2004;60:971–973. doi: 10.1107/S0907444904004743. [DOI] [PubMed] [Google Scholar]
- 34.SYBYL molecular simulation package, 2004http://www.sybyl.com
- 35.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyland LJ, Tomaszek TA, Meek TD. Human immunodeficiency virus-1 protease. II. Use of Ph rate studies and solvent kinetic isotope effects to elucidate details of chemical mechanism. Biochemistry-US. 1991;30:8454–8463. doi: 10.1021/bi00098a024. [DOI] [PubMed] [Google Scholar]
- 37.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong GM, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang JM, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 38.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 39.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JAJ, Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, COshterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Baboul AG, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Gonzalez C, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Andres JL, Gonzalez C, Head-Gordon M, Replogle ES, Pople JA. Gaussian 98. Gaussian; Pittsburgh, PA: 1998. [Google Scholar]
- 40.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the Resp model. J Phys Chem-US. 1993;97:10269–10280. [Google Scholar]
- 41.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 42.Darden T, York D, Pedersen L. Particle mesh ewald—an NLog(N) method for ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 43.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 44.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-integration of cartesian equations of motion of a system with constraints— molecular-dynamics of N-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 45.Wang JM, Hou TJ, Xu XJ. Recent advances in free energy calculations with a combination of molecular mechanics and continuum models. Curr Comput-Aided Drug Design. 2006;2:287–306. [Google Scholar]
- 46.Kollman PA, Massova I, Reyes C, Kuhn B, Huo SH, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Accounts Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 47.Onufriev A, Bashford D, Case DA. Modification of the generalized Born model suitable for macromolecules. J Phys Chem B. 2000;104:3712–3720. [Google Scholar]
- 48.Weiser J, Shenkin PS, Still WC. Approximate solvent-accessible surface areas from tetrahedrally directed neighbor densities. Biopolymers. 1999;50:373–380. doi: 10.1002/(SICI)1097-0282(19991005)50:4<373::AID-BIP3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 49.Gohlke H, Kiel C, Case DA. Insights into protein—protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RaIGDS complexes. J Mol Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.