Abstract
The oxidative stress theory of aging postulates that aging results from the accumulation of molecular damage caused by reactive oxygen species (ROS) generated during normal metabolism. Superoxide dismutases (SODs) counteract this process by detoxifying superoxide. It has previously been shown that elimination of either cytoplasmic or mitochondrial SOD in yeast, flies, and mice results in decreased lifespan. In this experiment, we examine the effect of eliminating each of the five individual sod genes present in Caenorhabditis elegans. In contrast to what is observed in other model organisms, none of the sod deletion mutants shows decreased lifespan compared to wild-type worms, despite a clear increase in sensitivity to paraquat- and juglone-induced oxidative stress. In fact, even mutants lacking combinations of two or three sod genes survive at least as long as wild-type worms. Examination of gene expression in these mutants reveals mild compensatory up-regulation of other sod genes. Interestingly, we find that sod-2 mutants are long-lived despite a significant increase in oxidatively damaged proteins. Testing the effect of sod-2 deletion on known pathways of lifespan extension reveals a clear interaction with genes that affect mitochondrial function: sod-2 deletion markedly increases lifespan in clk-1 worms while clearly decreasing the lifespan of isp-1 worms. Combined with the mitochondrial localization of SOD-2 and the fact that sod-2 mutant worms exhibit phenotypes that are characteristic of long-lived mitochondrial mutants—including slow development, low brood size, and slow defecation—this suggests that deletion of sod-2 extends lifespan through a similar mechanism. This conclusion is supported by our demonstration of decreased oxygen consumption in sod-2 mutant worms. Overall, we show that increased oxidative stress caused by deletion of sod genes does not result in decreased lifespan in C. elegans and that deletion of sod-2 extends worm lifespan by altering mitochondrial function.
Author Summary
In this paper, we examine the oxidative stress theory of aging using C. elegans as a model system. This theory proposes that aging results from the accumulation of molecular damage caused by reactive oxygen species (ROS). To test this theory, we examined the effect of deleting each of the five individual superoxide dismutase (SOD) genes on lifespan and sensitivity to oxidative stress. Since SOD acts to detoxify ROS, the oxidative stress theory predicts that deletion of sod genes should increase oxidative stress and decrease lifespan. However, in contrast to yeast, flies, and mice, where loss of either cytoplasmic or mitochondrial SOD results in decreased lifespan, we find that none of the sod deletion mutants in C. elegans exhibits a shortened lifespan despite increased sensitivity to oxidative stress. Surprisingly, we find that sod-2 mutant worms have extended lifespan and even worms with the primary cytoplasmic, mitochondrial, and extracellular sod genes deleted can live longer than wild-type worms. By examining genetic interactions with other genes known to extend lifespan and by comparing the phenotype of worms lacking sod-2 to that of known long-lived mitochondrial mutants such as clk-1 or isp-1, we provide evidence that the loss of sod-2 extends lifespan through alteration of mitochondrial function.
Introduction
The oxidative stress theory of aging proposes that reactive oxygen species (ROS) generated by normal metabolism cause damage to macromolecules within the cell and that the accumulation of this damage over time leads to cellular dysfunction and eventually organismal death [1]–[3]. The majority of ROS present in the cell is thought to be generated in the mitochondria. In order to counteract this process, cells have a number of defense mechanisms which serve to detoxify ROS. Superoxide dismutase (SOD) is a detoxification enzyme that converts superoxide to hydrogen peroxide, which can subsequently be converted to water [4].
The oxidative stress theory of aging predicts that loss of SOD activity should result in increased sensitivity to oxidative stress, since the organism would be less able to detoxify ROS. This should, in turn, result in a shortened lifespan. This is essentially what is observed in yeast, flies and mice for both cytoplasmic SOD (SOD1, CuZnSOD) and mitochondrial SOD (SOD2, MnSOD). In yeast, knocking out sod1 has been shown to decrease clonal and replicative lifespan [5],[6] and accelerate chronological aging [7],[8]. In flies, knocking out Sod1 decreases lifespan [9]. In mice, targeted inactivation of Sod1 results in high oxidative stress and a 30% decrease in lifespan [10].
For SOD2, yeast knockouts show decreased chronological and replicative lifespan [6]–[8]. Reduction of Sod2 in flies by either RNA interference (RNAi) or genetic deletion results in marked reductions in lifespan [11],[12]. In mice, Sod2 knockouts exhibit high degrees of oxidative stress and neonatal or perinatal lethality [13],[14]. In contrast, loss of extracellular SOD (SOD3, EC-SOD) does not appear to impact lifespan despite an increased sensitivity to hyperoxia [15]. Thus, in support of the oxidative stress theory, the effect of deleting Sod1 or Sod2 in all three model species is increased oxidative stress and decreased lifespan or early lethality in the case of Sod2 mice.
In contrast, lifespan in C. elegans may be relatively unaffected by decreased sod expression. Using an RNAi approach to knockdown either sod-1 or sod-2, Yang et al. showed a mild decrease in lifespan with sod-1 RNAi but no effect of sod-2 RNAi, despite the fact that both knockdowns resulted in increased sensitivity to paraquat and an increase in oxidatively damaged proteins [16]. However, it is possible that if the RNAi did not completely abolish sod expression then the remaining low level of SOD activity is sufficient for normal lifespan.
Here, we examine the effect of eliminating SOD on lifespan and sensitivity to oxidative stress in C. elegans and thereby test the oxidative stress theory of aging. Whereas most organisms have only three SODs (one cytoplasmic, one mitochondrial and one extracellular), C. elegans has five sod genes [17]. sod-1, sod-2 and sod-4 encode the primary cytoplasmic, mitochondrial and extracellular SODs respectively [18]–[22] (equivalent to Sod1, Sod2 and Sod3 in mice). In addition, sod-3 is expressed in the mitochondrial matrix and sod-5 is expressed in the cytoplasm, thereby providing C. elegans with two cytoplasmic and two mitochondrial SODs [18],[23]. By examining C. elegans mutants with deletions in each of the five sod genes, we find that elimination of individual sod genes can increase sensitivity to oxidative stress but does not decrease lifespan. Furthermore, we find that sod-2 mutant worms are long-lived and propose that their lifespan extension is due to an alteration of mitochondrial function.
Results
Elimination of Individual sod Genes Does Not Reduce Lifespan
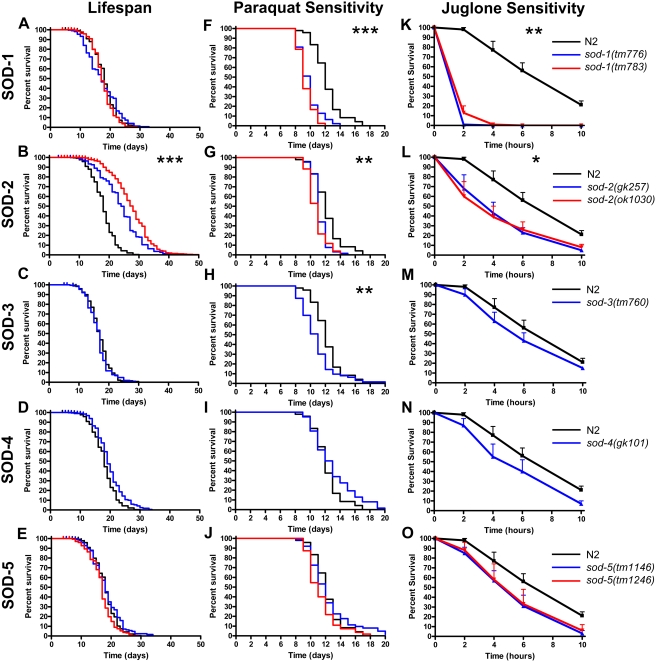
The oxidative stress theory of aging predicts that increasing oxidative stress should result in decreased lifespan. To test this hypothesis, we assessed the lifespan of worms lacking each of the five individual sod genes in C. elegans (the location and size of the mutation for each allele tested are shown in Figure S1). For sod-1, sod-2 and sod-5 we assessed two independent alleles. We found that lifespan was not affected by the disruption of sod-1, sod-3, sod-4 or sod-5 (Figure 1A,C–E; all lifespan data is included in Table S1). This is particularly surprising in the case of sod-1 since SOD-1 accounts for the majority of SOD activity in the cell [23]. In addition we found that deletion of sod-2 resulted in a significant increase in lifespan (Figure 1B). This is also a surprising result given that SOD-2 is the primary SOD present in the mitochondrial matrix and the mitochondria is a major site of superoxide production in the cell.
Figure 1. Deletion of Individual sod Genes Does Not Decrease Lifespan Despite Increasing Sensitivity to Oxidative Stress.
A–E) sod-1(A), sod-3(C), sod-4(D) and sod-5(E) mutant worms live as long as wild-type N2 worms while sod-2 mutant worms (B) live significantly longer than wild-type worms. F–J) sod-1 (F), sod-2 (G) and sod-3 (H) mutant worms showed decreased survival on 4 mM paraquat plates compared to N2 worms, while sod-4 (I) and sod-5 (J) mutant worms survived as long as N2 worms. K–O) Similarly, sod-1 (K) and sod-2 (L) mutant worms showed decreased survival on 240 µM juglone plates compared to N2 worms, while the survival of sod-3 (M), sod-4 (N) and sod-5 (O) mutant worms was not significantly different than N2 worms. Overall, sod deletion increases sensitivity to oxidative stress but does not decrease lifespan. * p<0.05, ** p<0.01, ***p<0.001.
To ensure that the lifespan extension in sod-2 mutant worms resulted from the deletion of the sod-2 gene we generated heteroallelic mutants. This was accomplished by crossing sod-2(gk257) males with sod-2(ok1030) hermaphrodites and following lifespan in the male offspring (since these must be cross progeny) and by crossing sod-2(gk257) males with either dpy-17 (control) or sod-2(ok1030);dpy-17 hermaphrodites and following the lifespan of the resulting non-dumpy hermaphrodite offspring. In both cases, we found that heteroallelic sod-2(gk257)/sod-2(ok1030) mutant worms lived significantly longer than their corresponding controls (Figure S2).
Elimination of Individual sod Genes Results in Increased Sensitivity to Oxidative Stress
Since the loss of individual SODs failed to decrease lifespan, we next sought to determine whether the deletion of individual sod genes had an impact on oxidative stress. As it is currently not possible to accurately quantify the levels of ROS in worms, we used paraquat and juglone to assess sensitivity to oxidative stress as has been described in previous experiments [24]–[26]. Both of these compounds are reduced upon entry into the cell and are thought to induce oxidative stress by generating superoxide from oxygen during their subsequent reoxidation [27],[28].
To assess paraquat sensitivity, we examined the survival of 7 day old adult worms on plates containing 4 mM paraquat. We found that sod-1 mutant worms were very sensitive to paraquat with all of the worms dying within one or two days (Figure 1F). We also found that sod-2 and sod-3 mutant worms were more sensitive to paraquat than wild-type worms although not as sensitive as sod-1 mutant worms (Figure 1G,H). In contrast, sod-4 and sod-5 mutant worms showed similar survival to wild-type worms (Figure 1I,K)
In order to confirm our observation of increased sensitivity to oxidative stress, we assessed sensitivity to juglone. One day old worms were transferred to plates containing 240 µM juglone and survival was monitored for the following 10 hours. As with the paraquat assay, both sod-1 deletion strains showed markedly increased sensitivity to oxidative stress as no worms survived to the 4 hour time point (Figure 1L). The sod-2 deletion strains also showed increased sensitivity to juglone which was not as severe as the sod-1 mutants (Figure 1M). In contrast, deletion of sod-3, sod-4 or sod-5 did not make worms significantly more sensitive to juglone-induced oxidative stress (Figure 1N–P).
Next, we assessed sensitivity to paraquat during development by exposing eggs to plates containing 0.2 mM paraquat and determining the latest developmental stage attained for each strain. While exposure to paraquat slowed development in all strains, including wild-type N2 worms, we found that all of the sod deletion mutants except for sod-2 were able to develop to adulthood (Figure S3). The sod-2 mutants were found to arrest at the L1 stage. Thus, sod-2 mutant worms are the most sensitive of all of the sod deletion mutants to oxidative stress during development while sod-1 mutant worms are the most sensitive in adulthood.
Mild Compensatory Upregulation of sod mRNA in sod Deletion Mutants
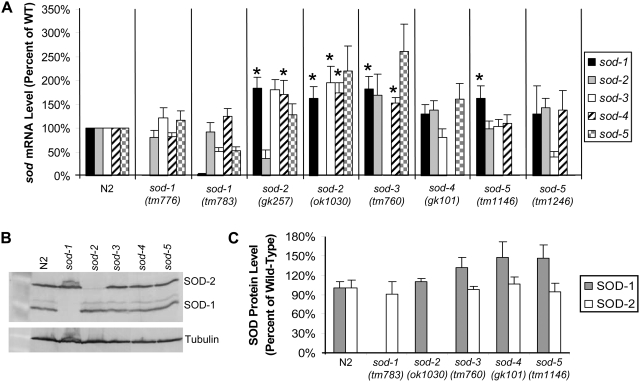
In order to confirm the absence of individual sod expression in the deletion strains, we assessed the levels of each of the five sod mRNAs by qRT-PCR (quantitative real time reverse transcription polymerase chain reaction). Importantly, we also sought to determine whether the deletion of individual sod genes is compensated for by the upregulation of other sod genes. In all strains, the deletion mutation resulted in decreased levels of the corresponding mRNA (in most cases no mRNA was detected) (Figure 2A). In both sod-2 mutant strains, sod-1, sod-3 and sod-4 mRNAs were significantly elevated and there was a trend towards increased expression of sod-5 (Figure 2A). Similarly, sod-3 mutant worms showed increased expression of sod-1 and sod-4 mRNA and a trend towards increased expression of sod-2 and sod-5 (Figure 2A). One of two sod-5 mutants showed significantly increased expression of sod-1 mRNA, while sod-1 and sod-4 mutant worms showed no significant changes in mRNA levels of the other four sod mRNAs (Figure 2A). Overall, we observed some compensatory upregulation of other sod mRNAs in the sod deletion strains but the degree of upregulation was small, generally 2-fold or less. The fact that sod-3 mutant worms showed a similar upregulation of sod mRNA as sod-2 mutant worms suggests that the lifespan extension in the sod-2 mutants does not result from the observed upregulation of sod mRNA.
Figure 2. Deletion of Individual sod Genes Results in Mild Compensatory Upregulation of sod mRNA.
A) sod mRNA levels for each of the five sod genes was examined in the sod deletion mutants by quantitative real-time RT-PCR. While compensatory upregulation was observed in sod-2, sod-3 and sod-5 mutant worms, the magnitude was small, generally 2-fold or less. No sod mRNA upregulation in sod-1 or sod-4 mutant worms. B) Examination of SOD-1 and SOD-2 protein levels by Western blotting reveals no SOD-1 expression in sod-1 mutant worms and no SOD-2 expression in sod-2 mutant worms. C) Quantification of SOD protein levels reveals no significant upregulation of SOD-1 or SOD-2 proteins in any of the sod deletion mutants. * p<0.05.
Since a compensatory increase in SOD expression could also occur at the translational level, we examined the level of SOD-1 and SOD-2 protein in the sod deletion mutants (antibodies to the other SOD proteins are currently not available). We observed no SOD-1 protein in sod-1 deletion mutants or SOD-2 protein in sod-2 deletion mutants (Figure 2B). As with mRNA expression we did not observe a dramatic upregulation of SOD-1 or SOD-2 in any of the sod deletion mutants (Figure 2C).
Increased Sensitivity to Oxidative Stress Does Not Decrease Lifespan in sod-sod Double Mutants
Although the magnitude of compensatory upregulation of other sod genes, when present, was small, it is possible that this could have accounted for the normal or extended lifespan we observed in the sod single deletion mutants. To investigate this possibility, we sought to determine whether elimination of a second sod gene would shorten the lifespan of the sod single deletion mutants. Accordingly, we generated a panel a sod-sod double mutants consisting of all of the double mutants for sod-1 and sod-2, since these are the major contributors to SOD activity in the cytosol and mitochondria respectively, and sod-3; sod-5 (this mutant lacks both of the “extra” sod genes found in C. elegans).
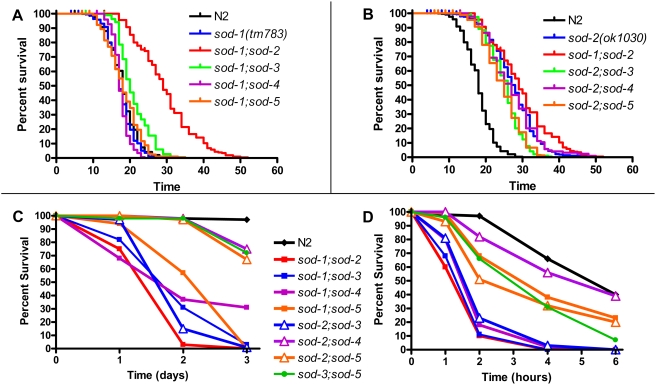
Examining the lifespan of sod-1 double deletion mutants revealed that deletion of sod-3, sod-4 or sod-5 did not shorten the lifespan of sod-1 mutant worms (Figure 3A). In contrast, sod-1;sod-2 mutant worms lived significantly longer than wild-type N2 worms (Figure 3A). Among the sod-2 double deletion mutants, all of the worms maintained the extended lifespan seen in sod-2 single deletion mutants indicating that in no case is the upregulation of another sod gene entirely responsible for the long life observed in sod-2 mutant worms (Figure 3B). Finally, we found that sod-3;sod-5 mutant worms had a similar lifespan to wild-type worms (not shown).
Figure 3. sod-sod Double Deletion Mutants Do Not Exhibit Decreased Lifespan Despite Increased Sensitivity to Oxidative Stress.
A) None of the sod-1 double deletion mutants showed a decreased lifespan compared to wild-type or sod-1 mutant worms. sod-1;sod-2 mutant worms show extended longevity. B) All of the sod-2 double deletion mutants exhibited extended lifespan compared to wild-type worms. C) Despite exhibiting a normal or extended lifespan, all of the sod-1 double deletion mutants as well as sod-2;sod-3 mutant worms were very sensitive to paraquat-induced oxidative stress. D) As with the paraquat assay, sod-1 double deletion mutants and sod-2;sod-3 mutant worms showed the greatest sensitivity to juglone-induced oxidative stress. Overall, sod-sod double deletion mutants show increased sensitivity to oxidative stress but exhibit a normal or extended lifespan.
Next, we examined the sensitivity to oxidative stress among the sod-sod double mutants using both paraquat and juglone. Examining the survival of one day old adult worms on 4 mM paraquat plates, we found that all of the sod-1 double mutants, including the long-lived sod-1;sod-2 mutant worms, had decreased survival compared to N2 worms (Figure 3C). Among the sod-2 double mutants, sod-2;sod-3 mutant worms were hypersensitive to paraquat, while sod-2;sod-4 and sod-2;sod-5 mutant worms appeared to be only mildly more sensitive than N2 worms (Figure 3C).
A similar pattern of sensitivity to oxidative stress was observed on juglone plates. All of the sod-1 double mutants as well as sod-2;sod-3 mutant worms were more sensitive to juglone than N2 worms (Figure 3D). There was also a trend towards decreased survival in the remaining double mutant strains (Figure 3D). Overall, the sod-sod double mutants showed increased sensitivity to oxidative stress but normal or extended longevity. Thus, we did not observe any correlation between sensitivity to oxidative stress and lifespan.
We also examined sod mRNA expression levels in the sod-sod double mutant worms (Figure S4). As with the sod single deletion mutants, we observed some compensatory upregulation of other sod genes but the magnitude of this increase was small and failed to rescue the observed increase in sensitivity to oxidative stress.
Mutants With Deletions in Three sod Genes Exhibit Normal or Extended Lifespan
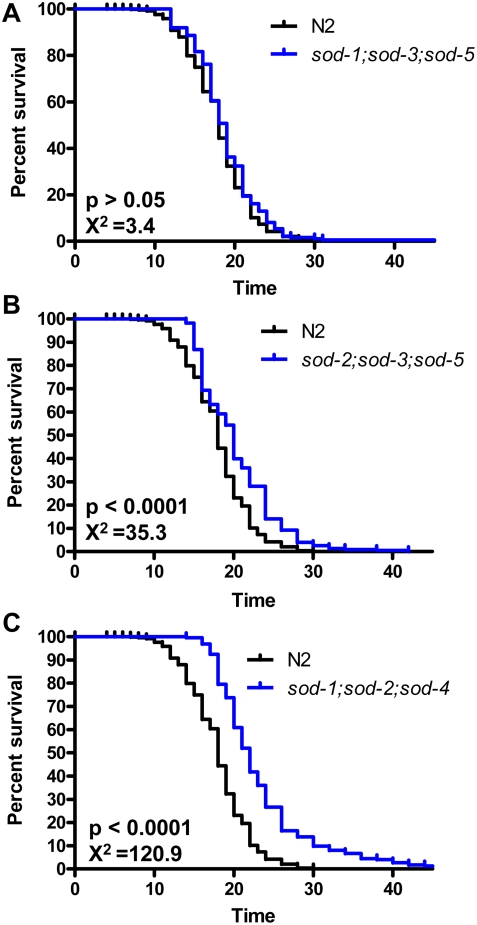
Based on our finding that even the elimination of two sod genes together does not shorten the lifespan of C. elegans, we assayed lifespan in a selection of sod triple mutants. To eliminate the possibility that the reason why sod-1 and sod-2 mutants of C. elegans do not show decreased lifespan is because C. elegans has duplicate SODs in the cytoplasm and mitochondria, we generated sod-1;sod-3;sod-5 and sod-2;sod-3;sod-5 triple mutants to model sod-1 and sod-2 knockouts in species with only three sod genes. We also generated sod-1;sod-2;sod-4 worms which lack the primary cytoplasmic, mitochondrial and extracellular sod genes. Examination of worm lifespan revealed that sod-1;sod-3;sod-5 mutant worms live as long as wild-type worms while both sod-2;sod-3;sod-5 and sod-1;sod-2;sod-4 mutant worms live significantly longer than wild-type (Figure 4A–C). This clearly indicates that the normal lifespan observed in sod-1 worms does not result from the overlapping expression of sod-3 in the mitochondria or sod-5 in the cytoplasm. Since sod-2;sod-3;sod-5 triple mutant worms do not survive as long as sod-2 single deletion mutants, it is possible that the mild upregulation of sod-3 and sod-5 may contribute to the increased lifespan of sod-2 mutant worms. However, the fact that similar upregulation of sod mRNA in sod-3 mutant worms does not result in extension of lifespan and that upregulation of sod-3 and sod-5 in sod-2 mutant worms is insufficient to prevent increased levels of oxidative damage (see below) suggests that other mechanisms are involved in the long life of sod-2 mutant worms.
Figure 4. Worms Lacking Combinations of Three sod Genes Show Normal or Extended Lifespan.
To model sod-1 and sod-2 knockouts in species which have only three sod genes, sod-1;sod-3;sod-5 (A) and sod-2;sod-3;sod-5 (B) mutant worms were generated respectively. sod-1;sod-2;sod-4 (C) mutant worms which lack the primary cytoplasmic, mitochondrial and extracellular sod genes were also generated. Surprisingly, none of the sod triple deletion mutants showed decreased lifespan compared to wild-type worms and the two triple mutants bearing the sod-2 allele lived significantly longer than wild-type. Thus, C. elegans is able to compensate for the loss of multiple sod genes to achieve a normal or extended lifespan.
Interaction of sod-2 Deletion With Lifespan Extending Mechanisms in Known Genetic Pathways
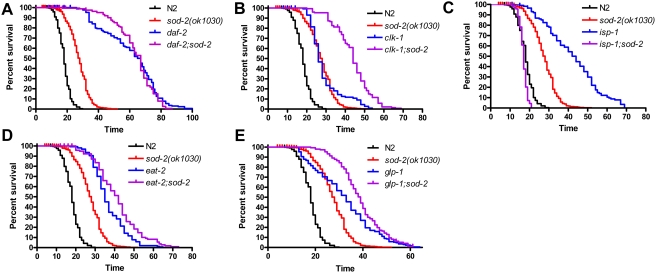
In order to investigate possible mechanisms of lifespan extension in sod-2 mutant worms, we generated double mutants with genes known to extend lifespan which are representative of different lifespan extending mechanisms including daf-2 (insulin/IGF signaling)[29], clk-1 (decreased mitochondrial function)[30], isp-1 (decreased mitochondrial function)[25], eat-2 (dietary restriction)[31] and glp-1 (germ-line ablation)[32]. Deletion of sod-2 did not extend the lifespan of daf-2 worms (Figure 5A). clk-1 worms showed a marked extension of lifespan when sod-2 was deleted (Figure 5B). In contrast, sod-2 deletion greatly shortened the lifespan of isp-1 worms such that isp-1;sod-2 worms had a shorter lifespan than wild-type N2 worms (Figure 5C). eat-2;sod-2 mutant worms showed a small increase in lifespan compared to eat-2 worms (Figure 5D). Finally, deletion of sod-2 in glp-1 worms resulted in a modest increase of mean but not maximum lifespan (Figure 5E). Overall, we found that sod-2 deletion had the greatest impact on the lifespan of mutants which exhibit extended longevity as a result of alterations in mitochondrial function.
Figure 5. sod-2 Deletion Alters the Lifespan of Extended Longevity Mitochondrial Mutants.
To gain insight into the mechanism of lifespan extension in sod-2 mutant worms, we examined the effect of sod-2 deletion on other genes known to increase longevity. A) sod-2 deletion did not affect lifespan in daf-2 worms (insulin/IGF1 signaling). B) sod-2 deletion markedly increases lifespan in clk-1 worms (decreased mitochondrial function). C) sod-2 deletion shortens lifespan in isp-1 worms (decreased mitochondrial function) such that isp-1;sod-2 worms do not survive as long as wild-type worms. D) sod-2 deletion moderately extends mean and maximum lifespan in eat-2 worms (dietary restriction). E) sod-2 deletion extends mean but not maximum lifespan of glp-1 worms (germ-line ablation). Thus, sod-2 deletion has the greatest impact on the lifespan of clk-1 and isp-1 worms, which extend lifespan by decreasing mitochondrial function.
sod-2 Mutant Worms Exhibit Phenotypes Characteristic of Extended Longevity Mitochondrial Mutants
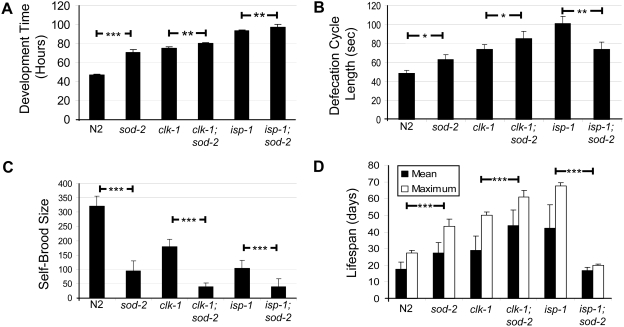
Based on our finding that sod-2 deletion interacts with long-lived mutants with altered mitochondrial function and the fact that SOD-2 is localized to the mitochondria, we hypothesized that deletion of sod-2 extends lifespan by decreasing mitochondrial function. In C. elegans a number of genes have been identified that affect mitochondrial function and at the same time increase lifespan [25],[30],[33],[34]. Although these genes do not necessarily interact and the exact mechanism of lifespan extension is unclear, these mutants are generally grouped together since it is believed that the alteration of mitochondrial function is the key to their long life. In addition to impaired mitochondrial function and extended longevity, these mutants, sometimes referred to as Mit mutants, are characterized by slow development, slow defecation rate and decreased brood size. Accordingly, we quantified the development, brood size and defecation rate of sod-2 mutant worms and compared this with two prototypes of this class of mutants - clk-1 and isp-1 worms [25],[30],[35]. We also examined clk-1;sod-2 and isp-1;sod-2 double mutants to determine if the loss of sod-2 enhanced the phenotypes observed in clk-1 and isp-1 worms.
Examination of post-embryonic development (PED) revealed that sod-2, clk-1 and isp-1 worms all developed slower than wild-type worms (Figure 6A). On a clk-1 and isp-1 background, sod-2 deletion resulted in further increase in PED time (Figure 6A). Examination of defecation cycle length revealed a slow rate of defecation in sod-2, clk-1 and isp-1 worms compared to wild-type N2 worms (Figure 6B). Deletion of sod-2 had opposite effects on defecation cycle length in clk-1 and isp-1 worms. sod-2 deletion further lengthened the defecation cycle of clk-1 worms but shortened the defecation cycle length of isp-1 worms (Figure 6B). Self-brood size was decreased in sod-2, clk-1 and isp-1 worms and deletion of sod-2 further decreased the brood size in clk-1 and isp-1 worms (Figure 6C). Finally, a comparison of lifespan between these strains revealed that isp-1;sod-2 worms were short-lived, sod-2 and clk-1 worms were long-lived and clk-1;sod-2 and isp-1 worms were very long-lived (Figure 6D). Clearly, sod-2 deletion mutants exhibit the key characteristics of extended longevity mitochondrial mutants and modulate these phenotypes in clk-1 and isp-1 worms.
Figure 6. sod-2 Mutant Worms Exhibit the Hallmark Features of Extended Longevity Mitochondrial Mutants in C. elegans.
Similar to extended longevity mitochondrial mutants such as clk-1 and isp-1, sod-2 mutant worms show slow post-embryonic development (A), slow defecation rate (B), decreased self-brood size (C) and long life (D). In clk-1 worms, the deletion of sod-2 slows development, slows defecation rate, decreases brood size and increases lifespan. In isp-1 worms, sod-2 deletion slows development, increases defecation rate, decreases brood size and decreases lifespan. *p<0.05, **p<0.01, ***p<0.001.
Oxygen Consumption Is Decreased in sod-2 Mutant Worms
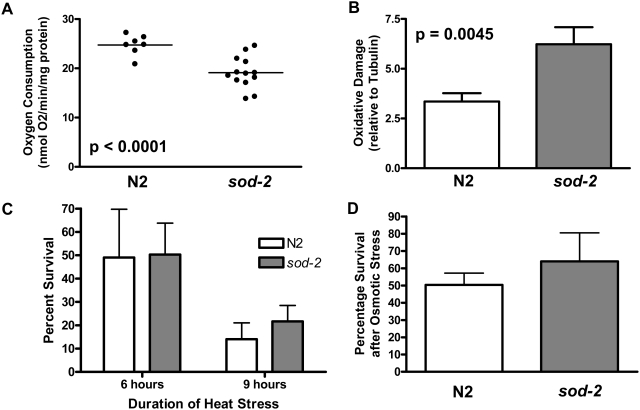
The phenotypic similarity of sod-2 mutant worms to extended longevity mitochondrial mutants as well as the ability of sod-2 deletion to alter these characteristic phenotypes in clk-1 and isp-1 worms suggests that sod-2 extends lifespan through a similar mechanism. Based on this hypothesis, we would predict that mitochondrial function would be altered in sod-2 mutant worms. To assess this, we measured whole worm oxygen consumption, which has previously been shown to be decreased in both clk-1 and isp-1 worms [16],[25]. We found that oxygen consumption in one day old adult worms was significantly decreased in sod-2 mutant worms compared to wild-type worms (Figure 7A).
Figure 7. sod-2 Mutant Worms Show Decreased Oxygen Consumption and Increased Oxidative Damage But Normal Tolerance to Heat and Osmotic Stress.
A) In order to assess mitochondrial function in sod-2 mutant worms, whole worm oxygen consumption was measured and compared to wild-type N2 controls. Oxygen consumption was significantly decreased in sod-2 mutant worms compared to wild-type worms. B) Examination of carbonylated proteins by Oxyblot reveals increased oxidative damage in sod-2 mutant worms compared to wild-type worms (N = 10 N2, 8 sod-2). Nonetheless, sod-2 mutant worms survive both heat stress (C) and osmotic stress (D) as well as wild-type worms (stress results represent three independent trials).
Oxidative Damage Is Increased in sod-2 Mutant Worms
We have previously reported that both clk-1 and isp-1 worms exhibit decreased levels of oxidatively damaged proteins [16]. Here, we find that sod-2 worms are hypersensitive to oxidative stress, suggesting that there may be an increase in oxidatively damaged proteins in these worms. To determine the level of oxidative damage in sod-2 mutant worms, we quantified carbonylated proteins in sod-2 mutant worms and wild-type worms. We found that sod-2 mutant worms had significantly more oxidative damage than wild-type worms (Figure 7B).
sod-2 Mutant Worms Are Not Hypersensitive to Heat or Osmotic Stress
In order to determine whether the sensitivity of sod-2 mutant worms to paraquat and juglone was the result of a specific sensitivity to oxidative stress or a sign of general weakness, we examined the ability of sod-2 worms to withstand heat stress [36] or osmotic stress [37]. After exposure to 35 degree Celsius heat stress for a period of 6 hours or 9 hours, we found that sod-2 mutant worms survived as well as wild-type worms (Figure 7C). Similarly, exposing sod-2 mutant worms to osmotic stress on 500 mM NaCl NGM plates for 20 hours revealed that sod-2 mutant worms survive osmotic stress as well as wild-type N2 worms (Figure 7D). Combined these results suggest that the sensitivity of sod-2 mutant worms to oxidative stress is a specific sensitivity resulting from their decreased ability to detoxifying ROS.
Discussion
In this paper we test the oxidative stress theory of aging in C. elegans. Examination of lifespan and sensitivity to oxidative stress in mutants with deletions in one, two or three sod genes reveals normal or extended lifespan in worms with markedly increased sensitivity to oxidative stress. In contrast to observations in other species, deletion of sod-2 results in long life despite increased levels of oxidative damage. The lifespan extending mechanism of sod-2 interacts most strongly with that of clk-1 and isp-1 mutations, which extend lifespan by decreasing mitochondrial function. The phenotypic similarity of sod-2 mutant worms with these mitochondrial mutants, the mitochondrial localization of SOD-2 and the decreased oxygen consumption in sod-2 mutant worms suggest that sod-2 deletion extends lifespan through alterations in mitochondrial function.
Oxidative Stress Theory of Aging
Since its origins in 1956, the oxidative stress theory of aging has been extensively tested in multiple organisms both by observing variations in natural populations and through genetic intervention [1]–[3]. Thus far there have been many experiments that support this theory, but also experiments which challenge the notion that molecular damage from ROS leads to aging (reviewed in [38]).
In this paper, we find that the effect of sod deletion on lifespan in C. elegans is unique from other organisms. In concordance with the oxidative stress theory of aging, yeast, flies and mice lacking either cytoplasmic or mitochondrial SOD show either decreased lifespan or lethality (in the case of Sod2 knockout mice) [5], [7], [9], [10], [12]–[14] while mice lacking extracellular SOD live as long as wild-type mice [15]. Here, we demonstrate that none of the sod deletion mutants in C. elegans show decreased lifespan. One possible explanation for this discrepancy is the fact that C. elegans has five sod genes, rather than three, including two cytoplasmic SODs and two mitochondrial SODs. To eliminate this explanation, we show that the lifespan of sod-1;sod-3;sod-5 and sod-2;sod-3;sod-5 triple mutants, which model sod-1 and sod-2 deficient organisms in species with only three sod genes, is not decreased.
Another possible explanation for why C. elegans sod mutants exhibit a normal lifespan would be compensatory upregulation of other sod genes. In support of this hypothesis, we observed sod mRNA upregulation in sod-2 and sod-3 mutant worms as well as one of two sod-5 mutants. However, the magnitude of this upregulation was small (2-fold or less) and we observed no significant sod upregulation in sod-1 or sod-4 mutant worms which also exhibit a normal lifespan. These results are in general agreement with studies of Sod knockouts in flies and mice where either no change in other SOD activity is reported [12], [15], [39]–[41] or changes with magnitudes of less than 50% [10],[14],[42].
Although the compensatory upregulation of other sod genes was small in magnitude and not present in all sod deletion mutants, it is possible that this small increase contributed to the normal or extended lifespans observed in these strains. It is also possible that changes in SOD protein levels or activity contributed to the preservation of lifespan in these strains. To investigate these possibilities, we used the genetic approach of generating sod double and triple mutants. The loss of an additional sod gene did not decrease the lifespan in sod-1 mutant worms, nor did it revert the lifespan of sod-2 mutant worms to wild-type. This indicates that the increased expression or activity of any single other sod gene is not responsible for the normal lifespan observed in sod-1 mutant worms or extended lifespan observed in sod-2 mutant worms. Similarly, all of the sod triple mutants were able to live at least as long as wild-type worms. The lack of lifespan shortening in worms with multiple sod genes deleted is in concordance with studies in mice where the loss of extracellular SOD [43] or the loss of glutathione peroxidase (another ROS detoxifying enzyme) and one copy of Sod2 [38] does not further decrease lifespan in Sod1 knockout mice. The lack of additive effects between different compartments can be explained by the inability of superoxide to cross biological membranes [44],[45]. A compartment specific effect of genes involved in ROS detoxification on lifespan has also been observed in C. elegans with genes encoding catalase, where deletion of peroxisomal catalase (ctl-2) results in decreased lifespan while deletion of cytoplasmic catalase (ctl-1) has no effect on lifespan [46].
A comparison of our results for lifespan and sensitivity to oxidative stress reveals no correlation. None of the sod single or double mutants exhibited a shortened lifespan despite many strains showing markedly increased sensitivity to oxidative stress. Most strikingly, sod-1;sod-2 mutants show the highest sensitivity to oxidative stress in combination with the longest lifespan. Similarly, our laboratory has recently shown that decreasing levels of sod-1 or sod-2 by RNAi increases paraquat sensitivity and oxidative damage to proteins in N2 as well as in multiple long-lived strains (daf-2, clk-1 and isp-1) yet does not decrease lifespan in these strains [16]. Initial experiments examining sensitivity to oxidative stress in long-lived worms indicated that increased resistance to oxidative stress occurs with increased lifespan [24],[47],[48]. It was also found that when longevity was selected for in flies, increased longevity was accompanied by resistance to oxidative stress [49]. More recently, in the reverse experiment examining the lifespan of worms that were resistant to paraquat-induced oxidative stress, it was found that only 84 of 608 RNAi treatments that increased stress resistance also increased lifespan [50]. Similarly, examination of the relationship between paraquat resistance and lifespan in 138 lines of flies revealed only a weak positive correlation [51]. In mice, Sod1 knockouts show increased sensitivity to oxidative stress and decreased lifespan [10],[39], mice heterozygous for the targeted inactivation of Sod2 showed a normal lifespan despite increased oxidative damage [52], while Sod3 knockout mice show increased sensitivity to oxidative stress and a normal lifespan [15]. Combined with our results, it appears that the correlation between sensitivity to oxidative stress and lifespan is weak at best.
Extended Longevity Resulting From sod-2 Deletion Is Unique to C. elegans
SOD2 is the primary, and normally sole, SOD present in the mitochondrial matrix. Since the mitochondria is a major source of superoxide within the cell and superoxide is not able to pass through membranes[44],[45], SOD2 may be the most critical SOD within the cell for decreasing superoxide-induced damage. This conclusion is supported by findings that decreasing or eliminating SOD2 expression affects lifespan more than elimination of SOD1 or the extracellular SOD. In flies, eliminating SOD1 reduces lifespan from about 60 days to 11.8 days [9] while eliminating SOD2 decreases lifespan to less than 1 day [12]. Similarly, Sod1 knockout mice show a 30% decrease in lifespan living an average of 20.8 months [10], while Sod2 knockout mice exhibit either peritnatal or neonatal lethality [13],[14].
In contrast to what is observed in other species, we find that sod-2 deletion in C. elegans results in extended lifespan. While these worms show small but significant increases in sod-1, sod-3 and sod-4 mRNA expression, deleting sod-1, sod-3 or sod-4 in sod-2 mutant worms does not revert their lifespan to wild-type suggesting that this upregulation of sod expression is not responsible for the lifespan increase in sod-2 mutant worms. Our observation of decreased lifespan in sod-2;sod-3;sod-5 mutant worms compared to sod-2 mutant worms suggests the possibility that upregulation of sod-3 and sod-5 partially contributes to the extended lifespan observed in sod-2 mutant worms. However, the fact that we observe similar upregulation of other sod genes in sod-3 mutant worms without the lifespan extension supports the conclusion that the mild compensatory upregulation of other sod genes is not responsible for the long life of sod-2 mutants. Furthermore, the fact that sod-2 mutant worms show increased oxidative damage indicates that the upregulation of sod-3 and sod-5 is not sufficient to reduce mitochondrial oxidative stress in sod-2 mutant worms.
Increased Longevity through Alteration of Mitochondrial Function
To gain insight into the mechanism of lifespan extension in sod-2 mutant worms, we examined the effect of sod-2 deletion on other mutants with extended longevity. sod-2 deletion did not extend lifespan in daf-2 worms, which extend lifespan through the insulin-IGF1 pathway [29] but did result in a modest extension of lifespan in eat-2 worms, which extend lifespan through caloric restriction [31] and glp-1 worms, which extend lifespan through the germ-line ablation [32]. In clk-1 worms, which extend lifespan by decreasing mitochondrial function [30],[53], deletion of sod-2 resulted in a 15 day increase in mean lifespan. In contrast, sod-2 deletion decreased the lifespan of isp-1 worms by 25 days despite the fact that isp-1 also extends lifespan through via a decrease in mitochondrial function [25].
The clear interaction of sod-2 deletion with mutants that extend lifespan through alterations in mitochondrial function suggested the possibility that sod-2 also increases longevity through a similar mechanism. In C. elegans a number of mutants have been identified by genetic deletion or RNAi that affect mitochondrial function and extend lifespan [25],[30],[33],[34],[54]. In addition to decreased mitochondrial function and extended lifespan, the group of mitochondrial mutants also share a number of characteristic phenotypes such as slow rate of development, slow rate of defecation and decreased brood size [25],[35],[55]. Phenotypic characterization of sod-2 mutant worms demonstrates that sod-2 mutants exhibit all of the phenotypes of the extended longevity mitochondrial mutants including slow development, slow defecation rate, decreased brood size, decreased mitochondrial function and increased lifespan.
While we have previously shown that clk-1 and isp-1 worms have decreased levels of oxidatively damaged proteins [16], here we find that sod-2 mutant worms exhibit an increase in oxidative damage. The fact that all three strains have a long lifespan suggests that both high and low levels of oxidative damage are compatible with long life. Moreover, the fact that oxidative damage in clk-1 and isp-1 worms can be increased to a level that is significantly greater than wild-type worms without diminishing the long life of these two strains suggests that the low levels of oxidative damage in clk-1 and isp-1 worms does not contribute to their extended longevity [16].
In addition to those genes which impair mitochondrial function and increase lifespan, there are at least two mutations, mev-1 [56] and gas-1 [57], which decrease mitochondrial function and decrease lifespan. While it is currently uncertain why these mutations have a different effect on lifespan compared to the extended longevity mitochondrial mutants, it appears that there are at least two ways in which decreasing mitochondrial function can lead to decreased lifespan. First, the severity of the mutation can be incompatible with long life. This has recently been demonstrated using an RNAi dilution series against genes involved in mitochondrial function [55]. These authors find that RNAi against the same gene can increase lifespan at low concentration (i.e. mildly inhibited mitochondrial function) and decrease lifespan at high concentration (i.e. severely inhibited mitochondrial function). In our work, we hypothesize that isp-1;sod-2 worms are another example whereby the overall mitochondrial function in the double mutant worm is severely affected leading to a shortened lifespan. Second, the decreased lifespan can be the result of the way in which mitochondrial function is altered. For example, RNAi targeted against any of the four subunits of electron transport chain complex II results in decreased lifespan [58] while RNAi targeted against proteins in any other complex of the electron transport chain can result in increased lifespan [33]. Furthermore, recent work examining the effect of an RNAi dilution series against mev-1 indicates that it is not the severity of this mutation that prevents it from extending lifespan, since mev-1 RNAi failed to increase the lifespan of wild-type worms at any concentration [55].
Modulation of clk-1 and isp-1 Phenotypes by Deletion of sod-2
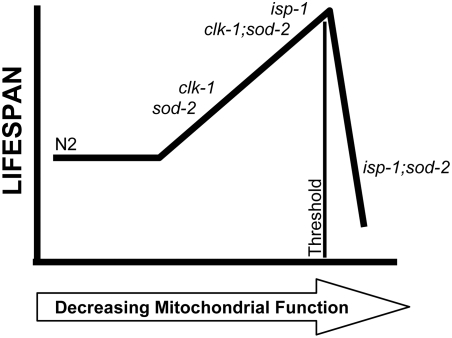
Examination of clk-1;sod-2 double mutants shows that sod-2 deletion enhances all of the mitochondrial mutant phenotypes of clk-1 worms. However, a different pattern is observed with isp-1 worms, where sod-2 deletion further slows development and decreases brood size but quickens defecation towards wild-type and decreases lifespan below wild-type N2 worms. We propose that the reason for the different effects of sod-2 deletion on clk-1 and isp-1 worms results from differences in the initial degree of mitochondrial function. Based on previous measurements of oxygen consumption, respiration is only mildly impaired in clk-1 worms [16],[53] while it is more than 50% reduced in isp-1 worms [25]. By comparing the other phenotypes of N2, clk-1 and isp-1 worms it can be seen that as mitochondrial function decreases, development time gets longer, defecation gets slower, self brood size decreases and lifespan increases. However, according to mitochondrial threshold theory, once a certain threshold of mitochondrial dysfunction is reached, the cell is no longer able to compensate and lifespan decreases [59]. This theory was recently explored in C. elegans through the use of an RNAi dilution series to show that progressively decreasing mitochondrial function resulted in increased lifespan only until a certain threshold after which lifespan began to decrease [55]. Based on these findings, we propose a model in which the shortened lifespan that we observe in isp-1;sod-2 worms results from the sod-2 deletion pushing mitochondrial function past the threshold at which the organism is able to compensate for the lost mitochondrial function and accordingly lifespan is decreased (Figure 8). Similarly, we propose that the increased lifespan in clk-1;sod-2 worms results from the sod-2 deletion reducing the mitochondrial function to a level similar to isp-1 worms. In line with our demonstration of decreased oxygen consumption in sod-2 mutant worms, deletion of sod-2 has also been shown to decrease mitochondrial function in mouse models [60]–[62].
Figure 8. Proposed Model Relating Mitochondrial Function and Lifespan in sod-2 Mutant Worms and sod-2 Double Mutants.
N2 exhibits normal mitochondrial function and a normal lifespan. In sod-2 and clk-1 worms, mitochondrial function is decreased leading to compensatory changes which increase lifespan. Deleting sod-2 in clk-1 worms further decreases mitochondrial function towards that of isp-1 worms with a coincident increase in lifespan. Deleting sod-2 in isp-1 worms decreases mitochondrial function past a threshold after which the organism is no longer able to compensate for the degree of mitochondrial impairment and lifespan decreases.
Although the first of the extended longevity mitochondrial mutants was identified more than a decade ago [30],[35],[54], the precise mechanism by which these mutants extend lifespan is still unresolved. Nonetheless, a number of potential mechanisms have been suggested [63]. Future studies will need to more precisely define how mitochondrial mutants, such as sod-2, extend lifespan and to determine how C. elegans is able to cope with reduced SOD activity. It will be particularly interesting to examine the interaction between SODs and other proteins involved in ROS detoxification (catalases, peroxidases, thioredoxins, peroxiredoxins) in order to obtain a more complete understanding of the relationship between oxidative stress and lifespan.
Materials and Methods
Strains
The following strains were used in these experiments: N2 (wild-type), sod-1(tm776), sod-1(tm783), sod-2(gk257), sod-2(ok1030), sod-3(tm760), sod-4(gk101), sod-5(tm1146), sod-5(tm1246), clk-1(qm30), eat-2(ad1116), daf-2(e1370), isp-1(qm150), glp-1(e2141). Strains obtained from external sources were outcrossed with our N2 worms for 5-10 generations. For these experiments the following double and triple mutant strains were generated: sod-1(tm783);sod-2(ok1030), sod-1(tm783);sod-3(tm760), sod-1(tm783);sod-4(gk101), sod-1(tm783);sod-5(tm1246), sod-2(ok1030);sod-3(tm760), sod-2(ok1030);sod-4(gk101), sod-2(ok1030);sod-5(tm1246), sod-3(tm760);sod-5(tm1246), clk-1(qm30);sod-2(ok1030), eat-2(ad1116);sod-2(ok1030), daf-2(e1370);sod-2(ok1030), isp-1(qm150);sod-2(ok1030), glp-1(e2141);sod-2(ok1030), sod-1(tm783);sod-2(ok1030);sod-4(gk101), sod-1(tm783);sod-3(tm760);sod-5(tm1246), and sod-2(ok1030);sod-3(tm760);sod-5(tm1246). All of the sod deletions were confirmed by PCR. All strains were maintained at 20°C.
Lifespan Analysis
Lifespan studies were completed at 20°C with a minimum of 3 independent trials and an initial number of 80 worms per strain per trial. Initial lifespan assays for sod single deletion mutants, sod-sod double deletion mutants and sod-2 double mutants with genes in known pathways of lifespan extension were completed on normal NGM plates. As some sod double mutant strains bagged extensively subsequent lifespan studies were completed on plates containing 100 µM FUDR (Sigma). Results obtained on NGM plates were all repeated and confirmed on FUDR plates. Survival plots shown represent pooled data from multiple trials on FUDR plates. For glp-1 and glp-1;sod-2 lifespan analyses worms were grown at 25°C and then transferred to 20°C at adulthood.
Paraquat and Juglone Sensitivity
Paraquat and juglone sensitivity assays were completed in triplicate with 30–40 worms per strain per trial at 20°C. To assay paraquat sensitivity, 7 day old adult worms were transferred to plates containing 4 mM paraquat (Sigma) and survival was monitored daily. Initially, paraquat assays were performed on 1 day old adult worms. However, by day 3 of adulthood, paraquat causes most of the worms to have internal hatching of progeny (bagging) such that more worms die of this than of paraquat toxicity.
Juglone sensitivity was assessed in 1 day old adult worms on plates containing 240 µM juglone (Sigma). For this assay, plates were made fresh on the day of the assay as the toxicity of juglone decreases rapidly over time. Survival was monitored for 6 to 10 hours. To assess the ability of worms to develop under oxidative stress, a minimum of 40 eggs were placed on plates containing 0.2 mM paraquat and seeded with OP50 bacteria.
Quantitative Real-Time RT-PCR
RNA was isolated from young adult worms using TRIZOL reagent (Invitrogen). Subsequently, 1 µg of RNA was converted to cDNA using the Quantitect Reverse Transcription kit (Qiagen). 1 µl of the resulting cDNA preparation was used for quantitative real-time PCR using the Quantitect SYBR Green PCR kit and a Biorad iCycler RT-PCR machine. Primer sequences for sod mRNAs were previously validated [64]. A combination of three control primer sets (cdc-42, pmp-3 and Y45F10D.4) were used as has been previously described [65]. Results represent the average of three independent biological samples, each of which was amplified in triplicate.
Post-Embryonic Development
Eggs were collected and allowed to hatch over a period of 3 hours. After 3 hours, L1 worms were transferred to a new plate and monitored for development to an adult worm. Results are the average of at least three independent trials with 20 worms per trial.
Defecation
Defecation cycle length in young adult worms was measured as the average time between consecutive pBoc contractions. Results represent a minimum of 3 trials with 10 worms per trial.
Self-Brood Size
To determine the average number of progeny produced by each strain, L4 worms were placed on individual NGM plates. Worms were transferred daily until egg laying ceased and the total number of live progeny produced was counted.
Oxygen Consumption
Gravid adult worms were collected from five to ten 100 mm NGM plates and bleached to recover eggs. Eggs were allowed to hatch in M9 buffer over a period of 5 days before L1 worms were transferred to NGM plates. At adulthood worms were collected in M9 buffer, washed free of bacteria and oxygen consumption was measured using a Clark electrode for a 10 minute period. Subsequently worms were pelleted and frozen for protein quantification. Proteins were quantified using a bicinchonic acid protein assay kit (Thermo Scientific) according to the manufacturer's protocol.
Western Blotting and Detection of Carbonylated Proteins
Western blotting for SOD proteins was completed as described previously [16]. Antibody dilutions were as follows: SOD-1 (1∶1000), SOD-2 (1∶1000), tubulin (1∶10,000). Levels of protein were compared in three independent samples of one day old adult worms. Oxidative damage was assessed using an Oxyblot assay kit (Millipore) to detect carbonylated proteins as previously described [16]. In this assay carbonyl groups are derivatized to 2,4-dinitrophenylhydrazone (DNP-hydrazone) which can then be detected by western blotting with a DNP specific antibody. The Oxyblot assay was completed according to the manufacturer's protocol using 10 samples of N2 worms and 8 samples of sod-2 mutant worms (Millipore). 9 µg of protein lysate was loaded in each lane. Quantification of carbonylated proteins was achieved by taking the ratio of DNP staining to tubulin staining.
Heat Stress and Osmotic Stress
Heat stress experiments were based on previously developed protocols [36]. Briefly, young adult worms on NGM plates were incubated at 35 degrees Celsius for a period of 6 or 9 hours. Worms were then transferred to a 20 degree Celsius incubator. Two days later the percentage of worms surviving was determined. Osmotic stress experiments were also done according to previously developed protocols [37]. Young adult worms were transferred to NGM plates containing 500 mM NaCl. After 20 hours, worms were washed off salt plates in M9 buffer containing 300 mM NaCl and transferred to normal NGM plates. After one day of recovery, the percentage of worms surviving was determined. Results for both stress assays are the average of three independent trials.
Statistical Analysis
Survival plots were compared using the log-rank test. The maximum lifespan of a given strain was measured as the average of the lifespan of the ten longest living worms. A student's t-test was used to compare maximum lifespan between strains. Significance between strains for paraquat and juglone sensitivity assays were assessed by ANOVA. Oxygen consumption results were compared by student's t-test. Error bars show standard deviation.
Supporting Information
Location of mutations in sod genes.
(0.02 MB PDF)
Heteroallelic sod-2 mutant worms show extended lifespan.
(0.02 MB PDF)
sod-2 mutant worms are sensitive to paraquat during development.
(0.02 MB PDF)
Mild compensatory upregulation of other sod mRNAs in sod-sod double deletion mutants.
(0.02 MB PDF)
Summary of mean and maximum lifespan.
(0.01 MB PDF)
Acknowledgments
We would like to thank Jingjing Li and Darius Camp for their initial observations on the sod deletion strains. We would like to thank the C. elegans knockout consortium, the National BioResource Project of Japan (Mitani laboratory) and the Caenorhabditis Genetics Center for providing strains used in this research.
Footnotes
The authors have declared that no competing interests exist.
JMVR is supported by the Canadian Institutes of Health Research, the Hereditary Disease Foundation and the McGill Tomlinson Fellowships. SH is supported by a grant from the Canadian Institutes of Health Research and McGill University, and holds the Campbell Chair of Developmental Biology and the Strathcona Chair of Zoology.
References
- 1.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 2.Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 3.Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 5.Wawryn J, Krzepilko A, Myszka A, Bilinski T. Deficiency in superoxide dismutases shortens life span of yeast cells. Acta Biochim Pol. 1999;46:249–253. [PubMed] [Google Scholar]
- 6.Unlu ES, Koc A. Effects of deleting mitochondrial antioxidant genes on life span. Ann N Y Acad Sci. 2007;1100:505–509. doi: 10.1196/annals.1395.055. [DOI] [PubMed] [Google Scholar]
- 7.Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 8.Longo VD, Liou LL, Valentine JS, Gralla EB. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch Biochem Biophys. 1999;365:131–142. doi: 10.1006/abbi.1999.1158. [DOI] [PubMed] [Google Scholar]
- 9.Phillips JP, Campbell SD, Michaud D, Charbonneau M, Hilliker AJ. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc Natl Acad Sci U S A. 1989;86:2761–2765. doi: 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, et al. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 11.Kirby K, Hu J, Hilliker AJ, Phillips JP. RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc Natl Acad Sci U S A. 2002;99:16162–16167. doi: 10.1073/pnas.252342899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duttaroy A, Paul A, Kundu M, Belton A. A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics. 2003;165:2295–2299. doi: 10.1093/genetics/165.4.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 14.Lebovitz RM, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, et al. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci U S A. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlsson LM, Jonsson J, Edlund T, Marklund SL. Mice lacking extracellular superoxide dismutase are more sensitive to hyperoxia. Proc Natl Acad Sci U S A. 1995;92:6264–6268. doi: 10.1073/pnas.92.14.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W, Li J, Hekimi S. A Measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–2074. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landis GN, Tower J. Superoxide dismutase evolution and life span regulation. Mech Ageing Dev. 2005;126:365–379. doi: 10.1016/j.mad.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Hunter T, Bannister WH, Hunter GJ. Cloning, expression, and characterization of two manganese superoxide dismutases from Caenorhabditis elegans. J Biol Chem. 1997;272:28652–28659. doi: 10.1074/jbc.272.45.28652. [DOI] [PubMed] [Google Scholar]
- 19.Giglio AM, Hunter T, Bannister JV, Bannister WH, Hunter GJ. The copper/zinc superoxide dismutase gene of Caenorhabditis elegans. Biochem Mol Biol Int. 1994;33:41–44. [PubMed] [Google Scholar]
- 20.Giglio MP, Hunter T, Bannister JV, Bannister WH, Hunter GJ. The manganese superoxide dismutase gene of Caenorhabditis elegans. Biochem Mol Biol Int. 1994;33:37–40. [PubMed] [Google Scholar]
- 21.Suzuki N, Inokuma K, Yasuda K, Ishii N. Cloning, sequencing and mapping of a manganese superoxide dismutase gene of the nematode Caenorhabditis elegans. DNA Res. 1996;3:171–174. doi: 10.1093/dnares/3.3.171. [DOI] [PubMed] [Google Scholar]
- 22.Fujii M, Ishii N, Joguchi A, Yasuda K, Ayusawa D. A novel superoxide dismutase gene encoding membrane-bound and extracellular isoforms by alternative splicing in Caenorhabditis elegans. DNA Res. 1998;5:25–30. doi: 10.1093/dnares/5.1.25. [DOI] [PubMed] [Google Scholar]
- 23.Jensen LT, Culotta VC. Activation of CuZn superoxide dismutases from Caenorhabditis elegans does not require the copper chaperone CCS. J Biol Chem. 2005;280:41373–41379. doi: 10.1074/jbc.M509142200. [DOI] [PubMed] [Google Scholar]
- 24.Vanfleteren JR. Oxidative stress and ageing in Caenorhabditis elegans. Biochem J. 1993;292(Pt 2):605–608. doi: 10.1042/bj2920605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 26.de Castro E, Hegi de Castro S, Johnson TE. Isolation of long-lived mutants in Caenorhabditis elegans using selection for resistance to juglone. Free Radic Biol Med. 2004;37:139–145. doi: 10.1016/j.freeradbiomed.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 27.Blum J, Fridovich I. Superoxide, hydrogen peroxide, and oxygen toxicity in two free-living nematode species. Arch Biochem Biophys. 1983;222:35–43. doi: 10.1016/0003-9861(83)90499-x. [DOI] [PubMed] [Google Scholar]
- 28.Dinis-Oliveira RJ, Duarte JA, Sanchez-Navarro A, Remiao F, Bastos ML, et al. Paraquat poisonings: mechanisms of lung toxicity, clinical features, and treatment. Crit Rev Toxicol. 2008;38:13–71. doi: 10.1080/10408440701669959. [DOI] [PubMed] [Google Scholar]
- 29.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 30.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 31.Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 33.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 34.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 35.Wong A, Boutis P, Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lithgow GJ, White TM, Hinerfeld DA, Johnson TE. Thermotolerance of a long-lived mutant of Caenorhabditis elegans. J Gerontol. 1994;49:B270–B276. doi: 10.1093/geronj/49.6.b270. [DOI] [PubMed] [Google Scholar]
- 37.Solomon A, Bandhakavi S, Jabbar S, Shah R, Beitel GJ, et al. Caenorhabditis elegans OSR-1 regulates behavioral and physiological responses to hyperosmotic environments. Genetics. 2004;167:161–170. doi: 10.1534/genetics.167.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Trends in oxidative aging theories. Free Radic Biol Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 39.Muller FL, Song W, Liu Y, Chaudhuri A, Pieke-Dahl S, et al. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic Biol Med. 2006;40:1993–2004. doi: 10.1016/j.freeradbiomed.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 40.Paul A, Belton A, Nag S, Martin I, Grotewiel MS, et al. Reduced mitochondrial SOD displays mortality characteristics reminiscent of natural aging. Mech Ageing Dev. 2007;128:706–716. doi: 10.1016/j.mad.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Remmen H, Salvador C, Yang H, Huang TT, Epstein CJ, et al. Characterization of the antioxidant status of the heterozygous manganese superoxide dismutase knockout mouse. Arch Biochem Biophys. 1999;363:91–97. doi: 10.1006/abbi.1998.1060. [DOI] [PubMed] [Google Scholar]
- 42.Huang TT, Carlson EJ, Gillespie AM, Shi Y, Epstein CJ. Ubiquitous overexpression of CuZn superoxide dismutase does not extend life span in mice. J Gerontol A Biol Sci Med Sci. 2000;55:B5–B9. doi: 10.1093/gerona/55.1.b5. [DOI] [PubMed] [Google Scholar]
- 43.Sentman ML, Granstrom M, Jakobson H, Reaume A, Basu S, et al. Phenotypes of mice lacking extracellular superoxide dismutase and copper- and zinc-containing superoxide dismutase. J Biol Chem. 2006;281:6904–6909. doi: 10.1074/jbc.M510764200. [DOI] [PubMed] [Google Scholar]
- 44.Gus'kova RA, Ivanov II, Kol'tover VK, Akhobadze VV, Rubin AB. Permeability of bilayer lipid membranes for superoxide (O2-.) radicals. Biochim Biophys Acta. 1984;778:579–585. doi: 10.1016/0005-2736(84)90409-7. [DOI] [PubMed] [Google Scholar]
- 45.Missirlis F, Hu J, Kirby K, Hilliker AJ, Rouault TA, et al. Compartment-specific protection of iron-sulfur proteins by superoxide dismutase. J Biol Chem. 2003;278:47365–47369. doi: 10.1074/jbc.M307700200. [DOI] [PubMed] [Google Scholar]
- 46.Petriv OI, Rachubinski RA. Lack of peroxisomal catalase causes a progeric phenotype in Caenorhabditis elegans. J Biol Chem. 2004;279:19996–20001. doi: 10.1074/jbc.M400207200. [DOI] [PubMed] [Google Scholar]
- 47.Larsen PL. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1993;90:8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson TE, Henderson S, Murakami S, de Castro E, Hegi de Castro S, et al. Longevity genes in the nematode Caenorhabditis elegans also mediate increased resistance to stress and prevent disease. J Inherit Metab Dis. 2002;25:197–206. doi: 10.1023/a:1015677828407. [DOI] [PubMed] [Google Scholar]
- 49.Arking R, Buck S, Berrios A, Dwyer S, Baker GT., III Elevated paraquat resistance can be used as a bioassay for longevity in a genetically based long-lived strain of Drosophila. Dev Genet. 1991;12:362–370. doi: 10.1002/dvg.1020120505. [DOI] [PubMed] [Google Scholar]
- 50.Kim Y, Sun H. Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell. 2007;6:489–503. doi: 10.1111/j.1474-9726.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- 51.Khazaeli AA, Nuzhdin SV, Curtsinger JW. Genetic variation for life span, resistance to paraquat, and spontaneous activity in unselected populations of Drosophila melanogaster: implications for transgenic rescue of life span. Mech Ageing Dev. 2007;128:486–493. doi: 10.1016/j.mad.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 53.Felkai S, Ewbank JJ, Lemieux J, Labbe JC, Brown GG, et al. CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. EMBO J. 1999;18:1783–1792. doi: 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ewbank JJ, Barnes TM, Lakowski B, Lussier M, Bussey H, et al. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- 55.Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishii N, Takahashi K, Tomita S, Keino T, Honda S, et al. A methyl viologen-sensitive mutant of the nematode Caenorhabditis elegans. Mutat Res. 1990;237:165–171. doi: 10.1016/0921-8734(90)90022-j. [DOI] [PubMed] [Google Scholar]
- 57.Kayser EB, Morgan PG, Sedensky MM. GAS-1: a mitochondrial protein controls sensitivity to volatile anesthetics in the nematode Caenorhabditis elegans. Anesthesiology. 1999;90:545–554. doi: 10.1097/00000542-199902000-00031. [DOI] [PubMed] [Google Scholar]
- 58.Ichimiya H, Huet RG, Hartman P, Amino H, Kita K, et al. Complex II inactivation is lethal in the nematode Caenorhabditis elegans. Mitochondrion. 2002;2:191–198. doi: 10.1016/s1567-7249(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 59.Letellier T, Heinrich R, Malgat M, Mazat JP. The kinetic basis of threshold effects observed in mitochondrial diseases: a systemic approach. Biochem J. 1994;302(Pt 1):171–174. doi: 10.1042/bj3020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams MD, Van Remmen H, Conrad CC, Huang TT, Epstein CJ, et al. Increased oxidative damage is correlated to altered mitochondrial function in heterozygous manganese superoxide dismutase knockout mice. J Biol Chem. 1998;273:28510–28515. doi: 10.1074/jbc.273.43.28510. [DOI] [PubMed] [Google Scholar]
- 61.Melov S, Coskun P, Patel M, Tuinstra R, Cottrell B, et al. Mitochondrial disease in superoxide dismutase 2 mutant mice. Proc Natl Acad Sci U S A. 1999;96:846–851. doi: 10.1073/pnas.96.3.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Remmen H, Williams MD, Guo Z, Estlack L, Yang H, et al. Knockout mice heterozygous for Sod2 show alterations in cardiac mitochondrial function and apoptosis. Am J Physiol Heart Circ Physiol. 2001;281:H1422–H1432. doi: 10.1152/ajpheart.2001.281.3.H1422. [DOI] [PubMed] [Google Scholar]
- 63.Rea SL. Metabolism in the Caenorhabditis elegans Mit mutants. Exp Gerontol. 2005;40:841–849. doi: 10.1016/j.exger.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 64.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 65.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Location of mutations in sod genes.
(0.02 MB PDF)
Heteroallelic sod-2 mutant worms show extended lifespan.
(0.02 MB PDF)
sod-2 mutant worms are sensitive to paraquat during development.
(0.02 MB PDF)
Mild compensatory upregulation of other sod mRNAs in sod-sod double deletion mutants.
(0.02 MB PDF)
Summary of mean and maximum lifespan.
(0.01 MB PDF)