Abstract
Human organic anion transporting polypeptides (OATP) 1B1 and 1B3 are multi-specific transporters that mediate uptake of amphipathic organic compounds into hepatocytes. The two OATPs contain twelve transmembrane domains (TMs) and share 80% amino acid sequence identity. Besides common substrates with OATP1B1, OATP1B3 specifically transports cholecystokinin octapeptide (CCK-8). To determine which structural domains/residues are important for the substrate selectivity of OATP1B3, we constructed a series of chimeric proteins between OATP1B3 and 1B1, expressed them in HEK293 cells and determined uptake of CCK-8 along with surface expression of the proteins. Replacing TM10 in OATP1B3 with TM10 of OATP1B1 resulted in dramatically reduced CCK-8 transport, indicating that TM10 is crucial for recognition and/or translocation of CCK-8. Using site-directed mutagenesis, we identified three key residues within TM10, namely Y537, S545 and T550. When we replaced these residues by the corresponding amino acid residues found in OATP1B1, CCK-8 transport was similarly low as for the replacement of the whole TM10. Kinetic experiments showedthat the Km values for CCK-8 transport in the TM10-replacement and triple mutant were only 1.3 and 1.1 μM, respectively as compared to 16.3 μM for wild-type OATP1B3. Similarly, the Vmax values dropped from 495.5 pmol/normalized mg/min for wild-type OATP1B3 to 13.3 and 19.0 for the TM10-replacement and triple mutant, respectively. Molecular modeling indicated that two of the three identified residues might form hydrogen bonds with CCK-8. In conclusion, we have identified three amino acid residues (Y537, S545 and T550) in TM10 of OATP1B3 that are important for CCK-8 transport.
The mammalian liver is the major detoxification organ of the body and the hepatic sinusoidal membrane is equipped with transporters that mediate influx and efflux of endogenous and xenobiotic compounds. Uptake transporters belong to the superfamily of solute carriers (SLC) (1) which mediate the first step of the hepatic elimination by facilitating hepatic uptake of those compounds from the portal vein. In humans, organic anion transporting polypeptides (OATPs, gene symbol SLCO) mediate the sodium-independent transport of a wide range of amphipathic organic compounds including numerous drugs and other xenobiotics (2). There are 11 human OATPs (3, 4) with the polyspecific OATP1B3 and 1B1 being predominantly expressed at the basolateral membrane of hepatocytes (5–9), indicating their significant roles in the hepatic clearance of amphipathic organic compounds. OATP1B3 shares 80% amino acid sequence identity with OATP1B1. Recent findings for rat Oatp1a1 suggest that all OATPs comprise twelve putative transmembrane domains (TMs) (10). As expected from their high sequence similarity, OATP1B3 and 1B1 exhibit overlapping transport activities for an array of substances including bile salts, unconjugated and conjugated bilirubin, bromosulfophthalein (BSP), steroid conjugates, thyroid hormones, eicosanoids, peptides, drugs and the natural toxins phalloidin and microcystin (4).
Although OATP1B3 shares a great number of substrates with OATP1B1, it also possesses its own specific substrates. OATP1B3 selectively transports cholecystokinin octapeptide (CCK-8), the anticancer drugs paclitaxel and docetaxel, and the cardiac glycosides digoxin and ouabain (11–13). So far, little is known about the molecular mechanism for substrate selectivity of OATP1B3. Therefore, in this study we focused on delineating the structural domains and molecular determinants responsible for CCK-8 transport of OATP1B3 by constructing a series of OATP1B1/1B3 chimeric transporters, performing site-directed mutagenesis, characterizing CCK-8 transport and constructing a molecular model.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Radiolabeled [3H]CCK-8 (92 Ci/mmol) was purchased from Amersham Biosciences (Piscataway, NJ) and unlabeled CCK-8 was obtained from Sigma-Aldrich (St. Louis, MO). Cell culture reagents, Lipofectamine 2000 and the vector pcDNA5/FRT were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum was obtained from Hyclone (Logan, UT). The site-directed mutagenesis kit QuikChange was from Stratagene (La Jolla, CA). Sulfo-N-hydroxysuccinimide-SS-biotin, streptavidin-agarose beads, and the BCA protein assay kit were from Pierce Chemical (Rockford, IL). Antibodies to detect the His-tag and the Na+/K+-ATPase a subunit were purchased from Abcam (Boston, MA). Poly-D lysine was purchased from Sigma-Aldrich (St. Louis, MO). The protease inhibitor cocktail was from Roche (Indianapolis, IN) and the enhanced chemiluminescence (ECL) plus western blotting detection kit was from Amersham Biosciences (Piscataway, NJ).
Construction of Chimeric Transporters and Mutants
A 6 His-tag was introduced at the C-terminal end of the open reading frames of human OATP1B1*1b (14) and OATP1B3 haplotype 1 (15) by PCR and the resulting constructs were cloned into the vector pcDNA5/FRT (Invitrogen, Carlsbad, CA) using NheI and NotI sites. To construct chimeric transporters between OATP1B1 and 1B3, the junction sites were determined based on the topological structures of OATP1B1 and 1B3 predicted by TMPred (http://www.ch.embnet.org/software/TMPRED_form.html), and overlapping PCR was employed. The lengths of overlapping regions were 20–23 bases. For site-directed mutagenesis, mutants were generated using the QuikChange kit (Stratagene, La Jolla, CA). The sequences of all constructs were confirmed by DNA sequencing.
Protein Expression in HEK293 cells
Human embryonic kidney (HEK293) cells were grown at 37 °C in a humidified 5% CO2 atmosphere in Dulbecco’s Modified Eagle Medium, containing 4.5 g/l D-glucose, 2 mM L-glutamine, 25 mM Hepes buffer and 110 mg/l sodium pyruvate, supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. Plasmids containing OATP1B1, OATP1B3 and their chimeric constructs were transiently transfected into HEK293 cells using Lipofectamine 2000 according to the manufacturer’s instruction. Transfected cells were incubated for 48 h at 37 °C and then used for protein surface expression and transport assays.
Cell Surface Biotinylation and Immunoblot Analysis
HEK293 cells were grown in poly-D lysine coated 6-well plates and transfected as described above. Biotinylation experiments were conducted 48 h post-transfection. Cells were washed twice with 2 ml of ice-cold phosphate-buffered saline (PBS) and then treated with 1 ml of sulfo-N-hydroxysuccinimide-SS-biotin (1 mg/ml in PBS) for 1 h at 4 °C. Then, cells were washed three times with 2 ml of ice-cold PBS containing 100 mM glycine and incubated for 10 min at 4 °C with the same buffer. After PBS washing for three times, cells were lysed with 700 μl of lysis buffer (10 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, and 1% Triton X-100, pH 7.4, containing protease inhibitors (Roche, Indianapolis, IN)) for 1 h at 4 °C with shaking. Lysates were centrifuged at 10,000 g for 2 min. Supernatants were incubated with 140 μl of streptavidin-agarose beads for 1 h at room temperature with constant agitation. Beads were then pelleted at 850 g for 1 min and washed three times with ice-cold lysis buffer. Cell surface proteins were recovered from the resin by incubating the beads with 150 μl of 2 × Laemmli buffer containing 100 mM dithiothreitol at room temperature for 30 min. Cell membrane proteins were subjected to SDS-polyacrylamide gel electrophoresis and immunoblot analysis. OATP1B1, 1B3 and chimeric proteins were detected with a rabbit anti-His polyclonal antibody (1:2,500 dilution), followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10,000 dilution). Protein loading was normalized by plasma membrane marker Na+/K+ ATPase detected with mouse anti-Na+/K+ ATPase α subunit antibody (1:5,000 dilution). Immunoblots were developed with ECL plus reagent (Amersham Biosciences, Piscataway, NJ) and detected with X-ray films. The intensities of the protein bands were quantified using the Quantity One analysis software (Bio-Rad Laboratories, Hercules, CA).
Functional Studies of Transporters in HEK293 cells
HEK293 cells were seeded in poly-D lysine coated 12-well plates and transfected with lipofectamine. Transport assays were performed 48 h post-transfection. Cells were washed three times with 2 ml of pre-warmed uptake buffer (100 mM NaCl, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM Hepes, pH adjusted to 7.4 with Trizma base) and uptake was started by adding 400 μl of uptake buffer containing radiolabeled substrate. After the incubation, uptake was stopped by removing the uptake solution and washing the cells four times with 2 ml of ice-cold uptake buffer. The cells were then solubilized with 500 μl of 1% Triton X-100 and 300 μl of the lysate was used for liquid scintillation counting (Research Products International Corp.) to quantify the radioactivity. Total protein concentration in each uptake well was measured using a BCA protein assay kit (Pierce) and the uptake in each well was normalized to its total protein concentration. In all studies, cells transfected with empty vector served as background control. Initial experiments showed that uptake was linear over at least 1 min. Therefore, all uptake and kinetic experiments were performed under these initial linear conditions at 1 min. Transporter-specific uptake was calculated by subtracting the background uptake of vector transfected cells (in pmol/mg total protein/min) from uptake of OATP-transfected cells (in pmol/mg total protein/min). This number then was normalized to OATP1B3 surface expression levels (surface expression of wild-type OATP1B3 was set to 1) and given as pmol/normalized mg/min as follows: pmol/(mg total protein × normalization ratio)/min, where normalization ratio = surface expression levelmutant/surface expression levelOATP1B3.
Structural Modeling of OATP1B3 and Molecular Docking of CCK-8 to OATP1B3
The fold recognition method of mGenTHREADER (16) was used to identify templates to model the structure of OATP1B3. Modeling was performed using the Homology module in the InsightII software package (17). First, the coordinates of the TMs of OATP1B3 were assigned according to the structure of EmrD (PDB entry 2GFP). Then the Loop_Search module was used to model the loop regions between the TMs and the whole structure was subjected to structural optimization with the Auto_Rotamer and Discover modules. The quality of the final model was assessed with several structural analysis programs such as Prostat to check bond lengths, angles and torsions and Procheck (18) for a stereochemical evaluation.
To identify the potential mode of interaction of CCK-8 with OATP1B3, molecular docking of CCK-8 to the OATP1B3 model was carried out using AutoDock 3.0 (19). The Lamarckian genetic algorithm (LGA) was used for docking with the following settings: a maximum number of 1,500,000 energy evaluations, an initial population of 50 randomly placed individuals, a maximum number of 37,000 generations, a mutation rate of 0.02, a crossover rate of 0.80 and an elitism value (number of top individuals that automatically survive) of 1. For the adaptive local search method, the pseudo-Solis and Wets algorithm was applied with a maximum of 300 iterations per search. The conformer with the lowest free binding energy and reasonable conformation was selected and subjected to a 500-step energy minimization within the binding site of OATP1B3 using Sybyl7.1 (20), and the obtained conformation was adopted as the final binding conformation of CCK-8 for OATP1B3. The interactions between CCK-8 and OATP1B3 were analyzed by Ligplot (21).
Data Analysis
Uptake experiments were performed in duplicate and repeated two to three times. Data with error bars represent the mean ± standard deviation. To analyze whether the groups were different from the control, ANOVA was performed followed by the Bonferroni t-test with SigmaStat 3.5 (Systat Software, Inc., San Jose, CA). The p value for statistical significance was set to be < 0.05. The kinetic parameters were obtained by nonlinear regression fitting with the Enzyme Kinetics Module of SigmaPlot (Systat Software, San Jose, CA).
RESULTS
Characterization of Surface Expression and Function of OATP1B1, OATP1B3 and the Chimeric Transporters T1-6 and T7–12
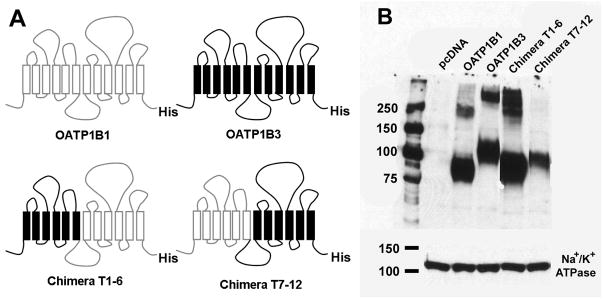
The full length human OATP1B1 and 1B3 comprise 691 and 702 residues, respectively. They share 80% amino acid sequence similarity and have twelve TMs with the N- and C-termini located intracellularly (3). Thus, we divided OATP1B1 and 1B3 into N- and C-terminal halves, each half consisting of six TMs. To determine which half is important for the transport of the OATP1B3 specific substrate CCK-8, we constructed two chimeric transporters. Chimera T1–6 contained the first half of OATP1B3 (residues 1–275) fused to the second half of OATP1B1 (residues 276–691) and chimera T7–12 contained the first half of OATP1B1 (residues 1–275) fused to the second half of OATP1B3 (residues 276–702) (Figure 1A). OATP1B1, 1B3 and the two chimeric transporters were transiently expressed in HEK293 cells. To quantitate surface expression of the two OATPs and the chimeras, we performed western blot analysis with an anti-His antibody after purifying surface proteins using biotinylation. As a loading control, we used the α-subunit of Na+/K+ ATPase (Figure 1B). The anti-His antibody detected a band at ~84 kDa for OATP1B1 and at ~120 kDa for OATP1B3 (Figure 1B), which is consistent with the reported molecular weights of their fully-glycosylated forms (8, 9). The apparent molecular weights of chimeras T1–6 and T7–12 were ~90 kDa and ~100 kDa, respectively. Bands with higher molecular weights were also detected for OATP1B1, 1B3 and chimera T1–6 and probably represent multimeric aggregates. The surface expression levels were normalized to OATP1B3 and were 1.04 for OATP1B1, 1.62 for chimera T1–6 and 0.53 for chimera T7–12.
FIGURE 1.
Schematic representation of human OATP1B1, 1B3 and chimeras T1–6 and T7–12, and their surface expression in HEK293 cells. (A) Chimera T1–6 consisted of TMs 1–6 (residues 1–275) of OATP1B3 and TMs 7–12 (residues 276–691) of OATP1B1, while chimera T7–12 consisted of TMs 1–6 (residues 1–275) of OATP1B1 and TMs 7–12 (residues 276–702) of OATP1B3. In order to detect all constructs with the same antibody, a His-tag was introduced at the C-terminal end of all constructs. (B) Western blot analysis of surface biotinylated proteins detected with an anti-His antibody. The plasma membrane marker Na+/K+ ATPase a-subunit was used as protein loading control.
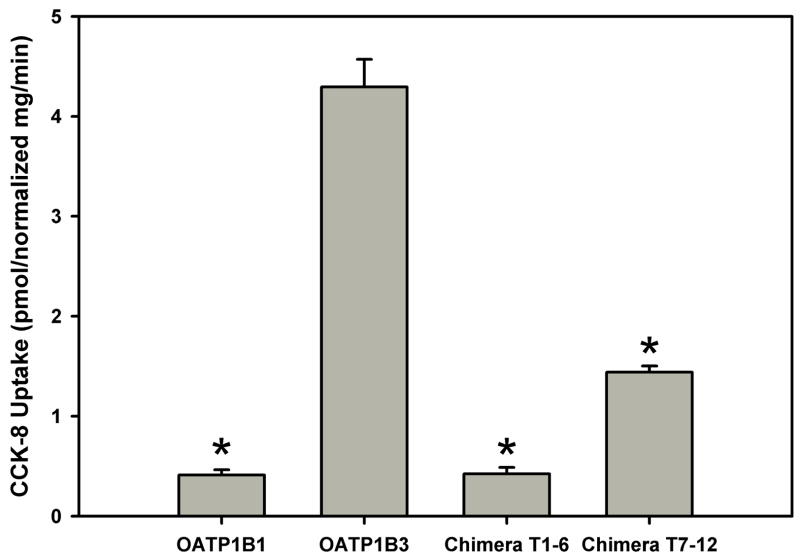
Figure 2 summarizes the functional results that we obtained with the four constructs. The expression system worked well as OATP1B3 had high transport activity for CCK-8 (4.3 pmol/normalized mg/min) which was 10-fold higher than that of OATP1B1 (0.4 pmol/normalized mg/min). The activities of chimeras T1–6 and T7–12 decreased significantly compared to OATP1B3, and were 10% and 34% of OATP1B3, respectively. These results indicate that both N- and C-terminal halves contribute to OATP1B3-mediated CCK-8 transport with the C-terminal half playing a more important role than the N-terminal half.
FIGURE 2.
Uptake of CCK-8 by HEK293 cells transfected with OATP1B1, 1B3, chimeras T1–6 and T7–12. Uptake of 0.1 μM CCK-8 was measured at 37 °C for 1 min with empty vector and OATP-expressing HEK293 cells. The net uptakes were obtained by subtracting the uptake of cells transfected with empty vector from the uptake of OATP-expressing cells. Final results were obtained by normalizing net uptakes to the surface expression levels of the respective proteins. Values were calculated as picomoles/(milligrams of total protein × normalization ratio)/minute, where normalization ratio = surface expression levelmutant/surface expression levelOATP1B3. * p < 0.05 significantly different from OATP1B3.
Twelve Chimeric Transporters Derived from OATP1B3 and their Transport Function
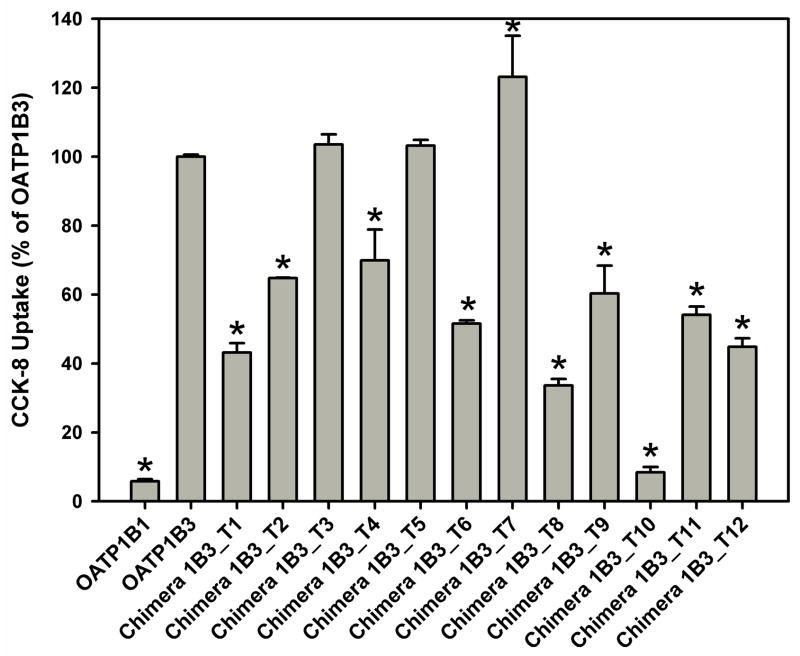
To determine which TM(s) is/are critical for the function of OATP1B3, we constructed twelve chimeras of OATP1B3 (chimera 1B3_T1, T2, T3, T4, T5, T6, T7, T8, T9, T10, T11 and T12) by replacing each TM together with the loop preceding it with the corresponding region of OATP1B1 (Figure 3).
FIGURE 3.
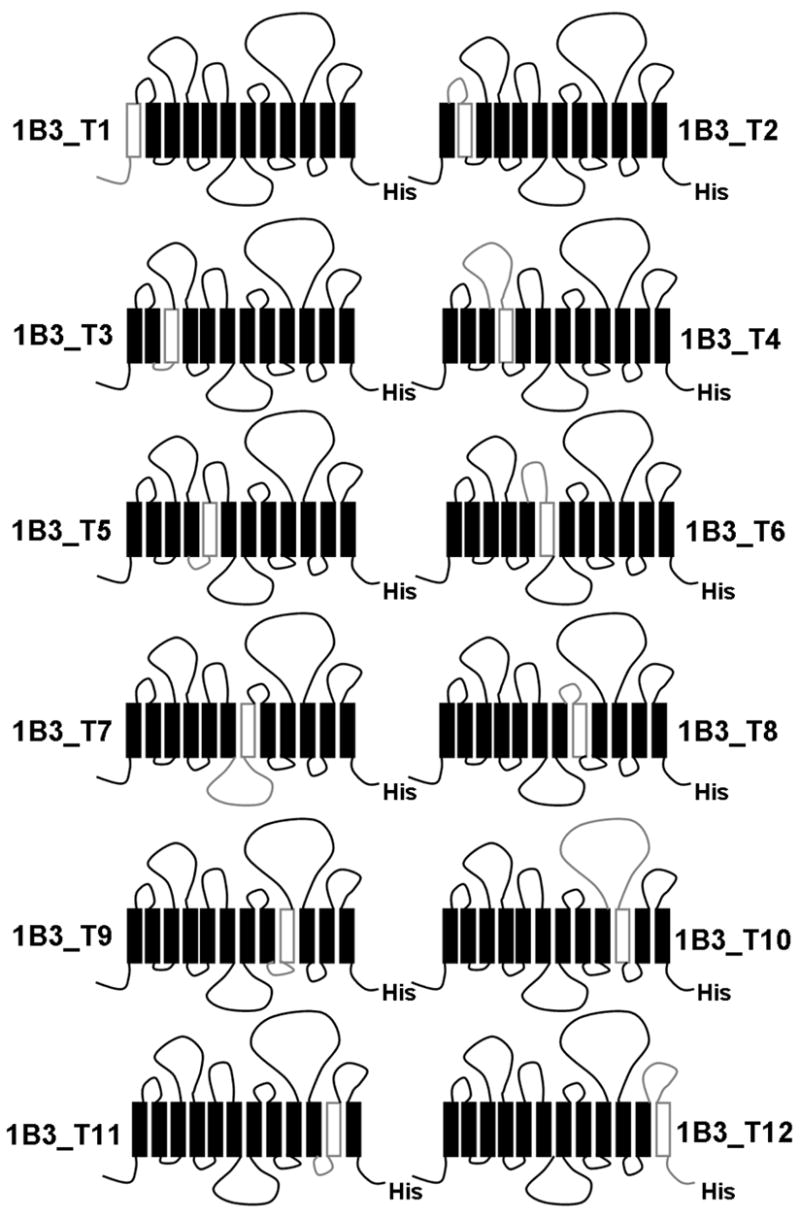
Schematic representation of twelve human OATP1B3-derived chimeric transporters. The chimeric transporters were constructed by replacing each TM domain together with the loop preceding it with its corresponding region of OATP1B1. The replaced residues for each chimera are: residues 1–48 for chimera 1B3_T1, residues 49–88 for 1B3_T2, residues 89–120 for 1B3_T3, residues 121–200 for 1B3_T4, residues 201–230 for 1B3_T5, residues 231–279 for 1B3_T6, residues 280–358 for 1B3_T7, residues 359–394 for 1B3_T8, residues 395–432 for 1B3_T9, residues 433–560 for 1B3_T10, residues 561–595 for 1B3_T11 and residues 596–691 for 1B3_T12.
Surface biotinylation and western blot experiments showed that all twelve chimeric transporters were properly expressed on the cell surface (data not shown). Figure 4 summarizes CCK-8 transport of the twelve chimeric transporters together with OATP1B1 and 1B3 after correction for surface expression. Transport mediated by chimeras 1B3_T1, T2, T4, T6, T8, T9, T10, T11 and T12 was reduced by 30 to 90% as compared to OATP1B3, while transport mediated by chimeras 1B3_T3 and T5 was similar to OATP1B3. Chimera 1B3_T7 showed a small increased transport in comparison with OATP1B3. We observed the greatest reduction for chimera 1B3_T10 whose activity was only 10% of OATP1B3. This suggests that the region of TM10 and extracellular loop 5 (EL5) is crucial for normal CCK-8 transport mediated by OATP1B3.
FIGURE 4.
Uptake of CCK-8 by HEK293 cells transfected with OATP1B1, 1B3 and chimeras 1B3_T1, T2, T3, T4, T5, T6, T7, T8, T9, T10, T11 and T12. Uptake of 0.1 μM CCK-8 was measured at 37 °C for 1 min with empty vector and OATP-expressing HEK293 cells. The net uptakes were obtained by subtracting the uptake of cells transfected with empty vector from the uptake of OATP-expressing cells. Final results were obtained by normalizing net uptakes to the surface expression levels of the respective proteins. * p < 0.05 significantly different from OATP1B3.
Chimeras 1B3_EL5 and 1B3_TM10 and their Transport Function
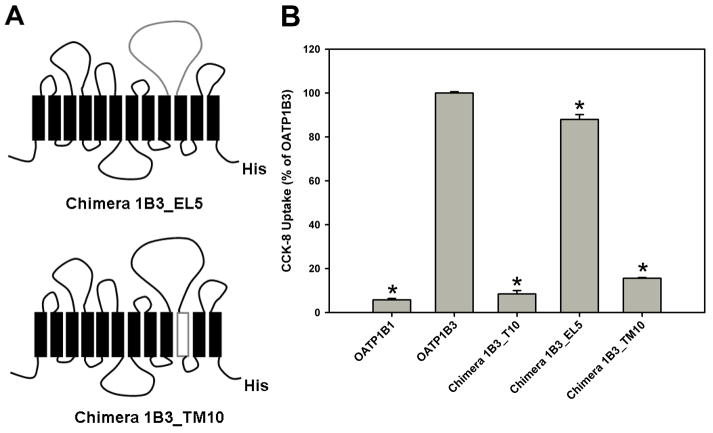
To narrow down the region of importance, we constructed two additional chimeras, 1B3_EL5 and 1B3_TM10. Chimera 1B3_EL5 was obtained by replacing EL5 (residues 433–527) of OATP1B3 with that of 1B1, and chimera 1B3_TM10 was obtained by replacing TM10 (residues 528–560) of OATP1B3 with that of 1B1 (Figure 5A). As shown in Figure 5B, chimera 1B3_EL5 maintained 88% transport activity of OATP1B3. In contrast, transport mediated by chimera 1B3_TM10 was reduced to 16% of the transport observed with OATP1B3, which is very similar to the transport of chimera 1B3_T10 (10% of OATP1B3). These results indicate that amino acid residues within TM10 (residues 528–560) but not EL5 are important for CCK-8 transport mediated by OATP1B3.
FIGURE 5.
Schematic representation of the chimeric transporters 1B3_EL5 and 1B3_TM10 and CCK-8 uptake of the constructs. (A) Chimeras 1B3_EL5 and 1B3_TM10 were constructed by replacing extracellular loop 5 (residues 433–527) and transmembrane domain 10 (residues 528–560) of OATP1B3 with their counterparts of OATP1B1. (B) Uptake of 0.1 μM CCK-8 was measured by OATP1B1, 1B3 and chimeras 1B3_T10, 1B3_EL5 and 1B3_TM10 in transfected HEK293 cells for 1 min at 37 °C. * p < 0.05 significantly different from OATP1B3.
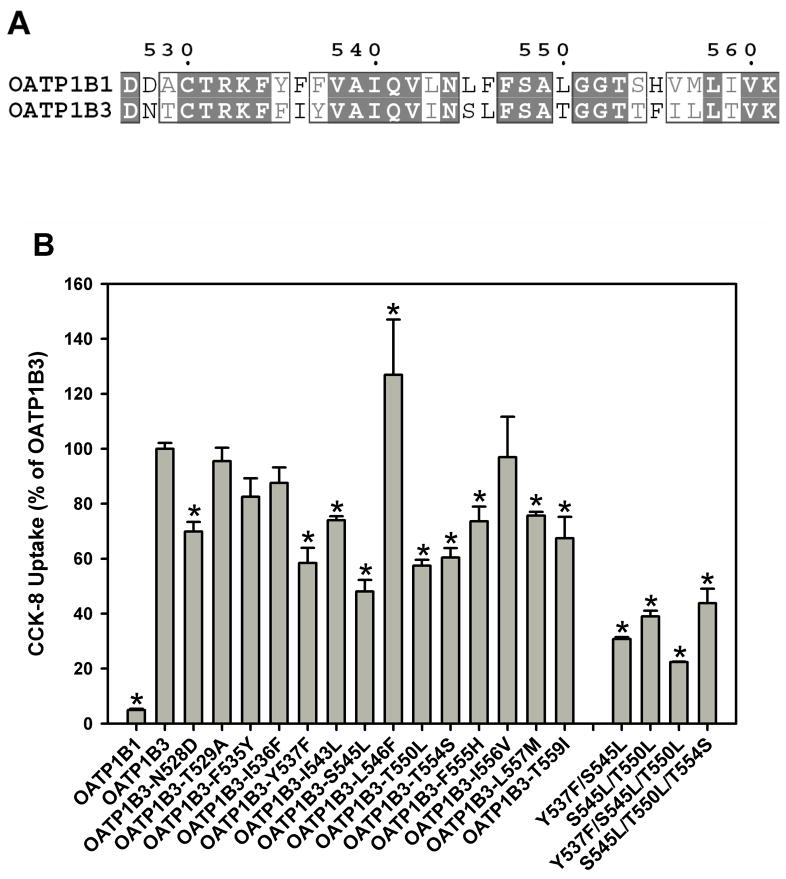
Sequence Alignment of TM10 and Site-directed Mutagenesis
Amino acid sequence alignment between OATP1B1 and 1B3 showed that there are fourteen residues that differ in TM10 between the two transporters (Figure 6A). To determine which of these fourteen amino acid residues would be important for CCK-8 transport of OATP1B3, we produced fourteen single point mutants by replacing each residue in OATP1B3 with its corresponding residue of OATP1B1. All fourteen mutants were expressed in HEK293 cells along with OATP1B3 and 1B1, and surface expression and CCK-8 uptake were determined and the data are summarized in Figure 6B. We observed the greatest reduction of transport activity for the mutants Y537F (58% of OATP1B3), S545L (48% of OATP1B3), T550L (57% of OATP1B3) and T554S (60% of OATP1B3). Based on these results, we constructed double mutants Y537F/S545L and S545L/T550L and triple mutants Y537F/S545L/T550L and S545L/T550L/T554S and measured their transport activity (Figure 6B). All double and triple mutants showed lower transport activity than single mutants. The triple mutant Y537F/S545L/T550L had the lowest transport activity (22% of OATP1B3), similar to mutant 1B3_TM10 where the whole TM10 was replaced (Figure 5B, 16% of OATP1B3).
FIGURE 6.
Sequence alignment of TM10 and uptake of CCK-8 by mutant OATP1B3. (A) Residues 528–560 of OATP1B1 and 1B3 were aligned and fourteen amino acids were identified as different between OATP1B3 and 1B1 in this domain: (1B3)N528D(1B1), T529A, F535Y, I536F, Y537F, I543L, S545L, L546F, T550L, T554S, F555H, I556V, L557M and T559I. (B) Uptake of 0.1 μM CCK-8 by OATP1B1, 1B3, fourteen OATP1B3 single point mutants, S545L/T550L double mutant and Y537F/S545L/T550L and S545L/T550L/T554S triple mutants in TM10 was measured at 37 °C for 1 min with empty vector and OATP-expressing HEK293 cells. The net uptakes were obtained by subtracting the uptake of cells transfected with empty vector from the uptake of OATP-expressing cells. Final results were obtained by normalizing net uptakes to the surface expression levels of the respective proteins. * p < 0.05 significantly different from OATP1B3.
Kinetic Studies of CCK-8 Transport by OATP1B3, Chimera 1B3_TM10 and Triple Mutant Y537F/S545L/T550L
Kinetics of CCK-8 transport were analyzed with different constructs. The obtained Km and Vmax values for OATP1B3, chimera 1B3_TM10 and the triple mutant Y537F/S545L/T550L are summarized in Table 1. OATP1B3 transported CCK-8 with an apparent Km of 16.3 μM and a Vmax of 495.5 pmol/normalized mg/min. Both chimera 1B3_TM10 and the triple mutant Y537F/S545L/T550L transported CCK-8 with dramatically decreased Vmax values of 13.3 and 19.0 pmol/normalized mg/min, respectively. Their apparent Km values also decreased from 16.3 μM to 1.3 μM for 1B3_TM10 and to 1.1 μM for the triple mutant Y537F/S545L/T550L. The overall transport efficiency characterized by Vmax/Km of OATP1B3 was decreased from 30.4 to 10.2 for 1B3_TM10 and to 17.3 for triple mutant Y537F/S545L/T550L. These results suggest that the reduced CCK-8 transport activity of chimera 1B3_TM10 and of triple mutant Y537F/S545L/T550L is mainly due to a reduction of Vmax.
Table 1.
Kinetic Parameters of CCK-8 Transport Mediated by OATP1B3, Chimera 1B3_TM10 and Triple Mutant Y537F/S545L/T550L
| Km (μM) | Vmax (pmol/normalized mg/min) | Vmax/Km | |
|---|---|---|---|
| OATP1B3 | 16.3 ± 2.4 | 495.5 ± 28.7 | 30.4 |
| Chimera 1B3_TM10 | 1.3 ± 0.1 | 13.3 ± 0.3 | 10.2 |
| Triple Mutant Y537F/S545L/T550L | 1.1 ± 0.1 | 19.0 ± 0.2 | 17.3 |
Structural Model of the OATP1B3/CCK-8 Complex and the Interactions between them
Due to the lack of crystal structures with high sequence similarity to OATP1B3, we used the fold recognition method incorporated in mGenTHREADER (16) which can detect similarities between three-dimensional structures that are not accompanied by significant sequence similarity to identify the template protein(s). Two proteins, glycerol-3-phosphate transporter GlpT (PDB entry 1PW4) (22) and multidrug transporter EmrD (PDB entry 2GFP) (23), were identify by mGenTHREADER with the highest scores. Both of them belong to the major facilitator superfamily (24) but unlike EmrD which, similar as OATP1B3, can transport a broad range of hydrophobic compounds, GlpT has a relatively narrow range of structurally related mainly hydrophilic substrates (23). Therefore, we selected EmrD as the template to model the structure of OATP1B3. The generated model was verified using the Prostat module of Insight II which shows that almost all values of the bond lengths, angles and torsions of the model are tolerable. Procheck was used to calculate the phi and psi angles. The percentage of residues within allowed regions in the Ramachandran plot was 94.7%. These results indicate that the model for OATP1B3 is structurally feasible.
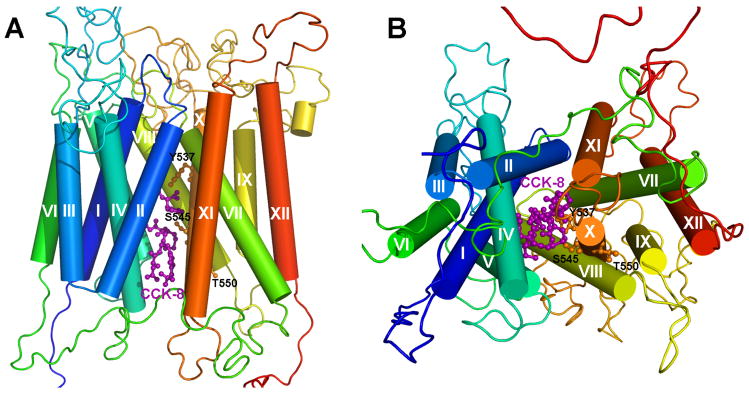
Transmembrane domains 1, 2, 4, 5, 7, 8, 10 and 11 of EmrD form the internal cavity that probably contains the substrate binding site or translocation pathway (23). Because this structure might reflect a general structure of multispecific transporters of the major facilitator superfamily (23) we used AutoDock 3.0 to dock CCK-8 to this internal cavity and Figure 7 shows the resulting OATP1B3/CCK-8 complex. The OATP1B3 model has twelve TMs with a large extracellular loop between TMs 9 and 10. The putative translocation pathway is formed by TMs 1, 2, 4, 5, 7, 8, 10 and 11, and CCK-8 is located in the middle of the translocation pathway. The side chains of Y537 and S545 in TM10 are facing towards the translocation pathway and might form hydrogen bonds with CCK-8, whereas T550 is located on the other side of TM10 far away from the pathway. Besides Y537 and S545, three additional residues, namely G184 in TM4, Y358 in TM7 and S548 in TM10 which are conserved between OATP1B3 and 1B1, could form hydrogen bonds with CCK-8. OATP1B3 also forms extensive hydrophobic interactions with CCK-8 (Figure 8). The estimated binding free energy ΔGbinding between CCK-8 and OATP1B3 was calculated using AutoDock3.0 to be −9.66 kcal/mol. The ΔGbinding between CCK-8 and triple mutant Y537F/S545L/T550L was also estimated to be −5.41 kcal/mol. These binding free energies are in good agreement with the respective Km values of the proteins.
FIGURE 7.
Three-dimensional model of the OATP1B3/CCK-8 complex. A side view (A) and a view from the intracellular side (B) are presented. Helical segments of the protein are shown in cylinders; the three residues Y537, S545 and T550 in TM10 are shown in ball-and-stick and in orange; CCK-8 is shown in ball-and-stick and in purple. CCK-8 is located in the putative translocation pathway which is formed by TMs 1, 2, 4, 5, 7, 8, 10 and 11. The side chains of Y537 and S545 are located on one side of TM10 facing towards the translocation pathway, while T550 is located on the other side of TM10.
FIGURE 8.
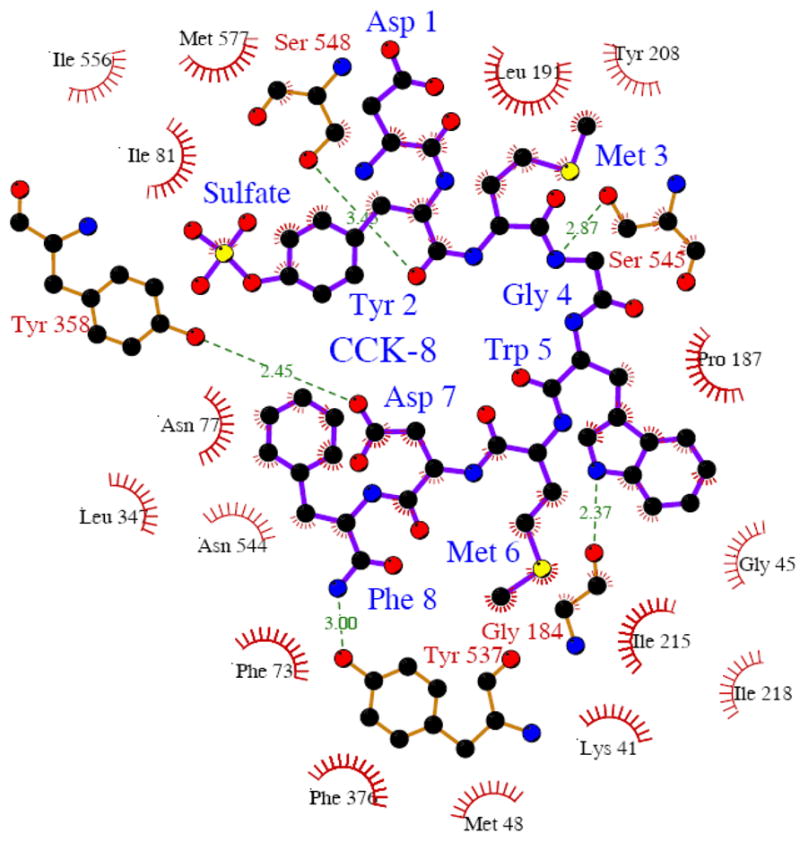
Schematic representations of hydrogen bond and hydrophobic interactions between CCK-8 and OATP1B3. Dashed lines represent hydrogen bonds between OATP1B3 and CCK-8, and spikes represent residues from OATP1B3 involved in hydrophobic contacts with CCK-8. This figure was made with the program Ligplot (21).
DISCUSSION
OATP1B3 and OATP1B1 are highly homologous proteins and share numerous substrates. However, CCK-8 is an OATP1B3-specific substrate. In order to determine which regions and residues are essential for OATP1B3-mediated CCK-8 transport, we constructed chimeric proteins between OATP1B3 and 1B1 and used site-directed mutagenesis to change individual amino acid residues in OATP1B3 to the corresponding residues in OATP1B1. Our results show that TM10 which is located in the C-terminal half of OATP1B3, is critical for CCK-8 transport. Furthermore, we have identified three amino acid residues in TM10, namely Y537, S545 and T550, that appear to be the most important residues for CCK-8 transport among the fourteen different residues between OATP1B3 and 1B1 in this region. Triple mutation of these three residues impairs OATP1B3-mediated CCK-8 transport nearly as strong as the whole TM10 mutation and results in almost identical kinetics. Molecular simulation predicts that TM10 contributes 3 out of 5 hydrogen bond interactions with CCK-8 (formed by Y537, S545 and S548), emphasizing its important role in CCK-8 binding/translocation.
Our observation that chimera T1–6 lost almost all of OATP1B3-mediated CCK-8 transport while chimera T7–12 maintained some of its activity (Figure 2) provides strong evidence that the C-terminal half of OATP1B3 contains the key domains for CCK-8 recognition/translocation. This conclusion is further supported by the results with the single TM replacements which demonstrate that chimera 1B3_T10 compromises CCK-8 transport function most dramatically (Figure 4). Evidence for the importance of TM10 in other OATPs has already been published. Pucci et al. (25) have reported that the C-terminal domain of the human prostaglandin transporter hPGT (OATP2A1) determines its binding affinity for prostaglandin E2. Based on cysteine-scanning mutagenesis, Chan et al. (26) proposed that TM10 of rat PGT forms part of its substrate-binding site. Pharmacogenetic studies demonstrated that patients with a non-synonymous polymorphism which results in the mutation L543W located in TM10 of OATP1B1 have a higher risk for pravastatin- or atrovastatin-induced myopathy (27). And finally, preliminary results from our lab indicate that this region is also critical for OATP1B1-mediated transport. Therefore, we conclude that TM10 might also be important for the function of other OATP family members.
Besides human OATP1B3, the liver-specific rat Oatp1b2 also transports CCK-8, whereas Oatp1a1, Oatp1a4 and Oatp1a5 do not show any measurable uptake (11). Sequence alignment analysis of the members of the OATP1 family shows that Y537 is conserved between OATP1B3 and both rat and mouse Oatp1b2, but replaced by a phenylalanine in all other OATP1 family members. This provides additional evidence for the importance of Y537 for CCK-8 transport. Serine at position 545 is also conserved between OATP1B3 and Oatp1b2, but is also found in several other OATP1 family members. These two residues might be important by forming hydrogen bonds with CCK-8 according to our molecular simulation results (Figure 8).
Mutations and polymorphisms can affect the activity of transporters by various mechanisms such as impairing their membrane localization, changing their binding affinity for substrates and altering their translocation capacity. In this study, all constructs were expressed on the plasma membrane, their expression levels were measured by surface biotinylation and western blot analysis and transport was corrected for surface expression. Hence, altered transport activities of the different chimeras investigated in this study reflect alteration of transport kinetics and not alterations of expression. Therefore, the dramatically reduced Vmax values of chimera 1B3_TM10 and of the triple mutant Y537F/S545L/T550L as compared to OATP1B3 (Table 1) suggest that the translocation capacity of OATP1B3 is greatly affected when TM10 or some of the amino acid residues in this region are changed.
To examine whether TM10 is also important for the transport of other substrates, we tested OATP1B3- and chimera 1B3_TM10-mediated uptake of [D-penicillamine2,5]enkephalin (DPDPE) and taurocholate. At low concentration, both of them exhibit higher transport in OATP1B3- than in OATP1B1-expressing HEK293 cells. Chimera 1B3_TM10 which does not transport CCK-8 (Figure 4) transports DPDPE to the same level of OATP1B3. Surprisingly, chimera 1B3_TM10 even showed increased taurocholate transport activity (data not shown). Thus, TM10 of OATP1B3 might play different roles in the transport of different substrates. Docking studies indicate that DPDPE forms a hydrogen bond with S548 and taurocholate forms a hydrogen bond with N544. Both S548 and N544 are conserved between OATP1B1 and 1B3. A similar phenomenon was described for a point mutation in TM11 of OATP1B3. When stably expressed in HEK293 cells, OATP1B3 with a glycine to glutamate mutation at position 583 showed partial transport activity for BSP and CCK-8, but no transport for estradiol-17β-glucuronide and dehydroepiandrosterone-3-sulfate (DHEAS) (28). One possible explanation might be that due to their structural diversity, different substrates interact with OATP1B3 through different residues in the substrate binding site. Furthermore, our previous studies on the interactions of clotrimazole with OATP1B3 indicated that OATP1B3 has distinct binding sites for its substrate estradiol-17β-glucuronide and clotrimazole, since clotrimazole can stimulate OATP1B3-mediated estradiol-17β-glucuronide transport (29). The molecular determinants for OATP1B3-mediated transport of other substrates remain to be elucidated.
In conclusion, our studies reveal that TM10 and in particular the three amino acid residues Y537, S545 and T550 are key structural and molecular elements for the transport of CCK-8 by OATP1B3. This information should be valuable for the elucidation of the structure-function relationship of OATP1B3.
Acknowledgments
We thank Dr. Bao Ting Zhu for sharing his computational resources with us.
Footnotes
This work was supported by National Institute of Health grants RR021940, and GM077336.
Abbreviations: BSP, bromosulfophthalein; CCK-8, cholecystokinin octapeptide; DHEAS, dehydroepiandrosterone-3-sulfate; DPDPE, [D-penicillamine2,5]enkephalin; EL, extracellular loop; HEK293, human embryonic kidney cells; OATP, organic anion transporting polypeptide; PBS, phosphate-buffered saline; SLC, solute carrier; TM, transmembrane domain.
References
- 1.Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 2.Hagenbuch B, Meier PJ. Organic anion transporting polypeptides of the OATP/SLC21 family: phylogenetic classification as OATP/SLCO superfamily, new nomenclature and molecular/functional properties. Pflugers Arch. 2004;447:653–665. doi: 10.1007/s00424-003-1168-y. [DOI] [PubMed] [Google Scholar]
- 3.Hagenbuch B, Meier PJ. The superfamily of organic anion transporting polypeptides. Biochim Biophys Acta. 2003;1609:1–18. doi: 10.1016/s0005-2736(02)00633-8. [DOI] [PubMed] [Google Scholar]
- 4.König J, Seithel A, Gradhand U, Fromm MF. Pharmacogenomics of human OATP transporters. Naunyn Schmiedebergs Arch Pharmacol. 2006;372:432–443. doi: 10.1007/s00210-006-0040-y. [DOI] [PubMed] [Google Scholar]
- 5.Abe T, Kakyo M, Tokui T, Nakagomi R, Nishio T, Nakai D, Nomura H, Unno M, Suzuki M, Naitoh T, Matsuno S, Yawo H. Identification of a novel gene family encoding human liver-specific organic anion transporter LST-1. J Biol Chem. 1999;274:17159–17163. doi: 10.1074/jbc.274.24.17159. [DOI] [PubMed] [Google Scholar]
- 6.Abe T, Unno M, Onogawa T, Tokui T, Kondo TN, Nakagomi R, Adachi H, Fujiwara K, Okabe M, Suzuki T, Nunoki K, Sato E, Kakyo M, Nishio T, Sugita J, Asano N, Tanemoto M, Seki M, Date F, Ono K, Kondo Y, Shiiba K, Suzuki M, Ohtani H, Shimosegawa T, Iinuma Nagura H, Ito S, Matsuno S. LST-2, a human liver-specific organic anion transporter, determines methotrexate sensitivity in gastrointestinal cancers. Gastroenterology. 2001;120:1689–1699. doi: 10.1053/gast.2001.24804. [DOI] [PubMed] [Google Scholar]
- 7.Hsiang B, Zhu Y, Wang Z, Wu Y, Sasseville V, Yang WP, Kirchgessner TG. A novel human hepatic organic anion transporting polypeptide (OATP2) J Biol Chem. 1999;274:37161–37168. doi: 10.1074/jbc.274.52.37161. [DOI] [PubMed] [Google Scholar]
- 8.König J, Cui Y, Nies AT, Keppler D. A novel human organic anion transporting polypeptide localized to the basolateral hepatocyte membrane. Am J Physiol. 2000;278:G156–G164. doi: 10.1152/ajpgi.2000.278.1.G156. [DOI] [PubMed] [Google Scholar]
- 9.König J, Cui Y, Nies AT, Keppler D. Localization and genomic organization of a new hepatocellular organic anion transporting polypeptide. J Biol Chem. 2000;275:23161–23168. doi: 10.1074/jbc.M001448200. [DOI] [PubMed] [Google Scholar]
- 10.Wang P, Hata S, Xiao Y, Murray JW, Wolkoff AW. Topological assessment of oatp1a1: a 12-transmembrane domain integral membrane protein with three N-linked carbohydrate chains. Am J Physiol. 2008;294:G1052–1059. doi: 10.1152/ajpgi.00584.2007. [DOI] [PubMed] [Google Scholar]
- 11.Ismair MG, Stieger B, Cattori V, Hagenbuch B, Fried M, Meier PJ, Kullak-Ublick GA. Hepatic uptake of cholecystikinin octapeptide by organic anion-transporting polypeptides OATP4 and OATP8 of rat and human liver. Gastroenterology. 2001;121:1185–1190. doi: 10.1053/gast.2001.28704. [DOI] [PubMed] [Google Scholar]
- 12.Smith NF, Acharya MR, Desai N, Figg WD, Sparreboom A. Identification of OATP1B3 as a high-affinity hepatocellular transporter of paclitaxel. Cancer Biol Ther. 2005;4:815–818. doi: 10.4161/cbt.4.8.1867. [DOI] [PubMed] [Google Scholar]
- 13.Kullak-Ublick GA, Ismair MG, Stieger B, Landmann L, Huber R, Pizzagalli F, Fattinger K, Meier PJ, Hagenbuch B. Organic anion-transporting polypeptide B (OATP-B) and its functional comparison with three other OATPs of human liver. Gastroenterology. 2001;120:525–533. doi: 10.1053/gast.2001.21176. [DOI] [PubMed] [Google Scholar]
- 14.Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J Biol Chem. 2001;276:35669–35675. doi: 10.1074/jbc.M103792200. [DOI] [PubMed] [Google Scholar]
- 15.Smith NF, Marsh S, Scott-Horton TJ, Hamada A, Mielke S, Mross K, Figg WD, Verweij J, McLeod HL, Sparreboom A. Variants in the SLCO1B3 gene: interethnic distribution and association with paclitaxel pharmacokinetics. Clin Pharmacol Ther. 2007;81:76–82. doi: 10.1038/sj.clpt.6100011. [DOI] [PubMed] [Google Scholar]
- 16.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 17.Accelrys. InsightII [molecular modeling package], Version 2005. Calif (USA): Molecular Simulation Inc; 2005. [Google Scholar]
- 18.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 19.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 20.Tripos. Sybyl [molecular modeling package], Version 7.1. St Louis (MO): [Google Scholar]
- 21.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 23.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pucci ML, Bao Y, Chan B, Itoh S, Lu R, Copeland NG, Gilbert DJ, Jenkins NA, Schuster VL. Cloning of mouse prostaglandin transporter PGT cDNA: species-specific substrate affinities. Am J Physiol. 1999;299:R734–741. doi: 10.1152/ajpregu.1999.277.3.R734. [DOI] [PubMed] [Google Scholar]
- 26.Chan BS, Satriano JA, Schuster VL. Mapping the substrate binding site of the prostaglandin transporter PGT by cysteine scanning mutagenesis. J Biol Chem. 1999;274:25564–25570. doi: 10.1074/jbc.274.36.25564. [DOI] [PubMed] [Google Scholar]
- 27.Morimoto K, Oishi T, Ueda S, Ueda M, Hosokawa M, Chiba K. A novel variant allele of OATP-C (SLCO1B1) found in a Japanese patient with pravastatin-induced myopathy. Drug Metab Pharmacokinet. 2004;19:453–455. doi: 10.2133/dmpk.19.453. [DOI] [PubMed] [Google Scholar]
- 28.Letschert K, Keppler D, Konig J. Mutations in the SLCO1B3 gene affecting the substrate specificity of the hepatocellular uptake transporter OATP1B3 (OATP8) Pharmacogenetics. 2004;14:441–452. doi: 10.1097/01.fpc.0000114744.08559.92. [DOI] [PubMed] [Google Scholar]
- 29.Gui C, Miao Y, Thompson L, Wahlgren B, Mock M, Stieger B, Hagenbuch B. Effect of pregnane X receptor ligands on transport mediated by human OATP1B1 and OATP1B3. Eur J Pharmacol. 2008;584:57–65. doi: 10.1016/j.ejphar.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]