SUMMARY
Nitric oxide (NO) regulates protein function by S-nitrosylation of cysteine to form nitrosothiols. Nitrosothiols are highly susceptible to nonenzymatic degradation by cytosolic reducing agents. Here we show that although most protein nitrosothiols are rapidly degraded by cytosolic reductants, a small subset form unusually stable S-nitrosylated proteins. Our findings suggest that stable S-nitrosylation reflects a protein conformation change that shields the nitrosothiol. To identify stable protein nitrosothiols, we developed a proteomic method for profiling S-nitrosylation. We examined the stability of over 100 S-nitrosylated proteins, and identified ten stable nitrosothiols. These proteins remained S-nitrosylated in cells after NO synthesis was inhibited, unlike most S-nitrosylated proteins. Taken together, our data identify a novel class of NO targets that form stable nitrosothiols in the cell and are likely to mediate the persistent cellular effects of NO.
INTRODUCTION
The primary mechanism by which NO exerts its diverse physiological effects is by chemical modification of specific effector proteins. A prominent NO modification is the nitrosylation of cysteine residues, called S-nitrosylation. This reversible modification can affect protein activity, localization, or stability (Hess et al., 2005). S-nitrosylation can be elicited nonenzymatically, for example by NO-derived nitrosating species, such as N2O3, or by transnitrosylation, in which a nitrosyl moiety of an existing nitrosothiol is transferred to a protein thiol (Foster et al., 2003; Mitchell and Marletta, 2005). Alternatively, examples of enzymatic pathways leading to the formation of protein nitrosothiols (Pawloski et al., 2001) or small molecule nitrosothiols (Inoue et al., 1999) have been described. The specificity of S-nitrosylation is governed by several mechanisms, including consensus sequences that increase the chemical reactivity of cysteine targets of NO (Stamler et al., 1997b) and adapter proteins that juxtapose NO synthase (NOS) isoenzymes to their targets (Paige and Jaffrey, 2007). Over 100 proteins have been reported to be S-nitrosylated in mammalian cells, likely accounting for many of the observed effects of NO in cells (Stamler et al., 2001).
Much less is known about how protein denitrosylation is elicited in cells. Denitrosylation reverses S-nitrosylation, and therefore affects the duration of NO signaling in cells. Additionally, the steady-state concentration of the nitrosylated form of a protein is determined by the rates of both S-nitrosylation and denitrosylation, making denitrosylation a critical determinant of the effectiveness of a NO signal. Since nitrosothiols are a reversible oxidative modification, they are predicted to be unstable in the highly reducing environment of the cytosol, which contains numerous small molecule reductants (Sen, 1998). Indeed, the levels of protein nitrosothiols in vivo are directly regulated by the intracellular concentration of the cytosolic reducing agent glutathione (GSH), which can be present at 0.5 – 10 mM (Kosower and Kosower, 1978; Mayer et al., 1995). Consistent with this prediction, small molecule nitrosothiols have half-lives of seconds to minutes in biological solutions (Kashiba-Iwatsuki et al., 1997; Singh et al., 1996). Since the chemical properties of nitrosothiols might be expected to be the same in small molecules and in the context of proteins, it is plausible that cytosolic protein S-nitrosylation is followed by rapid nonenzymatic denitrosylation due to cytosolic reductants.
Despite this prediction, two protein-mediated mechanisms have been identified for denitrosylation in cells. The first mechanism involves S-nitrosoglutathione (GSNO) reductase, a highly conserved enzyme capable of denitrosylating GSNO in vivo (Liu et al., 2001). Additionally, recent studies of caspase-3 have highlighted a second denitrosylation mechanism. In an in vitro study, Marletta and colleagues (Mitchell and Marletta, 2005) made the finding that the S-nitrosylated form of caspase-3 could not be reduced by in vitro application of GSH (Mitchell and Marletta, 2005). Instead, thioredoxin, a protein that can catalyze thiol reduction, was capable of eliciting caspase-3 denitrosylation (Mitchell and Marletta, 2005). Stamler and colleagues recently showed that thioredoxin reductase/thioredoxins are physiologic caspase-3 denitrosylases (Benhar et al., 2008). These studies indicate that protein nitrosothiols might exhibit different reactivity towards reductants than small molecule nitrosothiols. However, it remains unclear whether nitrosothiol stability is rare or a general property of protein nitrosothiols.
To understand how S-nitrosylation serves as an effector pathway of NO in cells, the chemical reactivity of protein nitrosothiols needs to be understood. To profile the chemical properties of S-nitrosylated proteins, we devised a proteomic approach to survey the reactivity of over 100 protein nitrosothiols. Our findings reveal that nitrosothiols in proteins can be classified as either “stable” or “unstable,” with the stable nitrosothiols appearing to result from conformational changes that reduce the accessibility of the nitrosothiol to solvent. We find that these chemically stable protein nitrosothiols are also stable in cells, thereby identifying a subset of S-nitrosylated proteins that can carry the NO signal even after NO signaling has ended and likely utilize specific denitrosylation pathways to terminate NO signaling.
RESULTS
A subset of protein nitrosothiols show significantly reduced GSH reactivity
To better understand the properties of nitrosothiols in proteins, we examined the ability of GSH to reduce different protein nitrosothiols. We first generated a population of S-nitrosylated proteins using GSNO, a physiological NO donor (Gaston et al., 1993; Kluge et al., 1997). GSNO was used because it is expected to catalyze the formation of a wide range of S-nitrosylated proteins (Hao et al., 2006; Jaffrey et al., 2001). As described previously, S-nitrosylated proteins were prepared by incubating brain cytosolic lysates with 40 μM GSNO and then removing the GSNO with a desalting column (Jaffrey et al., 2001).
To characterize the reactivity of these protein nitrosothiols, we assessed their ability to be denitrosylated following incubation with 1 mM GSH. Protein nitrosothiols were detected using the biotin switch method, which introduces a biotin moiety on cysteines that are S-nitrosylated (Figure 1A) (Jaffrey et al., 2001). Each of the bands detected in the biotin switch assay are dependent upon ascorbate treatment, consistent with the ascorbate-dependent reduction of nitrosothiol bonds (Figure S1A) (Jaffrey et al., 2001). GSH treatment resulted in rapid and essentially complete denitrosylation of most proteins after 5 min (Figure 1B). Surprisingly, a small subset of proteins remained S-nitrosylated even after 30 min of GSH exposure (Figure 1B and 1C). Higher concentrations of GSH or higher temperatures did not reduce the stability of these protein nitrosothiols (Figure S1B–S1E). Using densitometry measurements, we classified the degree of denitrosylation after 30 min into five categories: 0–20%, 21–40%, 41–60%, 61–80%, and 81–100%. Nearly all proteins exhibited either 81–100% or 0–20% reduction of S-nitrosylation, which we termed unstable nitrosothiols or stable nitrosothiols, respectively (Figure 1D). Together, these data indicate that, unlike small molecule nitrosothiols which are rapidly denitrosylated (Kashiba-Iwatsuki et al., 1997; Singh et al., 1996), protein nitrosothiols have different reactivities and that a subset of proteins form nitrosothiols which have significantly reduced susceptibility to GSH reduction.
Figure 1. A subset of protein nitrosothiols are resistant to GSH-mediated denitrosylation.
(A) Schematic diagram of the biotin switch method (Jaffrey et al., 2001). A theoretical protein is indicated with cysteines in either the free-thiol, disulfide or nitrosothiol conformation. In Step 1, all non-nitrosylated thiols are blocked with methyl methanethiosulfonate (MMTS) in the presence of SDS to ensure access of MMTS to internal cysteines. Under the conditions used, MMTS does not react with nitrosothiols or preexisting disulfide bonds. In Step 2, residual MMTS is removed by acetone precipitation and nitrosothiols bonds are selectively decomposed with ascorbate, which results in the reduction of nitrosothiols to thiols. In Step 3, the thiols that remain derive exclusively from formerly S-nitrosylated cysteines, and are then labeled with the thiol-specific biotinylation reagent N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide (biotin-HPDP).
(B) Detection of a subset of stably S-nitrosylated proteins. Lysates were exposed to 40 μM GSNO and then 1 mM GSH for different time intervals. Western blot bands are labeled as either GSH-unstable (open arrowheads) or GSH-stable (closed arrowheads).
(C) Expansion of boxed region in (B) shows two S-nitrosylated proteins that display significantly different levels of GSH stability even though both proteins originally display comparable levels of GSNO-induced S-nitrosylation.
(D) Densitometry measurements of the results in (B). Band intensity was compared at 0 and after 30 min of GSH treatment to quantify the degree of GSH-mediated denitrosylation. The number of S-nitrosylated proteins that exhibit the indicated degree of denitrosylation are shown. Proteins generally were either highly stable in the presence of GSH, i.e., exhibiting less than 20% denitrosylation, or they were highly susceptible to GSH, i.e., exhibiting greater than 80% denitrosylation. Most proteins were highly sensitive to GSH-mediated denitrosylation. Densitometry was averaged from three independent experiments.
(E) Protein conformation stabilizes nitrosothiols in GSH-stable proteins. Lysates were exposed to GSNO and then either 1 mM GSH or 2-ME, with or without 1% SDS in the buffer. The small reducing agent 2-ME exhibited more denitrosylation than GSH. Denitrosylation was highly effective in the presence of the denaturant SDS.
Stable nitrosothiols appear conformationally protected from reducing agents
To characterize the basis for the reduced reactivity of the GSH-stable nitrosothiols, we used 2-mercaptoethanol (2-ME), a considerably smaller reducing agent that should more easily gain access to buried nitrosothiols. Treatment of S-nitrosylated proteins with 2-ME did not result in substantially more denitrosylation than GSH (Figure 1E). However, inclusion of the protein denaturating agent sodium dodecyl sulfate (SDS) with 2-ME in the denitrosylation solution resulted in complete denitrosylation of all proteins (Figure 1E). In principle, a simple transnitrosation reaction is expected to exhibit complete chemical reversibility. The existence of S-nitrosylated proteins that are incapable of being reversibly denitrosylated implies additional thermodynamically favorable steps, such as conformational changes, that occur after S-nitrosylation in these proteins. The experiments above suggest that protein structure might limit the accessibility of the nitrosothiol to the solvent or otherwise reduces the reactivity of the nitrosothiol, accounting for the decreased ability of these nitrosothiols to be reduced.
A proteomic method for identification and quantification of S-nitrosylation
We next sought to identify proteins that have the capacity to form stable nitrosothiols. A small pool of stable protein nitrosothiols is found constitutively in cells, as exemplified by mitochondrial caspase-3 (Mannick et al., 1999) and more recently by the work of Chvanov and colleagues (Chvanov et al., 2006). Since previous studies focused on specific proteins, we sought to identify stable protein nitrosothiols without any biases towards their identity. Currently, there are no methods that permit the relative quantification of protein nitrosothiols on a proteome-wide scale. We decided to adapt the isotope-coded affinity tag (ICAT) technique (Gygi et al., 1999), which utilizes isotopically labeled protein-modifying agents in conjunction with liquid chromatography tandem mass spectrometry (LC-MS/MS) to identify and quantify expression level differences of proteins in two distinct samples. We synthesized SNOCAP (S-nitrosothiol capture) reagents based on N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide (biotin-HPDP), the activated mixed disulfide reagent used to selectively label nitrosylated thiols in the biotin switch method (Jaffrey et al., 2001). SNOCAP reagents were synthesized in “light” and “heavy” forms and comprise three parts: (i) a cysteine-reactive activated mixed disulfide; (ii) a linker that contains either zero (light) or four atoms (heavy) of the heavy isotopes of carbon (13C) and nitrogen (15N); and (iii) a biotin moiety that is used to recover peptides on immobilized neutravidin (Figure 2A). SNOCAP (Figure 2B and 2C) is used to compare two samples and is based on the biotin switch method, which was recently confirmed to be highly specific for nitrosothiols (Forrester et al., 2007).
Figure 2. SNOCAP method for quantitative nitrosoproteomics.
(A) Structure of the SNOCAP reagent. The SNOCAP reagent is comprised of three parts: a thiol-reactive activated mixed disulfide (green); an isotope-coded linker (red); and a biotin moiety used for recovery (blue). Carbon and nitrogen atoms that are incorporated into SNOCAP reagents in isotopically light or heavy forms are labeled (*).
(B) Schematic diagram of the SNOCAP method. A theoretical protein is indicated with cysteines in either the free-thiol, disulfide or nitrosothiol conformation. In step 1, free thiols are blocked by methylthiolation with MMTS. In step 2, MMTS is removed by acetone precipitation, and nitrosothiols are selectively reduced with ascorbate to reform the thiol. In step 3, newly formed thiols are isotopically labeled and biotinylated with the SNOCAP reagent. Proteins are then digested with trypsin in step 4 and biotinylated peptides are recovered using neutravidn in step 5.
(C) Strategy for use of the SNOCAP method to quantitatively compare S-nitrosylation between two samples.
In control experiments, we assessed whether the SNOCAP procedure labels non-nitrosylated proteins. To test this we treated pre-reduced brain lysates with either vehicle or GSNO, and then labeled the samples with light and heavy SNOCAP reagents, respectively. After processing the samples according to the SNOCAP protocol, over 100 S-nitrosylated proteins were identified by MS/MS (Table S2). Importantly, in each case, there was no peptide ion present from the vehicle-treated sample; all ions were detected as the heavy-labeled peptide derived from the GSNO-treated sample (Figure S2). These results indicate that SNOCAP, like the biotin switch method (Forrester et al., 2007), does not result in artifactual labeling from incomplete thiol blockage. Although biotinylated peptides increase the complexity of MS/MS spectra due to biotin-derived fragment ions (Tao and Aebersold, 2003), y- and b-ions derived from fragmentation of SNOCAP-labeled peptides were still readily detectable in MS/MS spectra (Figure 3A).
Figure 3. Quantitative MS identifies sites of S-nitrosylation and reveals differences in GSH-mediated denitrosylation of these sites.
(A) MS/MS spectra of SNOCAP-labeled peptides identifies sites of S-nitrosylation. Representative collision-induced dissociation spectra showing the MS/MS peaks of a SNOCAP-labeled peptide from CRMP-2 with the y-ion series (red), b-ion series (blue), and the precursor ion (green) labeled. The modified cysteine residue is also labeled (*). The spectrum also shows that SNOCAP tags do not substantially alter fragmentation patterns which might affect the fidelity of database-searching algorithms.
(B,C) MS spectra of isolated S-nitrosylated peptides reflect large differences in susceptibility to GSH-induced denitrosylation. The majority of S-nitrosylated peptides appear highly unstable in the presence of GSH, as seen with the MS spectra of a peptide derived from 14-3-3 θ showing 92.7% denitrosylation (B). A small subset of peptides appears highly stable to GSH, as seen by MS spectra of peptide isolated from GST-pi showing only 10.4% denitrosylation (C).
(D,E) Several proteins contained multiple cysteines that undergo S-nitrosylation, but only a single cysteine capable of stable nitrosylation. MS spectra of two peptides isolated from CRMP-2 are shown which differential stability to GSH. One peptide is highly susceptible to denitrosylation (D) while the other undergoes negligible denitrosylation (E).
To determine whether SNOCAP can accurately measure relative S-nitrosylated protein abundance in two different samples, we prepared mixtures of hemoglobin (Hb) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), two well-established targets of NO (Hara et al., 2005; Jia et al., 1996), at known but different concentrations. After nitrosylation, the mixtures were labeled with either the light or heavy SNOCAP reagents, and processed according to the SNOCAP protocol. The isolated peptides, corresponding to the original nitrosopeptides, were then quantified and sequenced by LC-MS/MS. The measured ratios of both the GAPDH and Hb-derived peptides were consistent with the ratios of each protein in the original two samples (Table S1).
Identification of stable nitrosoproteins by proteomic profiling of nitrosothiol reactivity
We next used the SNOCAP strategy to survey the reactivities of nitrosothiols in proteins. In these experiments, a preparation of S-nitrosylated proteins were prepared using GSNO. Since our goal was to monitor the denitrosylation of dozens of protein nitrosothiols simultaneously, we needed the cysteines of as many previously-described nitrosoproteins to be S-nitrosylated to a degree sufficient for detection. Although some proteins are more efficiently S-nitrosylated by specific transnitrosylating molecules other than GSNO, to the best of our knowledge, there is no protein nitrosothiol that cannot be elicited to some degree by GSNO. Nitrosylated proteins were incubated with either vehicle (GSNO→vehicle) or 1 mM GSH (GSNO→GSH) for 5 min. Next, the protein nitrosothiols in the GSNO→vehicle sample and the GSNO→GSH sample were labeled with light and heavy SNOCAP reagents, respectively.
MS/MS analysis of these samples identified 102 S-nitrosylated proteins, some of which exhibited multiple sites of S-nitrosylation (Table S2 and Figure S3). Of these proteins, 50 have previously been shown to be S-nitrosylated (Table S2). The sites of S-nitrosylation that we identified also matched previously reported sites in the few instances where the specific site of S-nitrosylation has already been identified and biochemically confirmed (Table S2). It should be noted that some proteins that have been described to be susceptible to S-nitrosylation were not detected in our screen. This may reflect the low abundance of these proteins in the cytosolic fraction of brain homogenates, such as ras (Mizoguchi et al., 1989) and thioredoxin (Holmgren and Luthman, 1978), and the low abundance of these proteins in brain compared to other tissues, such as caspase-3 (Krajewska et al., 1997). Additionally, disulfide bonds can impair trypsin cleavage, resulting in larger peptides and impaired peptide detection. Because the SNOCAP method does not involve a disulfide reduction step, cysteines that fall within tryptic peptides containing disulfide bonds might also be missed. Additionally, MS/MS analysis can be incomplete since certain peptides do not ionize or fragment efficiently (Elias and Gygi, 2007; Mallick et al., 2007), and the peptides identified can vary significantly depending on the MS instrument as well as the search algorithm used (Elias et al., 2005).
The majority of nitrosothiols identified were unstable in the presence of GSH, as evidenced by the marked reduction in the intensity of the heavy peak, which derives from the sample denitrosylated with GSH. In these cases, this peak was 20% or less than the intensity of the light peak, which derives from the sample treated with vehicle (Figure 3B). These results indicate that the nitrosothiol in these proteins were quantitatively reduced by GSH treatment. However, for eleven of the peptides, both the heavy and light peptide ions were nearly the same intensity, with the heavy peak being at least 80% the intensity of the light peak (Figure 3C). These peptides, which correspond to GSH-stable sites of S-nitrosylation, are listed in Table 1. Each of these proteins contain only one stably modified cysteine with the exception of GAPDH, which possessed two cysteines capable of stable S-nitrosylation. Interestingly, several of the proteins in Table 1 also contained S-nitrosylated cysteines that were not stable to GSH treatment (Figure 3D and 3E and Table S2), indicating that nitrosothiols within the same protein can have different reactivity. These data indicate that a small subset of protein nitrosothiols exhibit markedly reduced reactivity compared to the majority of protein nitrosothiols.
Table 1.
Proteins resistant to GSH-mediated denitrosylation
| Protein name | Peptide sequence identifieda | Percent reduction in S-nitrosylationb (Mean ± s.d.)c |
|---|---|---|
| α-tubulin | AYHEQLSVAEITNACFEPANQMVK | 11.8% ± 2.3 |
| β-tubulin | EIVHIQAGQCGNQIGAK | 15.2% ± 3.6 |
| Collapsin response mediator protein 2 | GLYDGPVCEVSVTPK | 11.8% ± 2.1 |
| Creatine kinase, B chain | FCTGLTQIETLFK | 16.3% ± 3.4 |
| Extracellular signal- regulated kinase 2 | DLKPSNLLLNTTCDLK | 14.8% ± 2.9 |
| Glutathione S-transferase pi | EAALVDMVNDGVEDLRCK | 16.6% ± 3.8 |
| Glyceraldehyde-3- phosphate dehydrogenase | VPTPNVSVVDLTCR IVSNASCTTNCLAPLAK |
7.3% ± 2.3 13.1% ± 2.3 |
| Hemoglobin, β chain | GTFAHLSELHCDK | 16.5% ± 4.8 |
| Pyruvate kinase | AGKPVICATQMLESMIK | 7.3% ± 3.6 |
| Peroxiredoxin-6 | DFTPVCTTELGR | 8.5% ± 3.7 |
Modified cysteine residues are highlighted by blue coloring and bold lettering.
Total denitrosylation in the presence of 1 mM GSH for 5 min.
These values represent the average of five experimental samples with each sample being reloaded and quantified by LC-MS/MS three to six times.
Biochemical confirmation and structural analysis of MS characterized S-nitrosylation sites
We next used in vitro denitrosylation assays to biochemically confirm the results of the MS experiments. Brain lysate was S-nitrosylated as above, and then denitrosylated by treatment with vehicle or 1 mM GSH for 5 min. The proteins that remained S-nitrosylated were then purified using the biotin switch method (Jaffrey et al., 2001). Proteins for which antibodies are readily available were selected for biochemical validation. The S-nitrosylated proteins that were identified by MS as GSH-unstable were essentially absent in GSH-treated samples, indicating that they were rapidly denitrosylated upon treatment with GSH (Figure 4A). In contrast, the S-nitrosylated proteins identified by MS as GSH-stable remained robustly nitrosylated under the same conditions (Figure 4B). In the case of β-tubulin and CRMP-2, the band seen for GSNO→GSH samples is slightly less intense than the band seen for GSNO→vehicle treated samples, which is consistent with our finding that only one of the several nitrosylation sites is stable on these proteins (see Figure 3D and 3E and Table S2).
Figure 4. Biochemical confirmation of MS-characterized S-nitrosylation.
(A) S-nitrosylated proteins identified by SNOCAP as being GSH-unstable are reduced by GSH in vitro. Brain lysates were nitrosylated with 40 μM GSNO and then treated with either vehicle (GSNO→vehicle) or 1 mM GSH (GSNO→GSH). After 5 min, S-nitrosylated proteins were then biotinylated using the biotin switch method, purified on immobilized neutravidin, and eluted using 2-ME. Eluates were blotted with the indicated antibodies.
(B) GSH-stable S-nitrosylated proteins identified by SNOCAP are resistant to reduction by GSH in vitro. S-nitrosylated proteins generated by GSNO treatment were treated with either vehicle (GSNO→Vehicle) or GSH (GSNO→GSH) and assayed as in (A). In each case, the proteins remained S-nitrosylated after GSH treatment.
Our biochemical experiments to probe the solvent accessibility of protein nitrosthiols suggest that S-nitrosylation is coupled to conformational changes that lead to reduced nitrosothiol accessibility.Thus, we next asked if the structural context could account for the reduced reactivity of the stable protein nitrosothiols. Examination of the crystal structures of some of the stably S-nitrosylated proteins in their unnitrosylated states suggests that a variety of structural mechanisms might affect nitrosothiol reactivity. In some cases, the cysteines that form stable nitrosothiols are fully solvent exposed (Figure S4–S6), and the solvent accessibility was indistinguishable from other cysteines that form unstable nitrosothiols (Figure S5 and S6). However, in other cases, the cysteine is found within grooves or between large mobile lobes of the proteins that could potentially undergo small conformational changes that would effectively shield a nitrosothiol (Figure S6 and S7).
GSH-stable nitrosothiols are also stable in cells
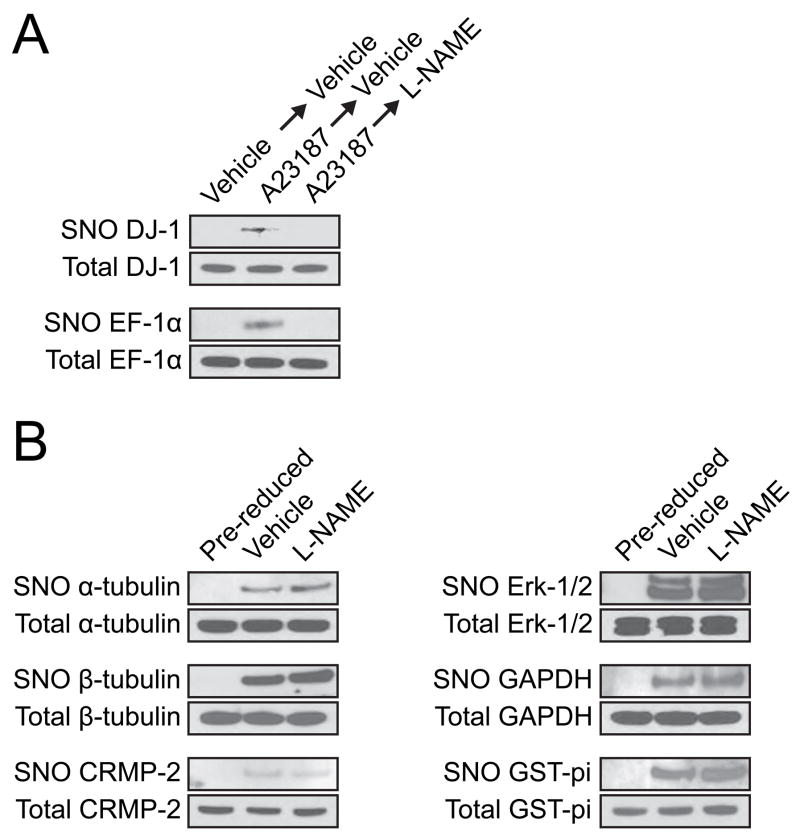
We next wanted to determine if the nitrosothiols that are stable to reduction by GSH in vitro are also stable in the complex reducing environment in living cells. To test the stability of S-nitrosylated proteins in cells, we used sEnd.1 murine endothelioma cells, which express eNOS and generate NO under basal conditions (Ghigo et al., 1995). Using the biotin switch method to label S-nitrosylated proteins in sEnd.1 cells, we found that proteins that were unstable to GSH in vitro were only minimally S-nitrosylated in unstimulated sEnd.1 cells (Figure 5A). S-nitrosylation could be more readily detected by pretreatment for 10 min with the calcium ionophore A23187, which activates eNOS (Figure 5A). The nitrosothiols in these proteins appeared to be unstable in cells as application of the NOS inhibitor N-nitro-L-arginine methyl ester (L-NAME) for 5 min to A23187-treated cells resulted in markedly reduced levels of S-nitrosylated protein compared to vehicle application to A23187-treated cells. Thus, these protein nitrosothiols were unstable to both in vitro application of GSH and to the physiologic reducing environment in cells.
Figure 5. Identification of stably S-nitrosylated proteins in cells.
(A) GSH-unstable proteins form labile nitrosothiols in sEnd.1 cells. sEnd.1 cells were treated with either vehicle (ethanol) or 10 μM A23187 for 10 min followed by either vehicle (media) or 1 mM L-NAME for 5 min. Cells were lysed and a total of 50 mg of protein was collected for each sample. S-nitrosylated (SNO) proteins were biotinylated using the biotin switch method, purified and eluted, and analyzed by Western blotting with indicated antibodies. GSH-unstable proteins were S-nitrosylated at a low stoichiometry in vehicle-treated cells (Vehicle→Vehicle), but readily detectable in cells pretreated for 10 min with calcium ionophore (A23187). Proteins remained S-nitrosylated if vehicle was added to the ionophore-treated cells (A23187→Vehicle), but were no longer S-nitrosylated after 5 min of L-NAME treatment (A23187→L-NAME), indicating that these proteins were not stably S-nitrosylated in cells after inhibition of NO synthesis. These effects do not reflect changes in expression levels in sEND.1 cells as the total protein levels were unaffected in the loading controls.
(B) GSH-stable proteins remain S-nitrosylated in cells after inhibition of NO synthesis. Proteins found to be GSH-stabile in in vitro experiments were readily found to be S-nitrosylated in vehicle (media)-treated sEnd.1 cells. These proteins remained S-nitrosylated even in cells that were treated with 1 mM L-NAME for 5 min. No protein labeling was detected in sEnd.1 samples that were pre-reduced with DTT.
We next examined the cellular stability of nitrosoproteins that were stable to GSH in vitro. Unlike the GSH-unstable proteins, these proteins were readily detectable as S-nitrosylated in sEnd.1 cells. Additionally, treatment of these cells with L-NAME did not substantially alter nitrosothiol levels in these proteins (Figure 5B and S8). As a control, in pre-reduced sEnd.1 lysates, nitrosothiols were undetectable, confirming the specificity of the biotin switch assay (Figure 5B). Taken together, these data indicate that endogenous nitrosothiol stability, even after NO synthesis is inhibited, is a feature of only a subset of S-nitrosylated proteins.
DISCUSSION
S-nitrosylation is a major mechanism by which NO signals in cells. Much of our current understanding of the properties of nitrosothiols comes from studies of low molecular weight nitrosothiols. These studies indicate that nitrosothiols are highly labile and susceptible to reduction by thiol-containing molecules, including molecules that are present at millimolar concentrations in the cytosol. Our studies indicate that there are certain instances in which nitrosothiols that form in proteins do not exhibit the predicted high degree of reactivity. We find that there are two separate types of nitrosothiols that can exist in proteins, i.e., stable and unstable nitrosothiols, and that the stabilization of nitrosothiols is conferred by conformational changes that shield the nitrosothiol from cytosolic reducing agents. Stably S-nitrosylated proteins may have selectively evolved to withstand the highly reducing cytosolic milieu, thus allowing them to transduce NO signals after ambient NO levels have dropped. Reduction of these stable nitrosothiols may involve enzymatic mechanisms such as the thioredoxin/thioredoxin reductase or GSNO reductase pathways (Benhar et al., 2008; Whalen et al., 2007).
Our studies suggest that stable and unstable nitrosothiols may have different roles in NO signaling. Unlike the small set of stable nitrosoproteins, most nitrosoproteins were not stable in the presence of physiological concentrations of GSH and were only minimally S-nitrosylated endogenously, consistent with studies of small molecule nitrosothiols (Kashiba-Iwatsuki et al., 1997; Singh et al., 1996). Although nitrosothiols in these proteins were undetectable under basal cellular conditions, they showed significant increases in S-nitrosylation when cellular levels of NO were high, suggesting that signaling by these proteins may be more transient and linked directly to the duration of NO synthesis. Alternatively, these nitrosothiols may accumulate under certain circumstances, such as during oxidative stress, when the reducing potential of the cell is compromised (Beltran et al., 2000). Our finding that a subset of proteins can exist as S-nitrosylated species in the presence of substantial concentrations of GSH in vitro and in cells after inhibition of endogenous NOS activity raises the possibility that these proteins may be capable of accumulating stoichiometric levels of nitrosothiols in cells, thereby quantitatively affecting protein function.
Interestingly, some studies have attempted to detect endogenously S-nitrosylated proteins from various cell types using proteomic approaches. However, these studies have resulted in the detection of surprisingly few endogenously S-nitrosylated proteins (J.S.P. and S.R.J., unpublished data and Gao et al., 2005; Hao et al., 2006; Martinez-Ruiz and Lamas, 2004), despite the large number of S-nitrosylated proteins that have been reported in the literature. These endogenously S-nitrosylated proteins include α-tubulin, β-tubulin, CRMP-2, and GAPDH. Notably, these proteins are also proteins that we have found are stably S-nitrosylated. This suggests that the reason that these proteins were detected was due to the stability of their nitrosothiols and the ability of these nitrosothiols to accumulate to levels that could be detected by MS methods. None of the proteins found to be endogenously S-nitrosylated in these prior studies were found to contain unstable nitrosothiols in our study. Together, these data provide further support to the idea that stable S-nitrosylation is a feature of certain proteins in vivo.
The basis for the stability of these protein nitrosothiols is not readily apparent from the crystal structures of the unnitrosylated forms of the proteins. In some cases the cysteines were partially buried, suggesting that minor conformational changes induced by S-nitrosylation could effectively shield the nitrosothiol. However, in several cases, the S-nitrosylated cysteine was situated in such a way that it is unclear how it could be shielded from reducing agents. Crystal structures of the S-nitrosylated forms of these proteins will be required to identify the S-nitrosylation-induced conformational changes that bury the nitrosothiol. The idea that S-nitrosylation promotes conformational changes that stabilize nitrosothiols is well demonstrated in Hb where S-nitrosylation of Cysβ93 promotes a conformational change in which the nitrosothiol is buried in a hydrophobic pocket where it is inaccessible to solvent (Singel and Stamler, 2005). Deoxygenation Hb reverses this conformational change increasing the reactivity of S-nitroso-Hb to GSH (Singel and Stamler, 2005). However, an additional mechanism that may account for nitrosothiol stability may be the presence of associated proteins that reduce solvent access to the nitrosothiol. In Erk-2, the nitrosylated cysteine is present on a surface that is associated with numerous protein-protein interactions (Tanoue et al., 2000). In agreement with this idea, S-nitrosylation of GAPDH results in increased interactions with other proteins (Hara et al., 2005). An additional factor that should be considered is the cis/trans conformation of the nitrosothiol, which could affect the overall rate of nitosothiol reduction (Stamler and Toone, 2002; Weichsel et al., 2007). Conceivably, nitrosothiol stability may be influenced by protein-mediated stabilization of specific cis/trans nitrosothiol isoforms.
The stably nitrosylated proteins identified in this study correspond to several proteins widely studied as NO effectors in cells, including GAPDH, creatine kinase, and Hb. S-nitrosylation of GAPDH has been shown to mediate NO-induced apoptotic death (Hara et al., 2005). Creatine kinase also undergoes S-nitrosylation leading to a decrease in Ca2+ uptake in the sacroplasmic reticulum (Wolosker et al., 1996), which is thought to contribute to inhibition of mitochondrial respiration (Stamler and Meissner, 2001). S-nitrosylation of Hb in cells has a well-established role in the regulation of blood flow (Stamler et al., 1997a) and platelet aggregation (Pawloski et al., 1998). We also identified several proteins which have not been rigorously linked to NO signaling in cells, such as α-tubulin, β-tubulin, CRMP-2 and Erk-2. However, these proteins are involved in pathways and processed which are affected by NO in cells. For example, NO has been shown to impair dendrite outgrowth (Inglis et al., 1998) and elicit cytoskeletal breakdown via microtubule destabilization in cerebellar granule cells (Bonfoco et al., 1996), which may reflect S-nitrosylation of tubulin subunits or CRMP-2, a microtubule regulatory protein. Similarly, we found Erk-2 to be stably nitrosylated which is consistent with the regulatory role of NO in several mitogen activated protein kinase pathways (Hess et al., 2005). Together, the stably nitrosylated proteins identified here represent both important physiological targets of NO as well as proteins that are involved in pathways mediated by NO in cells.
The degree to which a nitrosothiol accumulates on a cysteine residue in a protein is determined by the balance between the rates of S-nitrosylation and denitrosylation. Our data highlight the importance of denitrosylation pathways in regulating the steady-state level of nitrosothiols on proteins. For certain proteins, the critical factor determining the nitrosothiol stoichiometry at each cysteine may be the accessibility of the nitrosothoiol to physiologic reducing agents. S-nitrosylation by GSNO or nitrosating agents such as N2O3 may occur on numerous cysteine residues and then be rapidly reversed on all but the few cysteines that have the unusual property of undergoing stable modification. Conceivably, certain proteins evolved the ability to bury the modified cysteine in order to allow NO, an otherwise ephemeral signaling molecule, to have a long lasting effect in cells.
SIGNIFICANCE
Previous work established that caspase-3 forms a protein nitrosothiol that cannot be denitrosylated by GSH (Mitchell and Marletta, 2005). However, it was unclear from this earlier study whether nitrosothiols in other S-nitrosylated proteins exhibit this property. In this study we designed a quantitative nitrosoproteomic approach to screen dozens of previously characterized, as well as novel, protein nitrosothiols for their chemical reactivity with reducing agents. Our results indicate that although most S-nitrosylated proteins are rapidly degraded by reducing agents, there is a novel class of NO targets that are capable of forming stable nitrosothiols. These stable nitrosothiols also exhibit exceptional stability in vivo. Some of these proteins are well-established NO targets, suggesting that physiologic NO signaling utilizes stable nitrosothiols. The inability of these proteins to be reduced by GSH, as is the case with caspase-3, raises the possibility that these proteins will utilize highly specific intracellular denitrosylation pathways that regulate the circumstances and degree to which these nitrosothiols accumulate after NO signaling.
EXPERIMENTAL PROCEDURES
Denitrosylation assay
Freshly isolated rat cerebellum was homogenized in 10 volumes of HEN buffer (25 mM HEPES pH 7.7, 1 mM EDTA, and 10 μM neocuproine) and then centrifuged at 20,000g for 15 min at 4 °C. The supernatant typically contained 5 mg/ml protein determined by BCA assay. To 45 μL of protein supernatant was added 5 μL GSNO stock (400 μM in water) or vehicle to make the final concentration of NO donor in each sample 40 μM, and the mixture was incubated for 20 min at 25 °C. After this step, residual GSNO was removed by centrifuging the lysate through a MicroBioSpin6 column (Biorad) pre-equilibrated in HEN buffer. To the desalted protein lysate was added 5 μL GSH stock (11 mM in water) or vehicle to make the final concentration of GSH 1 mM, and the mixture was incubated at 25 °C for 5, 10, or 30 min. To the denitrosylated samples was then added four volumes of blocking buffer (nine volumes of 10X HEN buffer plus one volume 20% SDS, adjusted to 20 mM MMTS with a 2 M stock prepared in dimethylformamide (DMF)) at 50 °C for 20 min with frequent vortexing. The proteins were then precipitated with two volumes of acetone at −20 °C for 1 h. After centrifugation, the protein pellet was washed six times with cold acetone to remove residual MMTS. Pellets were then resuspended in 125 μL of HENS buffer (HEN with 1% SDS). To the resuspended proteins was added biotin HPDP prepared fresh as a 4 mM stock in dimethylsulfoxide (DMSO) from a 50 mM stock suspension in DMF. Sodium ascorbate was added in a final volume of 2 μL to a final concentration of 2 mM. After incubation for 1 h at 25 °C, SDS PAGE sample buffer was added and the samples (without reduction or boiling) were resolved by SDS PAGE and transferred for immunoblotting. All steps preceding SDS PAGE were carried out in the dark.
To evaluate the role of protein structure on denitrosylation, nitrosylated proteins were prepared with GSNO as described above. Next, to the lysates was added either 5 μL of GSH or 2-ME stock (11 mM) prepared in either water or 11% SDS, making the final concentration of GSH and 2-ME 1 mM, and the final SDS concentration 1%. The colorimetric thiol probe, DTNB, was used to confirm that stock solutions of GSH and 2-ME contained equal amounts of free thiol. Samples were then analyzed using the biotin switch method as described above.
SNOCAP analysis of standard S-nitrosylated protein mixtures
GAPDH and Hb (Sigma) were resuspended in HEN buffer to a concentration of 1 mg/ml. The protein solutions were then reduced with 50 mM DTT, dialyzed, and the two protein stocks were combined in two samples of different concentrations. To each sample was then added GSNO to a final concentration of 40 μM and the samples were incubated for 20 min at 25 °C. Residual GSNO was removed by centrifuging the suspension through a MicroBioSpin6 column pre-equilibrated in HEN buffer, and to the eluate was added four volumes of blocking buffer. After blocking and precipitation as described above, the samples were resuspended in HENS buffer with 2 mM sodium ascorbate and incubated with 1 mM of either light or heavy SNOCAP reagent for 1 h at 25 °C. Proteins were again acetone precipitated to remove SDS and pelleted by centrifugation. The two samples were then resuspended in HEN buffer with 8 M urea and combined. The combined sample was diluted to 1.2 M urea and digested with trypsin (1:25) overnight at 37 °C. Trypsin was inhibited with 1 mM phenylmethyl sulfonyl fluoride (PMSF) and 1 μM leupeptin, and the digest was passed over 250 μL of preequilibrated neutravidin resin (Pierce). The resin was washed with 2X PBS twice, 1X PBS four times, and 20% MeOH twice. Biotinylated peptides were eluted with 1 ml acetonitrile:water:trifluoroacetic acid (TFA) (50:50:0.1, vol/vol/vol). Samples were then concentrated to 100 μL under reduced pressure and approximately 1 pmole of peptide, as determined by fluorescamine assay, was injected and analyzed by nLC-MS/MS. The entire procedure (SNOCAP and LC-MS/MS) can be completed in 1–2 days, allowing for the rapid analysis of dozens or hundreds of nitrosoproteins compared to the time-consuming analysis of individual nitrosoproteins.
Profiling of stably S-nitrosylated proteins
600 mg of rat brain was homogenized as described above providing approximately 30 mg of protein. The homogenate was divided into two equal samples and incubated with 40 μM GSNO for 20 min. Residual GSNO was removed by gel filtration using MicroBioSpin6 columns. To the eluate was added GSH or vehicle to a final concentration of 1 mM and the mixture was incubated at 25 °C for 10 min. Proteins were then blocked, acetone precipitated and resuspended in HENS buffer as described above. Nitrosothiols were then reduced with sodium ascorbate (2 mM) and labeled with either 1 mM light SNOCAP reagent (GSNO→vehicle treated sample) or 1 mM heavy SNOCAP reagent (GSNO→GSH treated sample). Proteins were next digested, biotinylated peptides recovered, and analyzed using nLC-MS/MS as described above. This experiment was repeated five times and each sample was reloaded and analyzed by LC-MS/MS three to six times. In control experiments using “pre-reduced” lysates, the cytosolic lysate was adjusted to 20 mM dithiothreitol and then dialyzed against 1X HEN buffer two times prior to performing SNOCAP.
LC-MS/MS and quantitative analysis
LC-MS/MS was performed on an XCT ion trap mass spectrometer (Agilent) as described in the Supplementary Methods.
Peptide sequence identification
Analysis of MS/MS spectra for peptide identification was performed by protein database searching with SPECTRUM MILL software (Agilent Technologies). The threshold used for peptide identification was a SPECTRUM MILL score of >9.0 and an SPI% (the percentage of assigned spectrum intensity of total spectrum intensity) of >60%. This threshold was established by searching the same data against two different decoy databases. One decoy database contained reversed rat protein sequences while the other contained completely scrambled protein sequences. The scrambled database contained the same number of total proteins with same number of amino acids for each protein as found in the original database. The scrambled sequences were generated based on the relative abundance of each type of amino acids in the whole rat protein database. Scores of the hits obtained using these decoy databases identify the threshold scores needed to reduce the likelihood of false positives (Elias and Gygi, 2007). In these experiments, the false positive rate for a score of >9.0 and an SPI >60% was 1.72 %. All hits were validated by manual inspection of the raw MS/MS data. Additional details are provided in the Supplementary Methods.
Purification of S-nitrosylated proteins for biochemical confirmation
10 mg of brain lysates were nitrosylated with 40 μM GSNO and then treated with either vehicle or 1 mM GSH. Any S-nitrosylated cysteines residues were then labeled with biotin by the biotin switch method. Biotinylated proteins were poured over immobilized neutravidin, the resin was washed, and S-nitrosylated proteins were specifically eluted with 2-ME. Approximately 20% of each eluate was then analyzed by Western blotting using antibodies to β-tubulin (Developmental Studies Hybridoma Bank), GAPDH (Millipore), ERK-2 (Santa Cruz Biotechnologies), CRMP-2 (Millipore), GST-pi (Genscript), hnRNP (Santa Cruz Biotechnologies), 14–3-3 θ (Cell Signaling), EF-1α (Upstate).
Purification of endogenously S-nitrosylated proteins
Murine sEnd.1 cells were grown on plastic culture dishes in high-glucose DMEM (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen) for 24–48 h before lysing cells and labeling S-nitrosylated proteins with the biotin switch method. For these experiments, approximately 50 mg of protein was extracted from cells for each sample. Transiently S-nitrosylated proteins were labeled after treating cells with either vehicle or 10 μM A23187 for 10 min, followed by treatment with vehicle or 1 mM L-NAME for 5 min. Stably S-nitrosylated proteins were labeled after treating cells with either vehicle or 1 mM L-NAME for 5 min. Prior to biotin switch, some lysates were pre-reduced with 50 mM DTT, which was subsequently removed by dialysis. Biotinylated proteins were poured over immobilized neutravidin, the resin was washed, and S-nitrosylated proteins were specifically eluted with 2-ME. Approximately 20% of the volume from each eluate was then analyzed by Western blotting using antibodies described above as well as antibodies to α-tubulin (Developmental Studies Hybridoma Bank) and DJ-1 (Millipore).
Crystal structure and solvent accessible area analysis
The images of Erk-2, GST-pi, α-/β-tubulin and GAPDH crystal structures were made using PyMOL. The solvent accessibility area of the sulfurs was calculated using the program NACCESS (Hubbard et al., 1991) with a probe of 1.4 Å.
Supplementary Material
(A) Signals from the biotin switch assay detecting stable nitrosothiols are ascorbate-dependent. Lysates were exposed to 40 μM GSNO and then 1 mM GSH as in Figure 1. Protein nitrosothiols were then labeled using the biotin switch method in the presence or absence of 30 mM ascorbate. Proteins were then visualized by anti-biotin Western blotting.
(B) Characterization of stable protein nitrosothiols under a variety of conditions. Lysates were exposed to 40 μM GSNO and then GSH (either 1 or 10 mM) for time intervals up to 1 h. GSH-mediated reduction was performed at either 25°C or 37°C. Protein nitrosothiols were detected as in (A) and quantification is shown in (C–F).
(C) Densitometry measurements of bands remaining after treatment with 1 mM GSH for 5 min at 25°C. Band intensity was compared at 0 and 5 min of GSH treatment to quantify the degree of GSH-mediated denitrosylation as in Figure 1D. Proteins were classified as either GSH-stable (exhibiting less than 20% denitrosylation) or GSH-unstable (exhibiting greater than 80% denitrosylation) as described in Figure 1D. Most proteins were highly sensitive to GSH-mediated denitrosylation. Densitometry was averaged from three independent experiments.
(D) Densitometry measurements of bands remaining after treatment with 10 mM GSH for 5 min at 25°C. A similar profile of stable and unstable nitrosothiols were detected using this treatment compared to the 1 mM treatment used in Figure S1C. Densitometry was averaged from three independent experiments.
(E) Densitometry measurements of bands remaining after treatment with 1 mM GSH for 5 min at 37°C. The number of stable and unstable nitrosothiols detected using this treatment were similar to that found using the 25°C treatment in Figure S1C. Densitometry was averaged from three independent experiments.
Supplementary Figure 2. Quantitative MS reveals that only GSNO-treated samples produced SNOCAP-labeled peptides
Brain lysates was treated with vehicle or GSNO, and processed with light or heavy SNOCAP reagent, respectively. Representative MS spectrum of an S-nitrosylated peptide labeled with heavy SNOCAP reagent, originating from GSNO-treated sample, is shown. Absent is the peak that corresponds to peptide labeled with light SNOCAP reagent, originating from vehicle-treated sample, demonstrating that the SNOCAP protocol results in exclusive labeling of nitrosothiol-containing peptides.
Supplementary Figure 3. SPECTRUM MILL search results for all identified peptides
A table organizing the search results for each identified peptide is presented. At the top of the table is shown the top ranked peptide match for each particular spectrum. In some cases SPECTRUM MILL lists the same matched peptide several times because the same peptide could have originated from more than one gene. In all of these cases, all the genes listed produce nearly, if not completely, identical proteins. The peptide sequence is shown with lines between residues to indicate which fragmentation ions were identified. Backslashes indicate b ions, forward-slashes indicate y ions, and vertical lines indicate that both y and b ions were identified. For each matched peptide is also listed the score, SPI, backbone cleavage score (BCS), number of unmatched ions, precursor mass calculated from the product mass measured, precursor mass error, protein molecular weight, species, Swiss-Prot accession number, and the protein name. Below each reported match is a table listing the details of the fragment ions found for each peptide. The major fragment ions are listed at the top of the table followed by more specific information about each fragment ion including: fractional intensity of total ion count (% of TIC), relative intensity of base peak ions (% of BP), score, ion type, and error in the mass (Delta Da). A representative MS/MS spectra of each peptide is also presented with labeled y-ions (red), b-ions (blue), and precursor ions (green). In all spectra, the major y+, y++, b+, and b++ ions have been labeled with colored peaks and with annotation. In some cases b+-H2O and other major ions were labeled as well, and below each spectra shows a checklist of which type of ions were chosen for labeling.
Supplementary Figure 4. Crystal structure of Erk-2 reveals high solvent accessibility of stably S-nitrosylated cysteine
(A) The structure of Erk-2 (pdb: 1TVO). All cysteine residues are highlighted as spheres in atomic coloring (nitrogen: blue; oxygen: red; sulfur: yellow; carbon: grey). The cysteine that undergoes stable S-nitrosylation is circled.
(B) Close-up view of stably S-nitrosylated site shows a high degree of solvent accessibility. Cysteine 161 is shown as a stick model in atomic coloring as in (A) with its solvent accessible surface shown as teal mesh.
(C) All cysteines of Erk-2 are listed in order of decreasing solvent accessibility of the thiol, with the stably S-nitrosylated cysteine highlighted in brown.
Supplementary Figure 5. Crystal structure of GST-pi homodimer reveals stable S-nitrosylated cysteine to be on solvent-accessible dimer interface
(A) The structure of GST-pi dimer (pdb: 6GSS). The separate chains of the dimer are signified by either teal or pink backbone. All cysteine residues are highlighted as spheres in atomic coloring (nitrogen: blue; oxygen: red; sulfur: yellow; carbon: grey). The cysteine that undergoes stable S-nitrosylation is circled.
(B) Close-up view of stably S-nitrosylated site shows high degree of solvent accessibility at dimer interface. Cysteine 101 is shown as a stick model in atomic coloring as in (A).
(C) Cysteine 101 is the most solvent accessible cysteine in GST-pi dimer. All cysteines of GST-pi are listed in order of decreasing solvent accessibility of the thiol, with the stably S-nitrosylated cysteine highlighted in brown.
Supplementary Figure 6. Crystal structure of GAPDH reveals a high degree of variation in the solvent accessibility of stable S-nitrosylated cysteines
(A) The structure of the GAPDH monomer (pdb: 1ZNQ). All cysteine residues are highlighted as spheres in atomic coloring (nitrogen: blue; oxygen: red; sulfur: yellow; carbon: grey). The two cysteines that undergo stable S-nitrosylation are circled.
(B) The structure of the GAPDH in its native tetrameric form (pdb: 1ZNQ). The separate chains are signified by either pink, purple, orange, or teal chains.
(C) Close-up view reveals no solvent access for cysteine 247, shown as a stick model in atomic coloring as in (A).
(D) In contrast, a close-up view of cysteine 152 reveals a highly solvent accessible surface indicated by teal mesh.
(E) All cysteines of GAPDH are listed in order of decreasing solvent accessibility of the thiol, with the stably S-nitrosylated cysteines highlighted in brown.
Supplementary Figure 7. Crystal structure of α-/β-tubulin heterodimer illustrates reveals a high degree of variation in the solvent accessibility of stable S-nitrosylated cysteines
(A) The structure of the α-/β-tubulin heterodimer (pdb: 1TUB). All S-nitrosylated cysteine residues are highlighted as spheres in atomic coloring (nitrogen: blue; oxygen: red; sulfur: yellow; carbon: grey). The cysteine that undergoes stable S-nitrosylation on each monomer is circled.
(B) Close-up view of stably S-nitrosylated cysteine of β-tubulin reveals a highly solvent accessible sulfur atom. Cysteine 12 is shown as a stick model in atomic coloring as in (A) with its solvent accessible surface shown as pink mesh.
(C) Close-up view of stably S-nitrosylated cysteine of α-tubulin reveals very little solvent accessibility. Cysteine 295 is shown as a stick model in atomic coloring as in (A) with its solvent accessible surface shown as teal mesh.
(D) All cysteines of α-tubulin are listed in order of decreasing solvent accessibility of the thiol, with the stably S-nitrosylated cysteines highlighted in brown and the unstably S-nitrosylated cysteines highlighted in blue.
(E) All cysteines of β-tubulin are listed in order of decreasing solvent accessibility of the thiol, with the stably S-nitrosylated cysteines highlighted in brown and the unstably S-nitrosylated cysteines highlighted in blue.
Supplementary Figure 8. Endogenous stably S-nitrosylated proteins remain S-nitrosylated even after prolonged inhibition of NO synthesis.
sEND.1 cells were cultured, treated, and analyzed as in Figure 5. Cells were treated with either vehicle (media) or 1 mM L-NAME for 5 min, 30 min or 24 h. Cells were harvested, and cell lysates (50 mg protein) were subjected to the biotin switch method. Formerly S-nitrosylated (SNO) proteins were purified on neutravidin agarose, eluted with 2-ME, and analyzed by Western blotting with the indicated antibodies. As in Figure 5, GSH-stable proteins remained S-nitrosylated after 5 min; however, the proteins were also S-nitrosylated at longer time points, up to 24 h. These effects were not associated with changes in the levels of the assayed proteins, as indicated by the loading controls (Total protein).
Acknowledgments
Supported by the National Institute of Mental Health (R01-MH066204) and NARSAD (S.R.J.), a fellowship by the PhRMA foundation (J.S.P.), and training grant T32CA062948 from the National Cancer Institute (G.X.). This work was performed at the Weill Cornell Medical College Mass Spectrometry Facility using instrumentation supported by NIH RR19355.
Footnotes
Accession codes
Protein Data Bank (PDB) identifiers: human Erk-2, 1TVO; human GAPDH, 1ZNQ; Pig α-/β-tubulin dimer, 1TUB; human GST-pi dimer, 6GSS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beltran B, Orsi A, Clementi E, Moncada S. Oxidative stress and S-nitrosylation of proteins in cells. Br J Pharmacol. 2000;129:953–960. doi: 10.1038/sj.bjp.0703147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfoco E, Leist M, Zhivotovsky B, Orrenius S, Lipton SA, Nicotera P. Cytoskeletal breakdown and apoptosis elicited by NO donors in cerebellar granule cells require NMDA receptor activation. J Neurochem. 1996;67:2484–2493. doi: 10.1046/j.1471-4159.1996.67062484.x. [DOI] [PubMed] [Google Scholar]
- Chvanov M, Gerasimenko OV, Petersen OH, Tepikin AV. Calcium-dependent release of NO from intracellular S-nitrosothiols. Embo J. 2006;25:3024–3032. doi: 10.1038/sj.emboj.7601207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- Gao C, Guo H, Wei J, Mi Z, Wai PY, Kuo PC. Identification of S-nitrosylated proteins in endotoxin-stimulated RAW264.7 murine macrophages. Nitric Oxide. 2005;12:121–126. doi: 10.1016/j.niox.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Gaston B, Reilly J, Drazen JM, Fackler J, Ramdev P, Arnelle D, Mullins ME, Sugarbaker DJ, Chee C, Singel DJ, et al. Endogenous nitrogen oxides and bronchodilator S-nitrosothiols in human airways. Proc Natl Acad Sci USA. 1993;90:10957–10961. doi: 10.1073/pnas.90.23.10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo D, Arese M, Todde R, Vecchi A, Silvagno F, Costamagna C, Dong QG, Alessio M, Heller R, Soldi R, et al. Middle T antigen-transformed endothelial cells exhibit an increased activity of nitric oxide synthase. J Exp Med. 1995;181:9–19. doi: 10.1084/jem.181.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Holmgren A, Luthman M. Tissue distrubution and subcellular localization of bovine thioredoxin determined by radioimmunoassay. Biochemistry. 1978;17:4071–4077. doi: 10.1021/bi00612a031. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Campbell SF, Thornton JM. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J Mol Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- Inglis FM, Furia F, Zuckerman KE, Strittmatter SM, Kalb RG. The role of nitric oxide and NMDA receptors in the development of motor neuron dendrites. J Neurosci. 1998;18:10493–10501. doi: 10.1523/JNEUROSCI.18-24-10493.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Akaike T, Miyamoto Y, Okamoto T, Sawa T, Otagiri M, Suzuki S, Yoshimura T, Maeda H. Nitrosothiol formation catalyzed by ceruloplasmin: Implication for cytoprotective mechanism in vivo. J Biol Chem. 1999;274:27069–27075. doi: 10.1074/jbc.274.38.27069. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Jia L, Bonaventura C, Bonaventura J, Stamler JS. S-nitrosohaemoglobin: a dynamic activity of blood involved in vascular control. Nature. 1996;380:221–226. doi: 10.1038/380221a0. [DOI] [PubMed] [Google Scholar]
- Kashiba-Iwatsuki M, Kitoh K, Kasahara E, Yu H, Nisikawa M, Matsuo M, Inoue M. Ascorbic acid and reducing agents regulate the fates and functions of S-nitrosothiols. J Biochem (Tokyo) 1997;122:1208–1214. doi: 10.1093/oxfordjournals.jbchem.a021883. [DOI] [PubMed] [Google Scholar]
- Kluge I, Gutteck-Amsler U, Zollinger M, Do KQ. S-nitrosoglutathione in rat cerebellum: identification and quantification by liquid chromatography-mass spectrometry. J Neurochem. 1997;69:2599–2607. doi: 10.1046/j.1471-4159.1997.69062599.x. [DOI] [PubMed] [Google Scholar]
- Kosower NS, Kosower EM. The glutathione status of cells. Int Rev Cytol. 1978;54:109–160. doi: 10.1016/s0074-7696(08)60166-7. [DOI] [PubMed] [Google Scholar]
- Krajewska M, Wang HG, Krajewski S, Zapata JM, Shabaik A, Gascoyne R, Reed JC. Immunohistochemical analysis of in vivo patterns of expression of CPP32 (Caspase-3), a cell death protease. Cancer Res. 1997;57:1605–1613. [PubMed] [Google Scholar]
- Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- Martinez-Ruiz A, Lamas S. Detection and proteomic identification of S-nitrosylated proteins in endothelial cells. Arch Biochem Biophys. 2004;423:192–199. doi: 10.1016/j.abb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Mayer B, Schrammel A, Klatt P, Koesling D, Schmidt K. Peroxynitrite-induced accumulation of cyclic GMP in endothelial cells and stimulation of purified soluble guanylyl cyclase. Dependence on glutathione and possible role of S-nitrosation. J Biol Chem. 1995;270:17355–17360. doi: 10.1074/jbc.270.29.17355. [DOI] [PubMed] [Google Scholar]
- Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- Mizoguchi A, Ueda T, Ikeda K, Shiku H, Mizoguti H, Takai Y. Localization and subcellular distribution of cellular ras gene products in rat brain. Brain Res Mol Brain Res. 1989;5:31–44. doi: 10.1016/0169-328x(89)90015-6. [DOI] [PubMed] [Google Scholar]
- Paige JS, Jaffrey SR. Pharmacologic manipulation of nitric oxide signaling: targeting NOS dimerization and protein-protein interactions. Curr Top Med Chem. 2007;7:97–114. doi: 10.2174/156802607779318253. [DOI] [PubMed] [Google Scholar]
- Pawloski JR, Hess DT, Stamler JS. Export by red blood cells of nitric oxide bioactivity. Nature. 2001;409:622–626. doi: 10.1038/35054560. [DOI] [PubMed] [Google Scholar]
- Pawloski JR, Swaminathan RV, Stamler JS. Cell-free and erythrocytic S-nitrosohemoglobin inhibits human platelet aggregation. Circulation. 1998;97:263–267. doi: 10.1161/01.cir.97.3.263. [DOI] [PubMed] [Google Scholar]
- Sen CK. Redox signaling and the emerging therapeutic potential of thiol antioxidants. Biochem Pharmacol. 1998;55:1747–1758. doi: 10.1016/s0006-2952(97)00672-2. [DOI] [PubMed] [Google Scholar]
- Singel DJ, Stamler JS. Chemical physiology of blood flow regulation by red blood cells: the role of nitric oxide and S-nitrosohemoglobin. Annu Rev Physiol. 2005;67:99–145. doi: 10.1146/annurev.physiol.67.060603.090918. [DOI] [PubMed] [Google Scholar]
- Singh SP, Wishnok JS, Keshive M, Deen WM, Tannenbaum SR. The chemistry of the S-nitrosoglutathione/glutathione system. Proc Natl Acad Sci USA. 1996;93:14428–14433. doi: 10.1073/pnas.93.25.14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler JS, Jia L, Eu JP, McMahon TJ, Demchenko IT, Bonaventura J, Gernert K, Piantadosi CA. Blood flow regulation by S-nitrosohemoglobin in the physiological oxygen gradient. Science. 1997a;276:2034–2037. doi: 10.1126/science.276.5321.2034. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Lamas S, Fang FC. Nitrosylation the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Meissner G. Physiology of nitric oxide in skeletal muscle. Physiol Rev. 2001;81:209–237. doi: 10.1152/physrev.2001.81.1.209. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Toone EJ. The decomposition of thionitrites. Curr Opin Chem Biol. 2002;6:779–785. doi: 10.1016/s1367-5931(02)00383-6. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: translocation, regulation, and a consensus motif. Neuron. 1997b;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- Tao WA, Aebersold R. Advances in quantitative proteomics via stable isotope tagging and mass spectrometry. Curr Opin Biotechnol. 2003;14:110–118. doi: 10.1016/s0958-1669(02)00018-6. [DOI] [PubMed] [Google Scholar]
- Weichsel A, Brailey JL, Montfort WR. Buried S-nitrosocysteine revealed in crystal structures of human thioredoxin. Biochemistry. 2007;46:1219–1227. doi: 10.1021/bi061878r. [DOI] [PubMed] [Google Scholar]
- Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CD, Benhar M, Keys JR, Rockman HA, et al. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- Wolosker H, Panizzutti R, Engelender S. Inhibition of creatine kinase by S-nitrosoglutathione. FEBS Lett. 1996;392:274–276. doi: 10.1016/0014-5793(96)00829-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Signals from the biotin switch assay detecting stable nitrosothiols are ascorbate-dependent. Lysates were exposed to 40 μM GSNO and then 1 mM GSH as in Figure 1. Protein nitrosothiols were then labeled using the biotin switch method in the presence or absence of 30 mM ascorbate. Proteins were then visualized by anti-biotin Western blotting.
(B) Characterization of stable protein nitrosothiols under a variety of conditions. Lysates were exposed to 40 μM GSNO and then GSH (either 1 or 10 mM) for time intervals up to 1 h. GSH-mediated reduction was performed at either 25°C or 37°C. Protein nitrosothiols were detected as in (A) and quantification is shown in (C–F).
(C) Densitometry measurements of bands remaining after treatment with 1 mM GSH for 5 min at 25°C. Band intensity was compared at 0 and 5 min of GSH treatment to quantify the degree of GSH-mediated denitrosylation as in Figure 1D. Proteins were classified as either GSH-stable (exhibiting less than 20% denitrosylation) or GSH-unstable (exhibiting greater than 80% denitrosylation) as described in Figure 1D. Most proteins were highly sensitive to GSH-mediated denitrosylation. Densitometry was averaged from three independent experiments.
(D) Densitometry measurements of bands remaining after treatment with 10 mM GSH for 5 min at 25°C. A similar profile of stable and unstable nitrosothiols were detected using this treatment compared to the 1 mM treatment used in Figure S1C. Densitometry was averaged from three independent experiments.
(E) Densitometry measurements of bands remaining after treatment with 1 mM GSH for 5 min at 37°C. The number of stable and unstable nitrosothiols detected using this treatment were similar to that found using the 25°C treatment in Figure S1C. Densitometry was averaged from three independent experiments.
Supplementary Figure 2. Quantitative MS reveals that only GSNO-treated samples produced SNOCAP-labeled peptides
Brain lysates was treated with vehicle or GSNO, and processed with light or heavy SNOCAP reagent, respectively. Representative MS spectrum of an S-nitrosylated peptide labeled with heavy SNOCAP reagent, originating from GSNO-treated sample, is shown. Absent is the peak that corresponds to peptide labeled with light SNOCAP reagent, originating from vehicle-treated sample, demonstrating that the SNOCAP protocol results in exclusive labeling of nitrosothiol-containing peptides.
Supplementary Figure 3. SPECTRUM MILL search results for all identified peptides
A table organizing the search results for each identified peptide is presented. At the top of the table is shown the top ranked peptide match for each particular spectrum. In some cases SPECTRUM MILL lists the same matched peptide several times because the same peptide could have originated from more than one gene. In all of these cases, all the genes listed produce nearly, if not completely, identical proteins. The peptide sequence is shown with lines between residues to indicate which fragmentation ions were identified. Backslashes indicate b ions, forward-slashes indicate y ions, and vertical lines indicate that both y and b ions were identified. For each matched peptide is also listed the score, SPI, backbone cleavage score (BCS), number of unmatched ions, precursor mass calculated from the product mass measured, precursor mass error, protein molecular weight, species, Swiss-Prot accession number, and the protein name. Below each reported match is a table listing the details of the fragment ions found for each peptide. The major fragment ions are listed at the top of the table followed by more specific information about each fragment ion including: fractional intensity of total ion count (% of TIC), relative intensity of base peak ions (% of BP), score, ion type, and error in the mass (Delta Da). A representative MS/MS spectra of each peptide is also presented with labeled y-ions (red), b-ions (blue), and precursor ions (green). In all spectra, the major y+, y++, b+, and b++ ions have been labeled with colored peaks and with annotation. In some cases b+-H2O and other major ions were labeled as well, and below each spectra shows a checklist of which type of ions were chosen for labeling.
Supplementary Figure 4. Crystal structure of Erk-2 reveals high solvent accessibility of stably S-nitrosylated cysteine
(A) The structure of Erk-2 (pdb: 1TVO). All cysteine residues are highlighted as spheres in atomic coloring (nitrogen: blue; oxygen: red; sulfur: yellow; carbon: grey). The cysteine that undergoes stable S-nitrosylation is circled.
(B) Close-up view of stably S-nitrosylated site shows a high degree of solvent accessibility. Cysteine 161 is shown as a stick model in atomic coloring as in (A) with its solvent accessible surface shown as teal mesh.
(C) All cysteines of Erk-2 are listed in order of decreasing solvent accessibility of the thiol, with the stably S-nitrosylated cysteine highlighted in brown.
Supplementary Figure 5. Crystal structure of GST-pi homodimer reveals stable S-nitrosylated cysteine to be on solvent-accessible dimer interface
(A) The structure of GST-pi dimer (pdb: 6GSS). The separate chains of the dimer are signified by either teal or pink backbone. All cysteine residues are highlighted as spheres in atomic coloring (nitrogen: blue; oxygen: red; sulfur: yellow; carbon: grey). The cysteine that undergoes stable S-nitrosylation is circled.
(B) Close-up view of stably S-nitrosylated site shows high degree of solvent accessibility at dimer interface. Cysteine 101 is shown as a stick model in atomic coloring as in (A).
(C) Cysteine 101 is the most solvent accessible cysteine in GST-pi dimer. All cysteines of GST-pi are listed in order of decreasing solvent accessibility of the thiol, with the stably S-nitrosylated cysteine highlighted in brown.
Supplementary Figure 6. Crystal structure of GAPDH reveals a high degree of variation in the solvent accessibility of stable S-nitrosylated cysteines
(A) The structure of the GAPDH monomer (pdb: 1ZNQ). All cysteine residues are highlighted as spheres in atomic coloring (nitrogen: blue; oxygen: red; sulfur: yellow; carbon: grey). The two cysteines that undergo stable S-nitrosylation are circled.
(B) The structure of the GAPDH in its native tetrameric form (pdb: 1ZNQ). The separate chains are signified by either pink, purple, orange, or teal chains.
(C) Close-up view reveals no solvent access for cysteine 247, shown as a stick model in atomic coloring as in (A).
(D) In contrast, a close-up view of cysteine 152 reveals a highly solvent accessible surface indicated by teal mesh.
(E) All cysteines of GAPDH are listed in order of decreasing solvent accessibility of the thiol, with the stably S-nitrosylated cysteines highlighted in brown.
Supplementary Figure 7. Crystal structure of α-/β-tubulin heterodimer illustrates reveals a high degree of variation in the solvent accessibility of stable S-nitrosylated cysteines
(A) The structure of the α-/β-tubulin heterodimer (pdb: 1TUB). All S-nitrosylated cysteine residues are highlighted as spheres in atomic coloring (nitrogen: blue; oxygen: red; sulfur: yellow; carbon: grey). The cysteine that undergoes stable S-nitrosylation on each monomer is circled.
(B) Close-up view of stably S-nitrosylated cysteine of β-tubulin reveals a highly solvent accessible sulfur atom. Cysteine 12 is shown as a stick model in atomic coloring as in (A) with its solvent accessible surface shown as pink mesh.
(C) Close-up view of stably S-nitrosylated cysteine of α-tubulin reveals very little solvent accessibility. Cysteine 295 is shown as a stick model in atomic coloring as in (A) with its solvent accessible surface shown as teal mesh.
(D) All cysteines of α-tubulin are listed in order of decreasing solvent accessibility of the thiol, with the stably S-nitrosylated cysteines highlighted in brown and the unstably S-nitrosylated cysteines highlighted in blue.
(E) All cysteines of β-tubulin are listed in order of decreasing solvent accessibility of the thiol, with the stably S-nitrosylated cysteines highlighted in brown and the unstably S-nitrosylated cysteines highlighted in blue.
Supplementary Figure 8. Endogenous stably S-nitrosylated proteins remain S-nitrosylated even after prolonged inhibition of NO synthesis.
sEND.1 cells were cultured, treated, and analyzed as in Figure 5. Cells were treated with either vehicle (media) or 1 mM L-NAME for 5 min, 30 min or 24 h. Cells were harvested, and cell lysates (50 mg protein) were subjected to the biotin switch method. Formerly S-nitrosylated (SNO) proteins were purified on neutravidin agarose, eluted with 2-ME, and analyzed by Western blotting with the indicated antibodies. As in Figure 5, GSH-stable proteins remained S-nitrosylated after 5 min; however, the proteins were also S-nitrosylated at longer time points, up to 24 h. These effects were not associated with changes in the levels of the assayed proteins, as indicated by the loading controls (Total protein).