Summary
Proper nerve connections form when growing axons terminate at the correct postsynaptic target. Here I show Transforming growth factor-beta (TGFβ) signals regulate axon growth. In most contexts, TGFβ signals are tightly linked to Smad transcriptional activity. Although known to exist, how Smad-independent pathways mediate TGFβ responses in vivo is unclear. In Drosophila mushroom body (MB) neurons, loss of the TGFβ receptor Baboon (Babo) results in axon overextension. Conversely, misexpression of constitutively active Babo results in premature axon termination. Smad activity is not required for these phenotypes. This study shows that Babo signals require the Rho GTPases Rho1 and Rac, and LIM kinase1, which regulate the actin cytoskeleton. Contrary to the well-established receptor activation model, in which type 1 receptors act downstream of type 2 receptors, this study shows that the type 2 receptors Wishful thinking (Wit) and Punt act downstream of the Babo type 1 receptor. Wit and Punt regulate axon growth independently, and interchangeably, through LIMK1-dependent and -independent mechanisms. Thus, novel TGFβ receptor interactions control non-Smad signals and regulate multiple aspects of axonal development in vivo.
Keywords: neural development, signal transduction, cytoskeleton, Drosophila
Introduction
In developing neurons, axon and dendrite extensions are directed by specialised motile structures termed growth cones. These extensions are often long and intricate but once nerve growth cones have reached their targets, cell extensions stop and synaptogenesis begins. How this takes place in vivo is unclear. Extracellular cues often direct growth cone motility through cytoskeletal reorganisation. Many (if not all) axon guidance cues regulate the nerve cell cytoskeleton through Rho family GTPases (Luo, 2002). Multiple aspects of axonal development are regulated by Rho GTPases. Although Rac generally mediates axon extension and attractive responses, and Rho1 (also known as RhoA) generally mediates axon retraction and axon repulsion, these distinctions can be complex. For example, Drosophila genetic studies show axon outgrowth and attractive responses mediated by Netrin (Forsthoefel et al., 2005), and axon repulsive cues mediated by Robo (Fan et al., 2003; Matsuura et al., 2004) both of which depend on Rac subfamily GTPases. Similarly, Drosophila Rho1 signals can mediate axon retraction (Billuart et al., 2001) and axon attraction (Bashaw et al., 2001) in different neurons. These, and many other, studies highlight the key and complex roles Rho GTPases play in growth cone responses.
Studies on mushroom body (MB) neurons in the Drosophila brain show Rho proteins regulate axon growth through LIM kinase (LIMK)-dependent and LIMK-independent pathways, and hat they can act antagonistically (Ng and Luo, 2004). LIMK regulates actin filament turnover by phosphorylating, and thereby inactivating, an actin depolymerisation and severing factor, ADF/cofilin (Bamburg, 1999). LIMK1 misexpression in neurons, in vitro or in vivo, leads to axon growth inhibition. Consistent with a role in ADF/Cofilin regulation, this phenotype is suppressed by increasing cofilin activity, either by co-expressing wild type cofilin or a form of (S3A) that cannot be phosphorylated, or by expressing a cofilin phosphatase, Slingshot (Ssh) (Endo et al., 2003; Ng and Luo, 2004). In Drosophila, one homologue of ADF/cofilin exists, twinstar (tsr), and its inactivation results in growth cone morphology and axon growth defects. These results suggest cofilin phosphoregulation is essential for axon growth.
How extracellular cues pattern axons through Rho GTPase and cofilin regulation in vivo is unclear. Here I show components of the Transforming growth factor-beta (TGFβ) pathway are involved. The TGFβ pathway regulates many morphogenic events, including cell fate specification, cell migration, proliferation and apoptosis (Hogan, 1996; Massague et al., 2000; Raftery and Sutherland, 1999). The conserved TGFβ pathway consists of a core complex of type 1 and type 2 transmembrane receptor serine/threonine kinases, which are activated by secreted TGFβ ligands (bone morphogenetic proteins, BMPs, or TGFβ/Activins)(Feng and Derynck, 2005; Shi and Massague, 2003). The presence of ligand dimers triggers a signalling cascade involving the receptor complex. The following events are essential; phosphorylation of type 1 receptors by the type 2 receptor kinase; phosphorylation of receptor activated Smads (R-Smads) by the type 1 receptor kinase; R-Smad complex formation with a common Smad (co-Smad); translocation of Smad complexes into the nucleus to elicit gene transcription. In Drosophila, there are three type 1 receptors, Baboon (Babo), Thickveins (Tkv) and Saxophone (Sax), and two type 2 receptors, Wishful thinking (Wit) and Punt (Put). The activated receptors phosphorylate two R-Smads, Mad and Smad2 (also known as dSmad2 and Smox - Flybase), which form a trimeric complex with the co-Smad, Medea (Med). In most models, Smad activation is an obligate effector response upon ligand binding.
Although Smad-independent pathways are known (Derynck and Zhang, 2003; Moustakas and Heldin, 2005; Foletta et al., 2003; Lee-Hoeflich et al., 2004; Ozdamar et al., 2005), how they affect development in vivo is unclear. In many instances, Smad-independent pathways exhibit cross-regulatory effects, which either regulate Smads or are under Smad regulation. However, some TGFβ signals are Smad-independent events. In C. elegans, mutations in a TGFβ signal (unc-129) result in dorsal-ventral axon guidance defects (Colavita et al., 1998). Mutation analyses of other TGFβ components, such as receptors or Smads, do not reveal this phenotype suggesting that axon guidance in worms involves atypical TGFβ signalling mechanisms. TGFβ signals also regulate dorsal-ventral axon guidance in the developing mouse spinal cord. BMP-7 expression in the dorsal roof plate acts to repel spinal cord neurons and guide their projections ventrally (Augsburger et al., 1999; Butler and Dodd, 2003). Whether Smads are involved is unclear; nonetheless, the rapid axonal responses would seem to preclude transcriptional events.
Recent studies have shown that BMP-4 and BMP-7 treatment in mammalian non-neuronal and neuronal cell cultures, respectively, leads to LIMK activation, resulting in a rapid increase in cofilin phosphorylation (Foletta et al., 2003; Lee-Hoeflich et al., 2004). This requires a direct interaction between the COOH-terminal tail of a BMP receptor (BMPR2), which is dispensable for Smad signalling, and LIMK. Lee-Hoeflich et al. (Hoeflich et al., 2004) have further shown that the BMPR2 COOH-terminus is required for dendritogenesis in cultured cortical neurons. Mammalian BMPs also regulate growth cone turning responses in cultured Xenopus spinal neurons (Wen et al., 2007). BMP-7 exposure causes attractive or repulsive growth cone turning behaviours by regulating cofilin through LIMK1 or Ssh activities, respectively.
Drosophila LIMK1 is essential for synaptic stability controlled by BMPs. Genetic analysis of Drosophila neuromuscular junction (NMJ) reveals that the stability of presynaptic terminals requires a retrograde BMP-type signal Glass bottom boat (Gbb) that acts through Wit (the Drosophila homologue of BMPR2). Like BMPR2, Wit binds to LIMK1 via its COOH-terminal extension. Without this interaction, NMJ synapses can grow (through Wit signalling via the Drosophila Smads, Mad and Medea) but they have defects in synaptic stability (Eaton and Davis, 2005). How TGFβ receptor interactions regulate LIMK1 is unclear (Foletta et al., 2003; Lee-Hoeflich et al., 2004). Nor is it clear how LIMK1 regulates synapses, as cofilin phosphoregulation does not appear to be essential (Eaton and Davis, 2005).
Here, I show that TGFβ signals regulate distinct aspects of axonal development. Loss of Babo results in MB axon overextension, while in other neurons axon outgrowth and targeting defects are observed. The results show that Babo acts together with Wit and Put, but is independent of Smads. Babo signals depend on Rho1, Rac and LIMK1. Consistent with a role in LIMK1 regulation, babo and wit genetically interact with LIMK1. babo and LIMK1 gain-of-function phenotypes are similar, and both are suppressed by increasing cofilin activity. Contrary to the canonical receptor activation model, the type 2 receptors Wit and Put both act downstream of the Babo type1 receptor, and distinct LIMK1-dependent and -independent pathways are required.
Materials and Methods
Drosophila strains
LIMK, tsr, ssh, RhoGEF2, pbl, trio, sif, RhoGAP p190, Rac, Rho, Cdc42, Pak and Rok mutant and transgenic strains were previously described and are referenced therein (Ng and Luo, 2004). The following additional strains were used: babo32, babo52, UAS-activated baboa Q302D (CA babo) (Brummel et al., 1999); tkv4, tkv7 (Penton et al., 1994); tkv4a21 (Gibson and Perrimon, 2005); UAS-putΔI, UAS-tkv1ΔGSK (DN tkv), UAS-saxΔI (DN sax), UAS-tkv1A (HA) Q199D (CA tkv), UAS-saxA (HA) Q263D (CA sax) (Haerry et al., 1998); sax4 (Singer et al., 1997); saxP, UAS-put (Nellen et al., 1994); UAS-babo-a, UAS-babo-b::Flag, UAS-baboaΔI (DN babo) ((Zheng et al., 2006); a gift from M. O’Connor, HHMI/University of Minnesota, Minneapolis and Theo Haerry, Florida Atlantic University, Boca Raton); witA12, witB11, witG15, P{wit genomic} (P{wit+}), P{wit tailess} (P{witΔC}), UAS-wit, UAS-witΔC (Marques et al., 2002); UAS-witΔI (McCabe et al., 2003); put135, UAS-put (Ruberte et al., 1995); put62 (Simin et al., 1998); Mad12 (Sekelsky et al., 1995); Med13 (Hudson et al., 1998); Smad21 (Zheng et al., 2003); UAS-Dad (Tsuneizumi et al., 1997); UAS-MYC::tum (RacGAP 50C) (Goldstein et al., 2005); UAS-EcR-B1 (Lee et al., 2000); Df(1)HF368, UAS-RhoGEF2 (Bloomington Drosophila Stock Center). Constitutively active (CA) forms of type 1 receptors result from a conserved Gln (Q) mutation to a Glu (D) leading to constitutively-active kinase activity (Wieser et al., 1995). Dominant-negative (DN) forms of type 1 and type 2 receptors derive from cytoplasmic deletions, with the loss of intracellular domains (cited above). Genetic crossing schemes used in this study are available upon request.
MARCM and Gal4-UAS expression studies
Loss-of-function clones were generated using the MARCM method (Lee and Luo, 1999). Neuroblast and single-cell αβ clones were generated as previously described (Ng et al., 2002). Neurons were visualised using the Gal4-OK107 driver expressing UAS-mCD8::GFP. Misexpression studies were performed using the same driver. For CA and DN misexpression studies, unless indicated otherwise, multiple copies (2-4) of the UAS transgene were used to derive the strongest, possible phenotypes. The strength of CA Babo phenotypes was correlated with Babo expression levels, using one, two, or four copies of UAS-CA babo (data not shown; Figs 4, 5, 7, and see Figs. S2 and S6D in the supplementary material). The data shown in Figs 4, 5 and 7 were obtained using two copies (UAS lines 1B and 9B). MARCM clones were visualised by immunostaining using anti-CD8 (Caltag, clone CT-CD8a, 1:100) and anti-Fas2 (a gift from G. Tear, King’s College London; clone 1D4, 1:5) antibodies. In misexpression studies, neurons were visualised using epifluorescent CD8::GFP together with anti-Fas2 staining. Additional antibodies used were HA (Santa Cruz, Y11, 1:500), Babo (Abcam, ab14681, 1:50), Wit (a gift from H. Aberle, MPI Developmental Biology, Tübingen; clone 23C7, 1:10), and FLAG (Sigma, clone M5, 1:200). These were used to estimate the level and localization of ectopic Sax-HA, Tkv-HA, Babo, Wit and WitΔC-FLAG proteins (respectively) in neurons. Although endogenous Babo and Wit were detected throughout brain tissue, ectopic levels were distinguished using these antibodies. Drosophila brains were dissected, fixed and stained as previously described (Ng et al., 2002). Confocal images were generated with a Zeiss LSM510 confocal microscope, using Zeiss LSM510, Image J and Adobe Photoshop software.
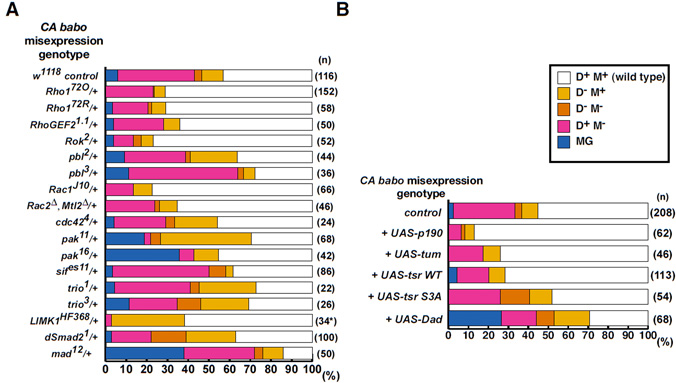
Fig. 4. CA babo misexpression resulted in MB axon truncations.
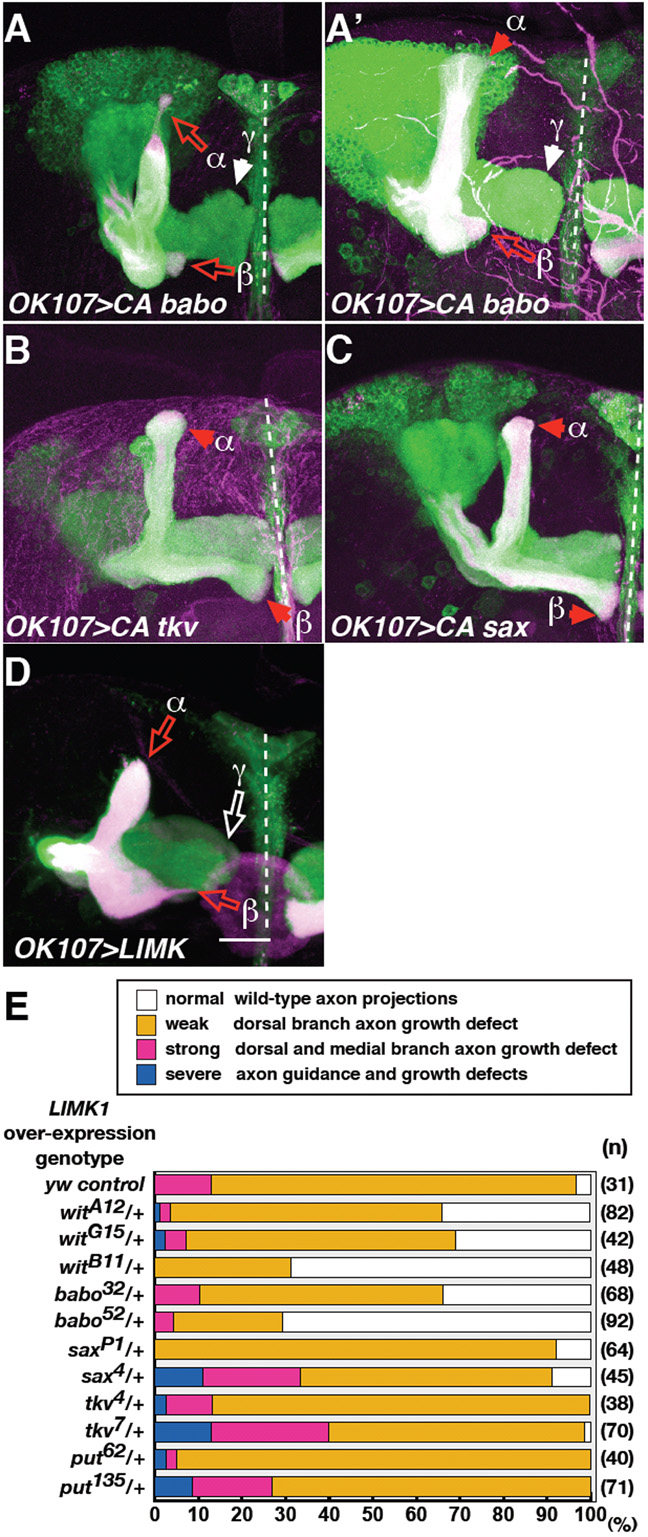
babo and wit genetically interact with LIMK1.
(A-D) Drosophila MB neurons misexpressing CA babo (A, A’), CA tkv (B), CA sax (C), or LIMK1 (D). Solid red or white arrowheads indicate normal α, β or γ lobe termination points, as indicated. Open red or white arrows indicate axon truncations in α, and β lobes, or in γ lobes, respectively. In D, the cell body section was removed to clearly reveal axon phenotypes. Scale bar: 20 μm.
(E) Quantification of axon growth defects in LIMK1-overexpressing neurons (using intermediate expression line F4) in the presence of control (y, w), or one copy of each TGFβ receptor mutant. Phenotypic quantifications were carried out as previously (Ng and Luo, 2004), and briefly summarised in the key. N; number of hemispheres examined.
Fig. 5. CA babo genetically interacts with the components of the Rho GTPase pathway.
(A) Quantification of CA Babo defects in the presence of control (w1118) or one mutant copy of Rho or Smad, as indicated. CA Babo phenotypes were classed according to the loss or truncation of dorsal (D-M+), medial (D+M-), or both lobes (D-M-). Axon fasciculation defects were also observed (classed as misguidance, ‘MG’; see Fig. S1 in the supplementary material). Based on the level of Babo expression (see Materials and methods), misguidance represents the strongest and loss of dorsal lobes represents the weakest phenotypes (MG > D+M- > D-M- > D-M+). The asterisk denotes CA Babo-induced β lobe overextensions upon the loss of one copy of LIMK1.
(B) Quantification of CA Babo defects in control (UAS-mCD8::GFP), or with one copy of the indicated transgene. n, number of hemispheres examined.
Fig. 7. Wit and Punt act downstream of Babo.
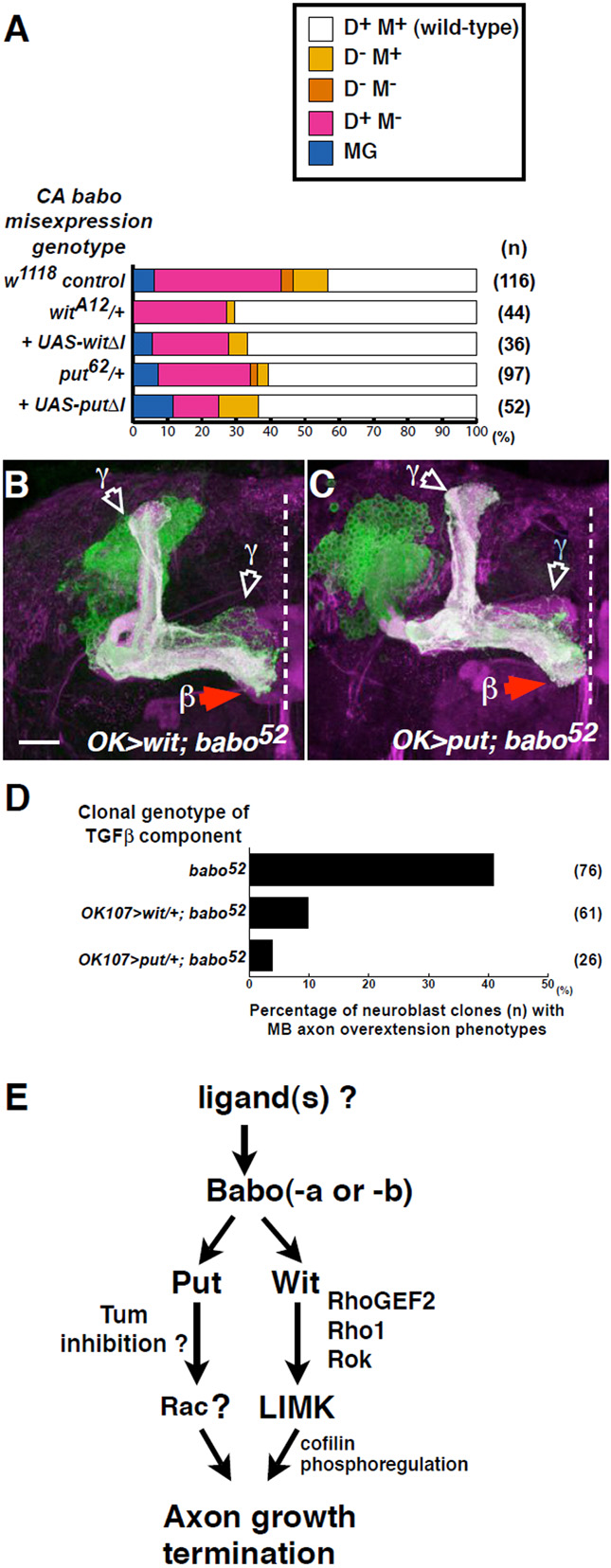
(A) Quantification of CA Babo defects in control (w1118), or with one mutant copy of wit, put, UAS-witΔI or UAS-putΔI, as indicated. n, number of hemispheres examined.
(B,C) babo null clones expressing either UAS-wit (B), or UAS-put (C). Wit or Punt expression suppresses the babo axon overextension but not the axon pruning phenotype. Scale bar: 20 μm
D) Quantification of babo axon growth phenotypes in the presence of one copy of UAS-wit, or UAS-put, as indicated.
(E) A model of Babo-regulated axon growth derived from data in this study.
Results
MB intrinsic neurons (‘Kenyon’ cells) in the Drosophila brain are well characterised with respect to their cell division, differentiation and projection patterns (Ito et al., 1997; Kurusu et al., 2002; Lee et al., 1999). There are three different sets of adult MB neurons (γ, α’β’ and αβ), which are born at different periods from common neuroblast progenitors and have distinct axonal projections (Lee et al., 1999) (Fig. 1A, B). Each neuron extends a primary neurite that gives rise to dendrites near the cell body, and a single axon that projects anterioventrally through the peduncle. Axons of α‘β’ or αβ neurons bifurcate to form a dorsal and a medial branch, whereas γ neurons extend only a medial branch (branches are also referred to collectively as ‘lobes’). All axons terminate either medially, close to the midline, or close to the anterior dorsal cortex (Fig. 1A, B).
Fig. 1. Babo inactivation results in axon overextension.
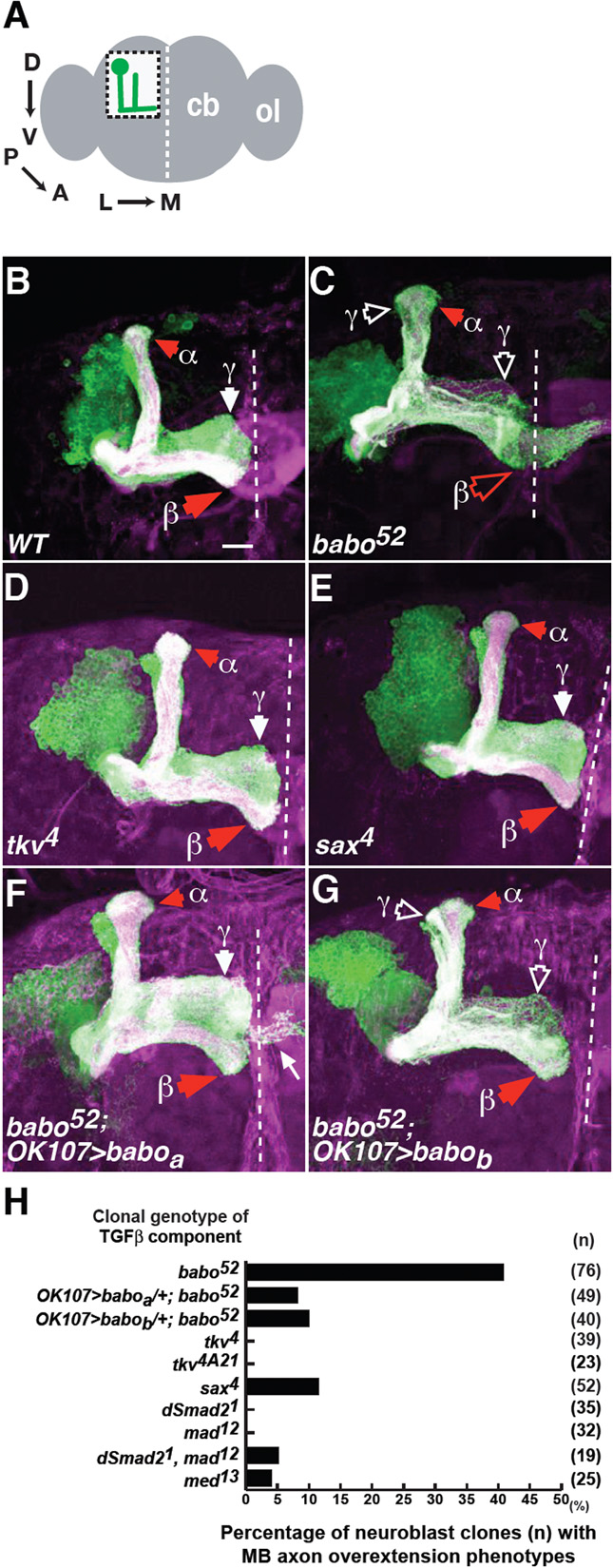
Babo regulates axon growth through Babo-a and -b isoforms.
(A) Schematic of the adult Drosophila whole brain. The boxed region shows mushroom body (MB) neurons in the left hemisphere of the central brain (cb). Arrows show the MB axon trajectory extending from posterior dorsal cell bodies, projecting anteroventrally and then turning towards the midline. The MB images shown are either from the left hemisphere in this orientation, or of the central brain, showing both hemispheres. Dashed white lines indicate the midline. ol, optic lobe. D, dorsal; V, ventral; P, posterior; A, anterior; L, lateral; M, medial.
(B) A wild-type MB neuroblast clone. Typical adult, wild-type clones generated from newly hatched larvae have axonal projections that terminate either in the dorsal anterior cortex or just prior to the midline. Only γ, α, and β projections are indicated.
(C-E) Representative images of babo52 (C), tkv4 (D) and sax4 (E) neuroblast clones. Note β lobe overextensions (open red arrowhead) across the midline in babo clones. Open white arrowheads indicate γ axon pruning defects, in this and subsequent figures.
(F,G) Representative images of babo52 neuroblast clones expressing either UAS-baboa (F), or UAS-babob (G). Many axons in the UAS-baboa rescue exhibited small protrusions that were not characteristic of any lobe (thin white arrow in F). These represent ectopic projections of a subclass of MB axons induced in the OK107>baboa genetic background. In these and subsequent figures, solid red or white arrowheads indicate normal α and β or γ lobe termination points, as indicated. All images in this and subsequent figures are z-projections of confocal sections. Green, expression of the marker mCD8::GFP on all MB, neuroblast or single-cell MARCM clones (sometimes multiple single cell clones); magenta, Fas2 staining of all MB γ (weakly stained) and αβ (strongly stained) axons (appearing as white when overlapping with mCD8::GFP). Dashed white line, midline. Scale bar: 20 μm.
(H) Quantification of axon overextension defects in the indicated genotype. n, number of neuroblast clones examined.
Babo inactivation results in MB axon overextension
To study the role of TGFβ signals in MB neurons, mutant clones were generated using strong, loss-of-function or null alleles of type 1 receptors babo, tkv and sax. babo-null (babo52) neuroblast clones had axon overextension phenotypes in αβ neurons, with β lobes overextending across the midline (Fig. 1, compare C with B, quantified in H). Consistent with previous studies (Zheng et al., 2003), babo clones also exhibited axon pruning defects, characterised by the presence of larval-stage dorsal and medial projections in adult brains (open white arrowheads in Fig. 1C). In wild-type adults, each γ neuron re-extends a single medial branch after axon pruning and the γ lobe appears more defasciculated along the dorsal-ventral axis (Fig. 1B). By mutant clonal analysis or by dominant-negative (DN) misexpression, loss of tkv or sax did not result in these defects (Fig. 1D, E, H; data not shown). These results suggest that Babo regulates axon growth, particularly of the β lobe.
Baboa and Babob isoforms regulate axon growth cell-autonomously
Recent data suggest that different Babo isoforms have distinct neural functions (Zheng et al., 2006). Expression of Baboa but not Babob isoform rescues the babo MB axon pruning phenotypes. In contrast, either isoform rescues the babo axon extension defects of dorsal cluster (DC) neurons in the optic lobe. To test whether different Babo isoforms regulate MB axon growth similar assays were performed. In a wild-type background, ectopically expressed Baboa or Babob was detected in all MB lobes and did not disrupt axonal projections (see Fig. S2 in the supplementary material). Baboa or Babob expression in babo52 neuroblast clones rescued the axon overextension defect, as most β lobes terminated correctly (Fig. 1F, G, H). Thus, either Babo isoform can regulate axon growth.
Consistent with a cell-autonomous role, Babo inactivation in single αβ neurons resulted in similar axon overextensions. Interestingly, non cell-autonomy was also observed, as single babo neurons caused heterozygote axons to similarly overextend across the midline (see Fig. S1 in the supplementary material).
Babo regulates MB axon growth independently of axon pruning
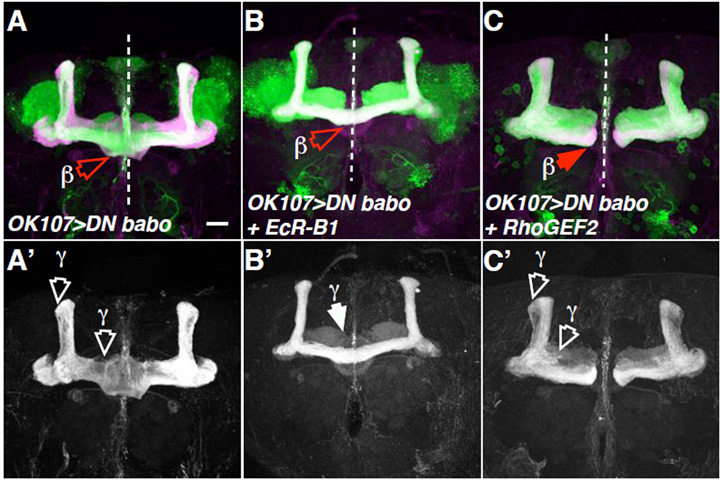
Using a different approach, a DN form of Babo was misexpressed in MB neurons. Like the null phenotype, axon pruning and overextension phenotypes were observed, with β lobes fusing at the midline (Fig. 2A,A’; 65.2% fusion defects, n=23 brains). To determine whether the axon overextension was secondary to axon pruning defects, DN babo was misexpressed together with the Ecdysone receptor B1 isoform (EcR-B1). Similar to previous results (Zheng et al., 2003), these axon pruning defects were suppressed by ectopic EcR-B1 (Fig. 2B’). However, β lobe fusions remained visible (64.5%, n=31; Fig. 2B). Therefore, the DN babo axon overextension was not secondary to the axon pruning defects. Conversely, nor were axon pruning defects a consequence of axon overextension, as UAS-babob expression rescued babo52 axon overextension but not the axon pruning defects (Fig. 1G). Similarly, RhoGEF2 co-expression also suppressed DN Babo axon overextension but not the axon pruning defects (Fig. 2C,C’; see below). Thus, Babo regulates axon pruning and axon growth independently.
Fig. 2. Babo regulates axon pruning and axon growth independently.
(A-C) Drosophila MB neurons misexpressing DN babo (A), DN babo plus EcR-B1 (B), or DN babo plus RhoGEF2 (C). Additional panels (A’,B’,C’) indicate the corresponding Fas2 (magenta in A, B and C) positive axon projections. Note β lobe overextensions (open red arrowheads) in A and B, but absent in C, and axon pruning phenotypes (open white arrowheads highlight the aberrant γ-dorsal and medial branches), which are visible in A’ and C’, but absent in B’. Cell body sections were removed from C to clearly show MB axons. Scale bar: 20 μm.
Babo regulates axon growth independently of Smads
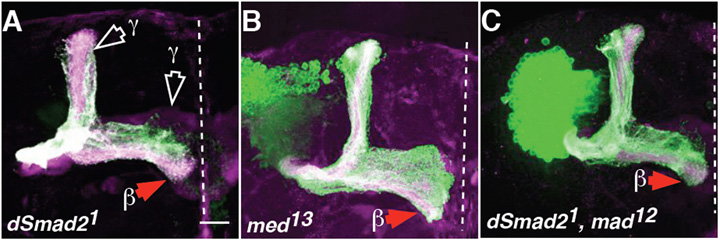
Babo functions through Smad2 (Brummel et al., 1999; Das et al., 1999; Zheng et al., 2003). When Smad2 strong, loss-of-function clones were analysed, axon overextension defects were not detected, although, consistent with previous data (Zheng et al., 2003), axon pruning defects were (Fig. 3A, quantified in Fig. 1H). Null clones of Medea (Med, the Drosophila homologue of the co-Smad/Smad4) also did not exhibit overextension defects (Fig. 3B and Fig. 1H). Recent data suggest that, under certain in vitro conditions, Babo can signal through Mad (Gesualdi and Haerry, 2007). When Mad null, or Smad2, Mad double mutant clones were analysed, axon overextensions were not observed either (data not shown; Figs 3C and 1H). Similarly, in a different strategy, misexpressing an inhibitory form of Smad, Dad, also did not perturb these axons (data not shown; 100% as wild-type, n=26 hemispheres). As Smads could play a redundant role, their role was tested in a sensitised background. Using a Babo gain-of-function phenotype, one mutant copy of either Smad2, Mad or UAS-Dad was introduced with constitutively active (CA) babo (Fig 5A,B; see below). Reducing Smad levels did not suppress CA Babo. In fact, loss of Mad, or Dad misexpression enhanced CA Babo phenotypes. Together, these results suggest that Babo regulates axon growth independently of Smads.
Fig. 3. Babo-regulated axon growth is Smad-independent.
(A-C) Drosophila dSmad21 (A), Med13 (B), and double Smad21, mad12 (C) neuroblast clones did not show β lobe overextensions. Scale bar: 20μm.
Expression of constitutively active Babo inhibits axon growth
To determine how Babo functions independently of Smads, a gain-of-function approach was taken. CA forms of type 1 receptors were misexpressed in MB neurons. CA Babo expression resulted in axon truncation phenotypes, with the loss of dorsal and/or medial branches (Fig. 4A,A’; for quantification see Fig. 5). Axon guidance defects were also observed; however, this phenotype represented a small fraction of animals [classed as misguidance (MG) in Figs 5, 7; see Fig. S2A,B in the supplementary material). To test whether CA Babo phenotypes were simply due to increased levels of Babo protein, ectopic wild type Babo levels were compared with CA Babo levels (see Fig. S2 in the supplementary material). The results showed that the dominant CA Babo phenotype is due to the Q302D mutation, which results in higher kinase activity. High levels of CA Tkv and CA Sax protein were detected in MB axons (data not shown). Nevertheless, these axon projections resembled those of the wild type (CA tkv, 100% as wild-type, n=26 hemispheres; CA sax, 92.1% as wild-type, n=38 hemispheres; Fig. 4B,C, respectively). These results again suggest that Babo, but not Tkv or Sax, regulates axon growth in vivo.
To determine whehter the truncation phenotypes reflect an initial failure of axon extension, as opposed to axons failing to stabilise and subsequently retracting, CA babo-misexpressing animals were developmentally staged and analysed from wandering L3 larvae (data not shown) through to puparium formation. The results suggest that CA Babo resulted in early axon extension defects in developing axons (see Fig. S3 in the supplementary material).
babo and wit genetically interact with LIMK1
LIMK1 misexpression results in similar MB axon phenotypes to those described above (Fig. 4, compare D with A) (Ng and Luo, 2004)). However, in contrast to LIMK1, which also led to γ lobe truncations, only αβ lobes were truncated in CA babo-misexpressing animals. Additionally, in CA babo, β lobes were predominantly disrupted (Fig. 4A’; see quantification in Fig. 5A,B).
To study the link between TGFβ and LIMK1, receptor mutants were introduced to determine if they could modify the LIMK1 misexpression phenotype (Fig. 4E). Loss of one copy of babo or wit suppressed the LIMK1 phenotype. LIMK1 misexpression was not suppressed by other type 1 receptors, such as tkv or sax, or by the other type 2 receptor put. These genetic assays suggest that Babo and Wit positively interact with LIMK1.
Babo-regulated axon growth requires components of the Rho1 and Rac pathway
Drosophila LIMK1 is regulated by Rho GTPases (Rho1, Rac and Cdc42) through the effector kinases, Rok and Pak (Ng and Luo, 2004). To determine whether Babo-regulated axon growth requires the Rho GTPase pathway, genetic interaction assays were performed using CA babo (Fig. 5A). Lowering the level of Rho1 signals, by loss of one copy of Rho1 or of the Rho1 activator RhoGEF2, resulted in a suppression of the CA Babo phenotype. Loss of the Rho1 effector kinase, Rok, also suppressed CA Babo.
When other Rho family members, Cdc42 and Rac (Rac1, Rac2 or Mtl), were tested, loss of Rac1 (using the hypomorphic allele J10), or a combined loss of one copy of Rac2 and Mtl (using null Δ alleles) also suppressed CA Babo (Fig. 5A). Stronger allelic combinations of Rac enhanced the CA Babo phenotypes (unpublished observations). This is expected, based on previous observations that Rac GTPases can play opposite roles in promoting and inhibiting MB axon growth (Ng and Luo, 2004; Ng et al., 2002). Loss of Cdc42 did not suppress CA Babo. Loss of the Cdc42/Rac effector kinase Pak also did not suppress CA Babo, but instead resulted in stronger CA Babo phenotypes. These results suggest, in addition to Rho1, CA Babo-mediated axon growth inhibition also requires Rac, but not Cdc42 or Pak.
In contrast to RhoGEF2, loss of pebble (pbl, another Rho1 activator) did not suppress CA Babo. Loss of the Rac activators, trio and still life (sif), also did not suppress CA Babo. This suggests that Babo regulates Rho1 through RhoGEF2. Whether Babo regulates axon growth via RacGEFs is unclear, although Sif and Trio are unlikely mediators.
Whether inhibiting Rho pathways through RhoGAPs affects CA Babo was then tested (Fig. 5B). In a wild-type background, single-copy expression of UAS-RhoGAPp190 or UAS-tumbleweed (tum, also known as RacGAP50C) did not disrupt normal axonal projections, although, as previously described, RhoGAPp190 caused a mild dorsal lobe overgrowth defect (Billuart et al., 2001; Goldstein et al., 2005). RhoGAPp190, which acts as a Rho1 inhibitor, strongly suppressed CA Babo (Figures 5B; data not shown). This is consistent with previous findings, where ectopic RhoGAPp190 also suppressed LIMK1 misexpression phenotypes (Ng and Luo, 2004). Tum expression also suppressed CA Babo (Figure 5B; data not shown). Drosophila tum genetically interacts with Rac1 in the wing and eye (Sotillos and Campuzano, 2000) and tum mutant clones exhibit MB axon extension defects (Goldstein et al., 2005).
Together, this suggests Babo-regulated axon growth requires the Rho1 and Rac GTPases and involves RhoGEFs (RhoGEF2) and RhoGAPs (RhoGAPp190 and Tum)(Fig. 7E; see below).
DN Babo-induced axon overextensions are suppressed by increased Rho1 activity
Based on these results, one would predict that DN Babo-induced axon overextensions (Fig. 2A; 65.2% fusion defects, n=23 brains) would be suppressed by increased Rho1 signals. Thus, when RhoGEF2 was coexpressed with DN Babo, axon overextension was suppressed (Fig. 2C; 8.7% fusion defects, n=46 brains). RhoGEF2 did not affect the DN Babo-axon pruning defect (Fig. 2C’). Similarly, Rok coexpression also suppressed the DN Babo-axon overextensions, but not the axon pruning phenotype (11.8%, n=34; data not shown).
Other RhoGEFs were tested, but none of these suppressed the DN babo-induced axon overextensions (UAS-pbl, 51.9%, n=77; UAS-trio, 63.3%, n=60; UAS-sif, 43.9%, n=41; data not shown). Taken together, these results suggest that Babo-regulated axon growth requires Rho1, through the activator RhoGEF2, and the effector kinase, Rok (Fig. 7E).
CA Babo is suppressed by loss of LIMK1 and by increased cofilin activity
Given their similar phenotypes, the link between CA Babo and LIMK1 was analysed further. Loss of one copy of LIMK1 (using the deficiency Df(1)HF368) strongly suppressed the CA babo axon truncation phenotype (Fig. 5A). Intriguingly, β lobe overextensions were observed in many CA babo brains (15 out of 17 brains; see Fig. S4 in the supplementary material), suggesting that CA Babo promotes axon extension under low LIMK1 levels. As the LIMK1 misexpression phenotype is inhibited by Drosophila cofilin (Tsr) (Ng and Luo, 2004), tsr was coexpressed with CA babo. Consistent with its predicted role in regulating LIMK1, Tsr (tsr WT) expression suppressed CA Babo (data not shown; Fig. 5B). However, the results suggest that Babo does not regulate cofilin phosphorylation alone (see Discussion).
type 2 receptors Wit and Put regulate axon growth independently and interchangeably
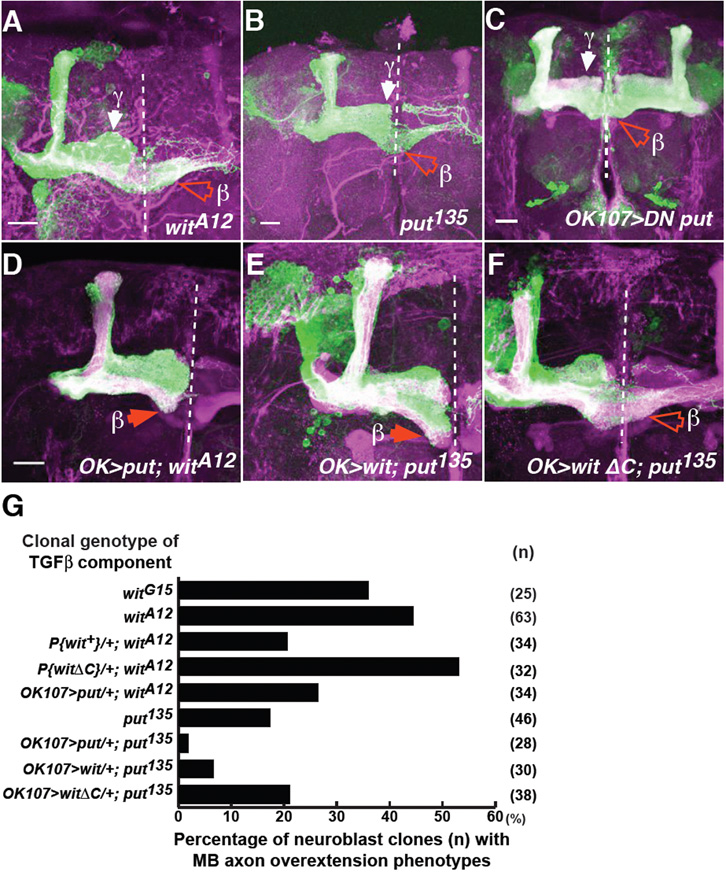
Whether TGFβ type 2 receptors regulate axon growth was tested. wit-null neuroblast clones exhibited β lobe overextensions similar to those of babo mutants (Fig. 6A,G, compared with Fig. 1C). Since the Wit C-terminal tail binds to LIMK1 (Eaton and Davis, 2005), the relevance of this region was analysed. Consistent with previous results, wit mutants are viable in the presence of the “tailess” genomic rescue transgene (P{witΔC}), which lacks the Wit C-terminal region but includes the kinase region (Marques et al., 2002) (data not shown). However, compared to the wild type full-length wit genomic construct (P{wit+}), the tailess wit transgene failed to suppress the wit-null overextensions (data not shown; Fig. 6G). This suggests that the C-terminal region is essential for Wit-regulated axon growth.
Fig. 6. type 2 receptors Wit and Punt regulate axon growth and can function interchangeably.
(A,B) witA12 (A), put135 (B) neuroblast clones show β lobe overextensions (open red arrowheads).
(C) DN put expressing neurons show similar overextensions.
(D-F) wit clones expressing UAS-put (D), or put clones expressing either UAS-wit (E), or UAS-witΔC (F).
(G) Quantification of these defects. n, number of neuroblast clones examined. Scale bars: 20 μm
put strong loss-of-function clones also exhibited (albeit to a lesser extent) axon overextensions (Fig. 6B,G). This was also observed when a DN form of Put (UAS-putΔI) was misexpressed (Fig. 6C; 45.5% fusion defects, n=44 brains).
To test whether type 2 receptors can function interchangeably, UAS-put was expressed in wit clones. wit axon overextensions were suppressed by Put expression (Fig. 6D,G). Conversely, put phenotypes were rescued by UAS-put or UAS-wit (data not shown; Fig. 6E,G). However, put phenotypes were not rescued by the tailess UAS-witΔC (Fig. 6F, G). These results suggest that although Wit and Put regulate axon growth independently, they can function interchangeably (Fig. 7E). However, distinct mechanisms are employed, involving LIMK1-dependent and -independent pathways (see Discussion).
The type 2 receptors Wit and Punt act downstream of type 1 receptor Babo
The results suggest that Babo, Wit and Put work together. In the canonical model of TGFβ signalling, type 1 receptors act downstream of type 2 receptors. Furthermore, activated type 1 receptors propagate Smad signals independently of ligands or type 2 receptors (Brummel et al., 1999; Wieser et al., 1995) and, in vivo, result in ectopic TGFβ responses independently of ligands (Haerry et al., 1998; Lecuit et al., 1996; Nellen et al., 1996). Using CA Babo, the relevance of this model was tested (Fig. 7A). Loss of one copy of wit suppressed CA Babo. Expression of a dominant-negative form of wit (UAS-witΔI), which alone did not disrupt MB axon projection (data not shown), also suppressed CA Babo. In similar assays, one mutant copy of put, or UAS-putΔI coexpression, also suppressed CA Babo. These results suggest that Babo regulates axon growth together with Wit and Put. However, contrary to the canonical model, CA Babo requires the presence of type 2 receptors.
To explore this further, genetic epistasis experiments were performed. Wit and Put were expressed in babo-null neurons (Fig. 7B,C, quantified in D). Ectopic Wit or Put suppressed the babo axon overextension but not the axon pruning phenotype (a Smad-dependent process). Collectively, these results suggest that in Babo-regulated axon growth, type 2 receptors act downstream of type 1 signals (Fig. 7E).
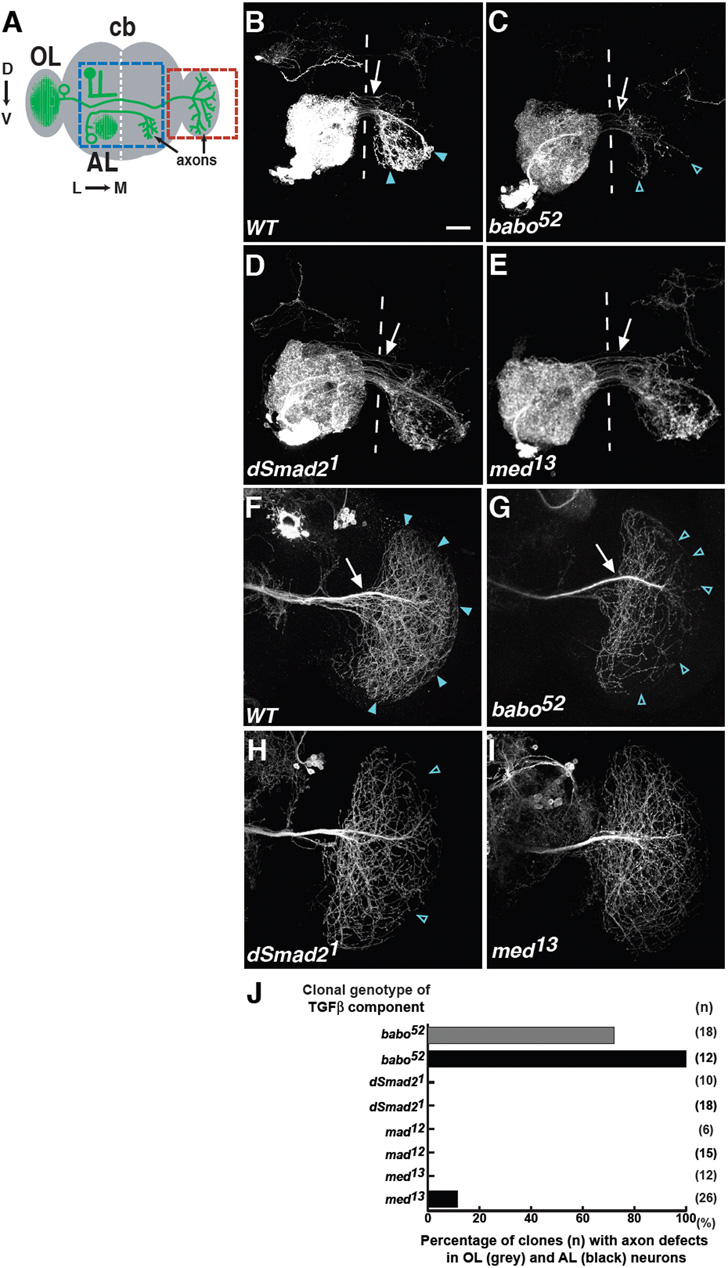
Babo regulates axon extension and targeting of AL and OL axons independently of Smads
To determine whether Babo regulates axon patterning of other neurons, antennal lobe (AL) and optic lobe (OL) contralateral projection neurons were analysed (Ng and Luo, 2004) (Fig. 8A,B,F). As previously described, these neurons extend axons contralaterally into the opposite AL (Fig. 8A,B), or OL (Fig. 8A,F), respectively. babo AL and OL clones showed axonal defects (Fig. 8C,G, quantified in J). babo AL axons were disrupted in the target area and fewer axons extended across the midline. babo OL axons displayed a subtler phenotype: although the number of babo OL axons projecting into the initial target area appeared normal, terminal branches were less elaborated and ‘gaps’ were observed in terminal zones (open blue arrowheads in Fig. 8G; see Fig. S5 in the supplementary material). No gross misprojections were observed. These results suggest that Babo regulates axon extension and targeting in AL neurons, but only axon targeting in OL neurons.
Figure 8. Babo regulates extension and targeting of AL and OL axons independently of Smads.
(A) Schematic of the adult Drosophila brain. Shown from the left hemisphere, antennal lobe (AL) contralateral projection neurons elaborate dendrites (green) ipsilaterally to one AL but project axons contralaterally to the opposite AL. The blue boxed region shows the location of all represented AL images. Also, from the left hemisphere, optic lobe (OL) contralateral projection neurons elaborate dendrites (green) ipsilaterally to one OL, but project axons contralaterally to the opposite OL. The red boxed region shows the orientation of all representative OL axons. Open green circles indicate cell bodies.
(B-I) Wild-type (B,F), babo52 (C,G), Smad21 (D,H), and Med13 (E,I) AL (B-E) and OL (F-I) contralateral projecting neurons. White arrows indicate wild-type number of axons that reach the target zone. Open white arrows indicate axon extension defects. Blue arrowheads indicate wild-type axon termination zones. Open blue arrowheads indicate targeting defects (‘gaps’). Scale bar: 20 μm. See also Fig. S5 in the supplementary material.
(J) Quantification of these OL (grey bars) and AL (black bars) phenotypes. n, number of clones analysed.
The relevance of Smads in AL and OL axonal development was also determined. Smad2 (Fig. 8D,H,J; see Fig. S5 in the supplementary material), Med (Fig. 8E,I,J), or Mad (data not shown; Fig. 8J) mutant clones did not reveal any gross AL or OL axon defects, although gaps similar to babo were occasionally observed in Smad2 OL axons. Thus, as with MB neurons, Babo regulates AL and OL axonal development independently of Smads.
Discussion
This study shows that non-canonical TGFβ signals play multiple roles in axonal development. Babo-regulated axon growth is Smad-independent but requires the type 2 receptors, Wit and Put. Contrary to the canonical receptor activation model, type 2 receptors act downstream of type 1 receptors in axon growth signalling. type 2 receptors work independently and interchangeably, requiring LIMK1-dependent (Wit) and -independent (Put) signals. The experiments show that TGFβ signals act through Rho1, Rac and LIMK1, in part by regulating cofilin. Finally, analysis of different neurons demonstrated that Babo signals do not simply restrict axon extension, but also promote axon extension and axon targeting.
Role of Smad-independent signals in neural connectivity
Once growing axons reach the correct postsynaptic target, axon outgrowth terminates and synaptogenesis begins. These studies suggest that TGFβ signals play a role. When Babo is inactivated, MB axon growth does not terminate properly and overextends across the midline. Consistent with this, CA Babo expression results in precocious termination, forming axon truncations. How Babo is spatially and temporally regulated remains to be determined. Analogous to the Drosophila NMJ, MB axon growth may be terminated through retrograde signalling. Target-derived TGFβ ligands could signal to Babo (on MB axon growth cones) and stop axons growing further. In an alternative scenario, TGFβ ligands may act as a positional cue that prevents MB axons from crossing the midline. Recent data have shown that Babo acting through Smad2 restricts individual R7 photoreceptor axons to single termini (Ting et al., 2007). Loss of Babo, Smad2, or the nuclear import regulator Importin-α3 (Karyopherin α3 - Flybase), results in R7 mutant axons invading neighbouring R7 terminal zones. With the phenotype described here, Babo could similarly be restricting MB axons to appropriate termination zones, and its loss resulting in inappropriate terminations on the contralateral side.
In contrast to MB neurons, Babo inactivation in AL and OL neurons resulted in axon extension and targeting defects. This might reflect cell-intrinsic differences in the response in different neurons to a common Babo signalling programme. This may be the case for MB axon pruning and DC axon extension, which require Babo/Smad2 signals (Zheng et al., 2006). Whether these differences derive from cell-intrinsic properties, or from Babo signal transduction, they underline the importance of Smad-independent signals in many aspects of axonal development.
Role of Rho GTPases in TGFβ signalling
The results suggest that Smad-independent signals involve Rho GTPases. One caveat in genetic interaction experiments is that the loss of any given gene may not be dosage-sensitive with a particular assay. Nevertheless, all the manipulations together suggest that Babo-regulated axon growth requires Rho1, Rac and LIMK1. How Babo signals involve Rho GTPases remains to be fully determined. In addition to LIMK1, which binds to Wit, one possibility, as demonstrated for many axon guidance receptors (Luo, 2002), is that the RhoGEFs, RhoGAPs and Rho proteins might be linked to the Babo receptor complex. Thus, ligand-mediated changes in receptor properties would lead to spatiotemporal changes in Rho GTPase and LIMK1 activities.
The data suggest that a RhoGEF2/Rho1/Rok/LIMK1 pathway mediates Babo responses (Fig. 7E). Whether Rac activators are required is unclear, as tested RacGEFs do not genetically interact with babo. In this respect, rather than through GEFs, Babo may regulate Rac through GAPs, by inhibiting Tum activity (Fig. 7E).
Do mutations in Rho1 and Rac components phenocopy babo phenotypes? β lobe overextensions are observed in Rok (Billuart et al., 2001), Rho1 and Rac mutant neurons (unpublished observations). In MB neurons, Rac GTPases also control axon outgrowth, guidance and branching (Ng et al., 2002). Rho1 also has additional roles in MB neurons (Billuart et al., 2001). Although Rho1 mutant neuroblasts have cell proliferation defects, single-cell αβ clones do show β lobe extensions (unpublished observations). RhoGEF2 strong, loss-of-function clones do not exhibit axon overextension (unpublished observations). As there are 23 RhoGEFs in the Drosophila genome (Adams et al., 2000; Hu et al., 2005), there might well be redundancy in the way Rho1 is activated. LIMK1 inactivation in MB neurons was previously reported (Ng and Luo, 2004). Axon overextensions were not observed as LIMK1 loss results in axon outgrowth and misguidance phenotypes. This suggests that LIMK1 mediates multiple axon guidance signals, of which TGFβ is a subset in MB morphogenesis.
Role of LIMK1 and cofilin phosphoregulation in Babo signalling
Although their phenotypes are similar, several lines of evidence indicate that CA Babo does not simply reflect LIMK1 misregulation in MB neurons. First, whereas LIMK1 genetically interacts with most Rho family members and many Rho regulators (Ng and Luo, 2004), CA babo is dosage-sensitive only to Rho1 and Rac and specific Rho regulators (this study), suggesting that Babo regulates LIMK1 only through a subset of Rho signals.
Second, the LIMK1 misexpression phenotype is suppressed by expression of wild type cofilin (Tsr), S3A Tsr, or by cofilin phosphatase Ssh (Ng and Luo, 2004). By contrast, only wild type Tsr, but not S3A Tsr or Ssh (Figure 5B; unpublished observations), suppresses CA Babo. The suppression by wild type Tsr may reflect a restoration of the endogenous balance or spatial distribution of cofilin-on (unphosphorylated) and -off (phosphorylated) states within neurons. Indeed, optimal axon outgrowth requires cofilin to undergo cycles of phosphorylation and dephosphorylation (Meberg and Bamburg, 2000; Ng and Luo, 2004). As S3A forms of cofilin cannot be inactivated and recycled from actin-bound complexes, wild type cofilin is more potent in actin cytoskeletal regulation.
CA Babo may not simply misregulate LIMK1 but also additional cofilin regulators. Recent data suggest that extracellular cues (including mammalian BMPs) can regulate cofilin through Ssh phosphatase (Endo et al., 2007; Nishita et al., 2005; Wen et al., 2007) and phospholipase Cγ activities (Mouneimne et al., 2006; van Rheenen et al., 2007). In different cell-types, cofilin phosphorylation and phospholipid-binding (which also inhibits cofilin activity) states vary and potently affect cell motility and cytoskeletal regulation. Whether a combination of LIMK1, Ssh, and phospholipid regulation affects cofilin-dependent axon growth remains to be determined.
Third, by phalloidin staining, LIMK1, but not CA Babo, misexpression results in a dramatic increase in F-actin in MB neurons (see Fig. S6 in the supplementary material). Thus, CA Babo does not in itself lead to actin misregulation. Fourth, Babo also regulates axon growth independently of LIMK1 (see below).
Role of Babo, Wit and Put in neuronal morphogenesis
This study differs significantly from the canonical model of Smad signalling (Feng and Derynck, 2005; Shi and Massague, 2003), in which type1 receptors function downstream of the ligand-type 2 receptor complex (Wieser et al., 1995). In this study, the gain- and loss-of-function results suggest type 2 receptors act downstream of type 1 signals. As ectopic Wit and Put only suppress the babo axon overextension phenotype, this implies that Smad-dependent and -independent signals have distinct type 1/type 2 receptor interactions. How these interactions propagate Smad-independent signals remains to be fully determined. Babo could act as a ligand-binding co-receptor with Wit and Put. In addition, Babo kinase activity could regulate type 2 receptor or Rho functions. The results suggest, however, that provided that Wit or Put signals are sufficiently high, Babo is not required. Whatever the mechanism(s), it is likely that Babo requires the Wit C-terminus-LIMK1 interaction to relay cofilin phosphoregulatory signals (Fig. 7E). How Put functions is unclear. As the put135 allele (used in this study) carries a missense mutation within the kinase domain, this suggests that kinase activity is essential. put does not genetically interact with LIMK1. As Put lacks the C-terminal extension of Wit that is necessary for LIMK1 binding, this suggests that Put acts independently of LIMK1. One potential effector is Rac, which in the context of Babo signalling, also appears to be Pak1- and thus LIMK1-independent (Fig. 7E).
In MB neurons, Wit and Put can function interchangeably. In other in vivo paradigms, type 2 receptors are not interchangeable (Marques et al., 2002). However, as the Wit COOH-terminal tail is required to substitute for Put, this suggests that Wit axon growth signals are independent of its kinase activity. Together, this suggests that Smad-independent signals involve LIMK1-dependent and -independent mechanisms.
Distinct roles of Babo in neuronal morphogenesis
This study, together with Zheng et al. (Zheng et al., 2003), shows that Babo mediates two distinct responses in related MB populations. How do MB neurons choose between axon pruning and axon growth? The babo rescue studies suggest that whereas Baboa or Babob elicits Smad-independent responses, only Baboa mediates Smad-dependent responses. As Babo isoforms differ only in the extracellular domain, differences in ligand-binding could determine Smad2 or Rho GTPase activation. However, it is worth noting that in DC neurons, either isoform mediates axon extension through Smad2 and Medea (Zheng et al., 2006). In addition, although expressed in all MB neurons, CA babo misexpression (which confers ligand-independent signals) perturbs only αβ axons (Fig. 4A,A’; see Fig. S2 in supplementary material). Thus cell-intrinsic properties may also be essential in determining Babo responses.
Many TGFβ ligands signal through Babo (Gesualdi and Haerry, 2007; Lee-Hoeflich et al., 2005; Parker et al., 2006; Serpe and O’Connor, 2006; Zheng et al., 2003; Zhu et al., 2008). For example, Dawdle, an Activin-related ligand, patterns Drosophila motor axons (Parker et al., 2006; Serpe and O’Connor, 2006), while Activin (Activin-β, Flybase) is required for MB axon pruning (Zheng et al., 2003). Whether these ligands regulate Babo MB, AL and OL axonal morphogenesis is unclear. Taken together, the evidence suggests that Babo signalling is varied in vivo and is involved in many aspects of neuronal development.
Smad-independent signals in cytoskeletal regulation and cell morphogenesis
TGFβ signals are responsible for many aspects of development and disease and, throughout different models, Smad pathways are closely involved. Although Smad-independent pathways are known, their mechanisms and roles in vivo are unclear. TGFβ signals often drive cell shape changes in vivo. During epithelial-to-mesenchymal transition (EMT), cells lose their epithelial structure and adopt a fibroblast-like structure that is essential for cell migration during development and tumour invasion (Grunert et al., 2003; Shook and Keller, 2003). TGFβ-mediated changes in the actin cytoskeleton and adherens junctions are necessary for EMT. Although Smads are crucial, TGFβ signals also involve the Cdc42/Par6 complex, resulting in cell de-adhesion and F-actin breakdown through Rho1 degradation (Ozdamar et al., 2005). In other studies, however, TGFβ-mediated EMT requires Rho1 (Bhowmick et al., 2001), which can be regulated by Smad activity (Levy and Hill, 2005).
Many TGFβ-driven events in Drosophila are Smad-dependent (Raftery and Sutherland, 1999). Whether Smad-independent roles exist beyond those identified in this study remains to be tested. Here I provide a framework to understand how non-Smad signals regulate cell morphogenesis during development.
Supplementary Material
Acknowledgements
I thank the Bloomington Drosophila Stock Center and many colleagues who provided reagents for this study, especially Theo Haerry and Michael O’Connor. Uwe Drescher, Theo Haerry, Greg Jefferis, Michael O’Connor and Rosa Gonzalez-Quevedo provided comments on the manuscript. Michael Fletcher and Coralie Moore provided technical assistance, and Guy Tear helped with laboratory equipment. The Wellcome Trust (078045) supported this study.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Augsburger A, Schuchardt A, Hoskins S, Dodd J, Butler S. BMPs as mediators of roof plate repulsion of commissural neurons. Neuron. 1999;24:127–41. doi: 10.1016/s0896-6273(00)80827-2. [DOI] [PubMed] [Google Scholar]
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Bashaw GJ, Hu H, Nobes CD, Goodman CS. A novel Dbl family RhoGEF promotes Rho-dependent axon attraction to the central nervous system midline in Drosophila and overcomes Robo repulsion. J Cell Biol. 2001;155:1117–22. doi: 10.1083/jcb.200110077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billuart P, Winter CG, Maresh A, Zhao X, Luo L. Regulating axon branch stability: the role of p190 RhoGAP in repressing a retraction signaling pathway. Cell. 2001;107:195–207. doi: 10.1016/s0092-8674(01)00522-0. [DOI] [PubMed] [Google Scholar]
- Brummel T, Abdollah S, Haerry TE, Shimell MJ, Merriam J, Raftery L, Wrana JL, O’Connor MB. The Drosophila activin receptor baboon signals through dSmad2 and controls cell proliferation but not patterning during larval development. Genes Dev. 1999;13:98–111. doi: 10.1101/gad.13.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SJ, Dodd J. A role for BMP heterodimers in roof plate-mediated repulsion of commissural axons. Neuron. 2003;38:389–401. doi: 10.1016/s0896-6273(03)00254-x. [DOI] [PubMed] [Google Scholar]
- Colavita A, Krishna S, Zheng H, Padgett RW, Culotti JG. Pioneer axon guidance by UNC-129, a C. elegans TGF-beta. Science. 1998;281:706–9. doi: 10.1126/science.281.5377.706. [DOI] [PubMed] [Google Scholar]
- Das P, Inoue H, Baker JC, Beppu H, Kawabata M, Harland RM, Miyazono K, Padgett RW. Drosophila dSmad2 and Atr-I transmit activin/TGFbeta signals. Genes Cells. 1999;4:123–34. doi: 10.1046/j.1365-2443.1999.00244.x. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Eaton BA, Davis GW. LIM Kinase1 controls synaptic stability downstream of the type II BMP receptor. Neuron. 2005;47:695–708. doi: 10.1016/j.neuron.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Endo M, Ohashi K, Mizuno K. LIM kinase and slingshot are critical for neurite extension. J Biol Chem. 2007;282:13692–702. doi: 10.1074/jbc.M610873200. [DOI] [PubMed] [Google Scholar]
- Endo M, Ohashi K, Sasaki Y, Goshima Y, Niwa R, Uemura T, Mizuno K. Control of growth cone motility and morphology by LIM kinase and Slingshot via phosphorylation and dephosphorylation of cofilin. J Neurosci. 2003;23:2527–37. doi: 10.1523/JNEUROSCI.23-07-02527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Labrador JP, Hing H, Bashaw GJ. Slit stimulation recruits Dock and Pak to the roundabout receptor and increases Rac activity to regulate axon repulsion at the CNS midline. Neuron. 2003;40:113–27. doi: 10.1016/s0896-6273(03)00591-9. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–93. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Foletta VC, Lim MA, Soosairajah J, Kelly AP, Stanley EG, Shannon M, He W, Das S, Massague J, Bernard O. Direct signaling by the BMP type II receptor via the cytoskeletal regulator LIMK1. J Cell Biol. 2003;162:1089–98. doi: 10.1083/jcb.200212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel DJ, Liebl EC, Kolodziej PA, Seeger MA. The Abelson tyrosine kinase, the Trio GEF and Enabled interact with the Netrin receptor Frazzled in Drosophila. Development. 2005;132:1983–94. doi: 10.1242/dev.01736. [DOI] [PubMed] [Google Scholar]
- Gesualdi S, Haerry TE. Distinct Signaling of Drosophila Activin/TGF-beta Family Members. Fly. 2007;1:212–221. doi: 10.4161/fly.5116. [DOI] [PubMed] [Google Scholar]
- Gibson MC, Perrimon N. Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science. 2005;307:1785–9. doi: 10.1126/science.1104751. [DOI] [PubMed] [Google Scholar]
- Goldstein AY, Jan YN, Luo L. Function and regulation of Tumbleweed (RacGAP50C) in neuroblast proliferation and neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:3834–9. doi: 10.1073/pnas.0500748102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4:657–65. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- Haerry TE, Khalsa O, O’Connor MB, Wharton KA. Synergistic signaling by two BMP ligands through the SAX and TKV receptors controls wing growth and patterning in Drosophila. Development. 1998;125:3977–87. doi: 10.1242/dev.125.20.3977. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins in development. Curr Opin Genet Dev. 1996;6:432–8. doi: 10.1016/s0959-437x(96)80064-5. [DOI] [PubMed] [Google Scholar]
- Hu H, Li M, Labrador JP, McEwen J, Lai EC, Goodman CS, Bashaw GJ. Cross GTPase-activating protein (CrossGAP)/Vilse links the Roundabout receptor to Rac to regulate midline repulsion. Proc Natl Acad Sci U S A. 2005;102:4613–8. doi: 10.1073/pnas.0409325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JB, Podos SD, Keith K, Simpson SL, Ferguson EL. The Drosophila Medea gene is required downstream of dpp and encodes a functional homolog of human Smad4. Development. 1998;125:1407–20. doi: 10.1242/dev.125.8.1407. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–71. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Kurusu M, Awasaki T, Masuda-Nakagawa LM, Kawauchi H, Ito K, Furukubo-Tokunaga K. Embryonic and larval development of the Drosophila mushroom bodies: concentric layer subdivisions and the role of fasciclin II. Development. 2002;129:409–19. doi: 10.1242/dev.129.2.409. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–93. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–76. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–61. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;28:807–18. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Lee-Hoeflich ST, Causing CG, Podkowa M, Zhao X, Wrana JL, Attisano L. Activation of LIMK1 by binding to the BMP receptor, BMPRII, regulates BMP-dependent dendritogenesis. Embo J. 2004;23:4792–801. doi: 10.1038/sj.emboj.7600418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Hoeflich ST, Zhao X, Mehra A, Attisano L. The Drosophila type II receptor, Wishful thinking, binds BMP and myoglianin to activate multiple TGFbeta family signaling pathways. FEBS Lett. 2005;579:4615–21. doi: 10.1016/j.febslet.2005.06.088. [DOI] [PubMed] [Google Scholar]
- Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor {beta} (TGF-{beta}) target genes and distinguishes TGF-{beta}-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol. 2005;25:8108–25. doi: 10.1128/MCB.25.18.8108-8125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol. 2002;18:601–35. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- Marques G, Bao H, Haerry TE, Shimell MJ, Duchek P, Zhang B, O’Connor MB. The Drosophila BMP type II receptor Wishful Thinking regulates neuromuscular synapse morphology and function. Neuron. 2002;33:529–43. doi: 10.1016/s0896-6273(02)00595-0. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Matsuura R, Tanaka H, Go MJ. Distinct functions of Rac1 and Cdc42 during axon guidance and growth cone morphogenesis in Drosophila. Eur J Neurosci. 2004;19:21–31. doi: 10.1046/j.1460-9568.2003.03084.x. [DOI] [PubMed] [Google Scholar]
- McCabe BD, Marques G, Haghighi AP, Fetter RD, Crotty ML, Haerry TE, Goodman CS, O’Connor MB. The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron. 2003;39:241–54. doi: 10.1016/s0896-6273(03)00426-4. [DOI] [PubMed] [Google Scholar]
- Meberg PJ, Bamburg JR. Increase in neurite outgrowth mediated by overexpression of actin depolymerizing factor. J Neurosci. 2000;20:2459–69. doi: 10.1523/JNEUROSCI.20-07-02459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G, DesMarais V, Sidani M, Scemes E, Wang W, Song X, Eddy R, Condeelis J. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr Biol. 2006;16:2193–205. doi: 10.1016/j.cub.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118:3573–84. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- Nellen D, Affolter M, Basler K. Receptor serine/threonine kinases implicated in the control of Drosophila body pattern by decapentaplegic. Cell. 1994;78:225–37. doi: 10.1016/0092-8674(94)90293-3. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–68. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Ng J, Luo L. Rho GTPases regulate axon growth through convergent and divergent signaling pathways. Neuron. 2004;44:779–93. doi: 10.1016/j.neuron.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Ng J, Nardine T, Harms M, Tzu J, Goldstein A, Sun Y, Dietzl G, Dickson BJ, Luo L. Rac GTPases control axon growth, guidance and branching. Nature. 2002;416:442–7. doi: 10.1038/416442a. [DOI] [PubMed] [Google Scholar]
- Nishita M, Tomizawa C, Yamamoto M, Horita Y, Ohashi K, Mizuno K. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J Cell Biol. 2005;171:349–59. doi: 10.1083/jcb.200504029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–9. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- Parker L, Ellis JE, Nguyen MQ, Arora K. The divergent TGF-beta ligand Dawdle utilizes an activin pathway to influence axon guidance in Drosophila. Development. 2006;133:4981–91. doi: 10.1242/dev.02673. [DOI] [PubMed] [Google Scholar]
- Penton A, Chen Y, Staehling-Hampton K, Wrana JL, Attisano L, Szidonya J, Cassill JA, Massague J, Hoffmann FM. Identification of two bone morphogenetic protein type I receptors in Drosophila and evidence that Brk25D is a decapentaplegic receptor. Cell. 1994;78:239–50. doi: 10.1016/0092-8674(94)90294-1. [DOI] [PubMed] [Google Scholar]
- Raftery LA, Sutherland DJ. TGF-beta family signal transduction in Drosophila development: from Mad to Smads. Dev Biol. 1999;210:251–68. doi: 10.1006/dbio.1999.9282. [DOI] [PubMed] [Google Scholar]
- Ruberte E, Marty T, Nellen D, Affolter M, Basler K. An absolute requirement for both the type II and type I receptors, punt and thick veins, for dpp signaling in vivo. Cell. 1995;80:889–97. doi: 10.1016/0092-8674(95)90292-9. [DOI] [PubMed] [Google Scholar]
- Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics. 1995;139:1347–58. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpe M, O’Connor MB. The metalloprotease tolloid-related and its TGF-beta-like substrate Dawdle regulate Drosophila motoneuron axon guidance. Development. 2006;133:4969–79. doi: 10.1242/dev.02711. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shook D, Keller R. Mechanisms, mechanics and function of epithelial-mesenchymal transitions in early development. Mech Dev. 2003;120:1351–83. doi: 10.1016/j.mod.2003.06.005. [DOI] [PubMed] [Google Scholar]
- Simin K, Bates EA, Horner MA, Letsou A. Genetic analysis of punt, a type II Dpp receptor that functions throughout the Drosophila melanogaster life cycle. Genetics. 1998;148:801–13. doi: 10.1093/genetics/148.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MA, Penton A, Twombly V, Hoffmann FM, Gelbart WM. Signaling through both type I DPP receptors is required for anterior-posterior patterning of the entire Drosophila wing. Development. 1997;124:79–89. doi: 10.1242/dev.124.1.79. [DOI] [PubMed] [Google Scholar]
- Sotillos S, Campuzano S. DRacGAP, a novel Drosophila gene, inhibits EGFR/Ras signalling in the developing imaginal wing disc. Development. 2000;127:5427–38. doi: 10.1242/dev.127.24.5427. [DOI] [PubMed] [Google Scholar]
- Ting CY, Herman T, Yonekura S, Gao S, Wang J, Serpe M, O’Connor MB, Zipursky SL, Lee CH. Tiling of r7 axons in the Drosophila visual system is mediated both by transduction of an activin signal to the nucleus and by mutual repulsion. Neuron. 2007;56:793–806. doi: 10.1016/j.neuron.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneizumi K, Nakayama T, Kamoshida Y, Kornberg TB, Christian JL, Tabata T. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997;389:627–31. doi: 10.1038/39362. [DOI] [PubMed] [Google Scholar]
- van Rheenen J, Song X, van Roosmalen W, Cammer M, Chen X, Desmarais V, Yip SC, Backer JM, Eddy RJ, Condeelis JS. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol. 2007;179:1247–59. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Han L, Bamburg JR, Shim S, Ming GL, Zheng JQ. BMP gradients steer nerve growth cones by a balancing act of LIM kinase and Slingshot phosphatase on ADF/cofilin. J Cell Biol. 2007;178:107–19. doi: 10.1083/jcb.200703055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser R, Wrana JL, Massague J. GS domain mutations that constitutively activate T beta R-I, the downstream signaling component in the TGF-beta receptor complex. Embo J. 1995;14:2199–208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Wang J, Haerry TE, Wu AY, Martin J, O’Connor MB, Lee CH, Lee T. TGF-beta signaling activates steroid hormone receptor expression during neuronal remodeling in the Drosophila brain. Cell. 2003;112:303–15. doi: 10.1016/s0092-8674(03)00072-2. [DOI] [PubMed] [Google Scholar]
- Zheng X, Zugates CT, Lu Z, Shi L, Bai JM, Lee T. Baboon/dSmad2 TGF-beta signaling is required during late larval stage for development of adult-specific neurons. Embo J. 2006;25:615–27. doi: 10.1038/sj.emboj.7600962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CC, Boone JQ, Jensen PA, Hanna S, Podemski L, Locke J, Doe CQ, O’Connor MB. Drosophila Activin- and the Activin-like product Dawdle function redundantly to regulate proliferation in the larval brain. Development. 2008;135:513–21. doi: 10.1242/dev.010876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.