SUMMARY
Centrosome duplication involves the formation of a single procentriole next to each centriole, once per cell cycle. The mechanisms governing procentriole formation and those restricting its occurrence to one event per centriole are poorly understood. Here, we show that HsSAS-6 is necessary for procentriole formation and that it localizes asymmetrically next to the centriole at the onset of procentriole formation. HsSAS-6 levels oscillate during the cell cycle, with the protein being degraded in mitosis and starting to accumulate again at the end of the following G1. Our findings indicate that APCCdh1 targets HsSAS-6 for degradation by the 26S proteasome. Importantly, we demonstrate that increased HsSAS-6 levels promote formation of more than one procentriole per centriole. Therefore, regulated HsSAS-6 levels normally ensure that each centriole seeds the formation of a single procentriole per cell cycle, thus playing a fundamental role in driving the centrosome duplication cycle and ensuring genome integrity.
INTRODUCTION
The centrosome is the main microtubule organizing center of animal cells and comprises a pair of centrioles surrounded by pericentriolar material (Chretien et al., 1997; Kuriyama and Borisy, 1981; Paintrand et al., 1992; Vorobjev and Chentsov, 1982). Proliferating cells are born with a single centrosome, which duplicates once per cell cycle to generate two centrosomes that direct bipolar spindle assembly during mitosis, thus ensuring faithful segregation of the genetic material. Failure of centrosome duplication can result in monopolar spindle assembly; excess centrosome duplication, in multipolar spindle assembly. Therefore, just like DNA replication, centrosome duplication must occur once per cell cycle to ensure genome integrity.
Centrosome duplication begins at the G1 to S transition when a single procentriole forms orthogonal to the proximal end of each centriole (Chretien et al., 1997; Kuriyama and Borisy, 1981; Paintrand et al., 1992; Vorobjev and Chentsov, 1982). Each growing procentriole remains tightly connected to its centriole until the end of mitosis when the two disorient from one another. Each daughter cell thus inherits two centrioles, which are dubbed the mother and the daughter centriole, and which for simplicity are referred to as centrioles in the remainder of this manuscript. The mechanisms that enable procentriole formation from the proximal end of each centriole are not understood. Furthermore, the mechanisms restricting the occurrence of procentriole formation to one event per centriole have remained elusive.
A few proteins have been reported to be required for procentriole formation in human cells, including the EF-hand protein centrin-2 and the Polo-like kinase Plk4 (Habedanck et al., 2005; Salisbury et al., 2002). Proteins of the centrin family localize to the distal lumen of centrioles and procentrioles throughout the cell cycle and are detected in procentrioles already in early S phase (Paoletti et al., 1996; Piel et al., 2000; Salisbury et al., 2002). However, it is not known whether the recruitment of centrin next to the centriole defines where procentriole formation occurs, or whether a distinct protein is recruited earlier to exert such an instructive function. Plk4 also associates with centrioles and procentrioles throughout the cell cycle and may be important for initiating procentriole formation, since overexpression of FLAG-tagged Plk4 results in the formation of several electron-dense patches on each centriole (Habedanck et al., 2005). These patches harbor centrin and presumably elongate further into mature procentrioles (Habedanck et al., 2005).
Another component that may be particularly important is HsSAS-6, a coiled-coil protein that belongs to an evolutionarily conserved family named after its founding member SAS-6, which is required for centriole formation in C. elegans (Dammermann et al., 2004; Leidel et al., 2005). Depletion of HsSAS-6 from U2OS cells results in an increased frequency of monopolar spindles, suggesting that HsSAS-6 is needed for an aspect of the centro some duplication cycle, although which one in particular is not known (Leidel et al., 2005). Conversely, overexpression of GFP-HsSAS-6 leads to multiple foci bearing centriolar markers and an increased frequency of multipolar spindles (Leidel et al., 2005). Although this could be due to overexpression of a fusion protein, which may have an altered function compared with the endogenous protein, these initial findings taken together suggest that HsSAS-6 may be important for proper progression through the centrosome duplication cycle, a possibility that we investigated in this study.
RESULTS
HsSAS-6 Is Required for Procentriole Formation
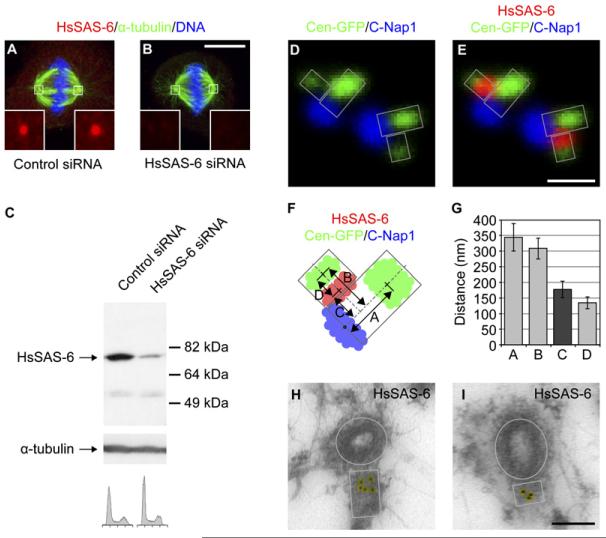
To determine the specific step in the centrosome duplication cycle at which HsSAS-6 is required, we depleted the protein from HeLa cells using siRNAs and analyzed the number of centrin foci in mitotic cells as a marker of centriole number. As anticipated, the vast majority (>90%) of cells treated with control siRNAs contained four centrin foci corresponding to two pairs of centriole/procentriole (Figures 1A and 1E). By contrast, 48 hr after transfection, 54% of cells treated with HsSAS-6 siRNAs contained only two separated centrin foci, likely corresponding to the two centrioles; such cells assembled a bipolar spindle (Figures 1B and 1E). The phenotype became more severe 72 hr after transfection, with 53% of cells containing a single centrin focus; over half (66%) of this subset of cells assembled a monopolar spindle (Figures 1C–1E). Analogous phenotypes were observed after depletion of HsSAS-6 in U2OS cells (see Figure S1 in the Supplemental Data available with this article online). We performed serial section electron microscopy (EM) to investigate whether the single centrin focus present in the monopolar configuration corresponds to a single centriole. As shown in Figure 1F, we found this to be the case and did not detect the presence of an associated procentriole or additional electron-dense material in the vicinity. Taken together, our findings demonstrate that HsSAS-6 is required for the step of procentriole formation in the centrosome duplication cycle.
Figure 1. HsSAS-6 Is Required for Procentriole Formation.

(A—D) HeLa cells transfected with control siRNAs or siRNAs directed against HsSAS-6, stained with antibodies against centrin-3 (red) and α-tubulin (green); DNA is shown in blue. Insets are magnified 5-fold. Scale bar, 10 μm. Cells were classified into the following categories: two pairs of centrin foci, bipolar spindle (A); two single centrin foci, bipolar spindle (B); one single centrin focus, bipolar spindle (C); one single centrin focus, monopolar spindle (D).
(E) Distribution of cells in the categories described above (n = 50 and n = 100 for 48 hr and 72 hr time points, respectively); Other: cells that could not be assigned to any of the four categories (∼5% of cells contained more than four centrin foci, for each experimental condition, and some cells contained a pair of juxtaposed centrin foci in the case of cells targeted with HsSAS-6 siRNAs). Note that monopolar spindle assembly was also observed with an independent siRNA (Leidel et al., 2005).
(F) HeLa cells expressing centrin-GFP were treated with siRNA directed against HsSAS-6 and observed 72 hr after transfection. A cell with a monopolar spindle and single centrin focus (arrow) was first imaged by DIC and fluorescence microscopy (scale bar, 10 μm), then fixed and analyzed by serial section EM (1—6: selected sections spanning the entire centriole; scale bar, 500 nm).
HsSAS-6 Localizes to the Proximal End of the Procentriole
To characterize the distribution of HsSAS-6, we raised and affinity-purified HsSAS-6 antibodies, which labeled one dot at each spindle pole, as well as the cytoplasm to a lesser extent (Figure 2A). These antibodies also recognized a single major band of the expected size of 74 kDa in whole-cell lysates (Figure 2C). The signals observed by immunofluorescence and western blot were significantly diminished in HsSAS-6 siRNA-treated cells (Figures 2B and 2C), indicating specificity. We used these antibodies to characterize the localization of HsSAS-6 using triplelabeling experiments. HeLa cells in the G2 phase of the cell cycle that expressed centrin-GFP (Piel et al., 2000) were labeled with GFP antibodies to mark the distal end of centrioles and procentrioles (Figure 2D, green) and with C-Nap1 antibodies to mark the base of centrioles (Fry et al., 1998)(Figure 2D, blue). This allowed us to estimate the position of centrioles and procentrioles (Figure 2D, gray lines). Strikingly, we found that HsSAS-6 was invariably located between the axis of the centriole and the distal end of the procentriole (Figure 2E, red). We found an analogous distribution in U2OS cells (Figure S2A). By measuring the distances between the signals in triply labeled HeLa cells (Figure 2F), we found that the focus of HsSAS-6 is located ∼175 nm away from the of the centriole, corresponding to the proximal part of the procentriole (Figure 2G, value C). Importantly, immunoelectron microscopy confirmed that HsSAS-6 localizes to the proximal part of the procentriole (Figure 2H) and established that the protein is present at that location already in the early stages of procentriole formation (Figure 2I).
Figure 2. HsSAS-6 Localizes to the Proximal Part of the Procentriole.
(A and B) HeLa cells transfected with control siRNAs or siRNAs directed against HsSAS-6 and stained with antibodies against α-tubulin (green) and HsSAS-6 (red); DNA is shown in blue. Insets are magnified 5-fold. Scale bar, 10 μm.
(C) Control siRNA and HsSAS-6 siRNA-treated HeLa whole-cell lysates were analyzed by western blot using HsSAS-6 antibodies (top); the blot was reprobed with antibodies against α-tubulin as a loading control (middle). Cells treated in the same manner were analyzed by FACS (bottom). Note that there is no significant difference in cell cycle distribution between cells treated with control and HsSAS-6 siRNAs.
(D and E) HeLa cell in G2 expressing centrin-GFP and stained with antibodies against GFP (green; shown in D and E), C-Nap1 (blue; shown in D and E), and HsSAS-6 (red; shown in E). The orientation of the centrioles (large rectangles) and the procentrioles (small rectangles) is illustrated in gray. Scale bar, 500 nm. (F and G) Distances between the center of the centrin-GFP, C-Nap1, and HsSAS-6 signals (schematically represented in F) were measured for ten centriole/procentriole pairs (imaged as in E) and are reported in (G). Error bars, 95% confidence intervals. Value C is highlighted in dark gray and corresponds to the distance separating the center of the HsSAS-6 signal from the axis of the centriole (see F).
(H and I) HeLa cells processed for immunoelectron microscopy and labeled with HsSAS-6 antibodies. The centriole (viewed in cross-section), oval contours, and the procentriole (viewed laterally), rectangular contours, are shown in gray, and all the gold particles are highlighted in yellow. The structure of procentrioles and centrioles is not fully preserved due to methanol fixation. Scale bar, 200 nm.
HsSAS-6 Protein Levels Are Regulated across the Cell Cycle
Next, we investigated the distribution of HsSAS-6 during the cell cycle. We determined the cell cycle stage of individual cells by staining them with a mixture of PCNA antibodies, which label discrete foci in S-phase nuclei (Leonhardt et al., 2000), and Cyclin B1 antibodies, which label the cytoplasm during G2 and until metaphase (reviewed by Peters, 2006) (Figure 3A, both antibodies visualized in green). We found that neither centriolar nor cytoplasmic HsSAS-6 was present in most G1 cells, which are negative for PCNA foci and cytoplasmic Cyclin B1 (Figure 3A). HsSAS-6 was first clearly detected at centrioles and in the cytoplasm of S-phase cells, and accumulated further in the cytoplasm as cells progressed through G2 and until mitosis (Figure 3A). We also observed that HsSAS-6 was no longer present at centrioles and in the cytoplasm at the end of mitosis and onward (Figure 3A). An identical distribution during the cell cycle was observed in U2OS cells (Figure S2B). Taken together, our results establish that the presence of HsSAS-6 at centrioles and in the cytoplasm is cell-cycle regulated.
Figure 3. HsSAS-6 Is Recruited to the Procentriole before Centrin in a Plk4-Independent Manner.
(A) HeLa cells were fixed and stained with a mixture of antibodies against PCNA, Cyclin B1 (both in green; top) and HsSAS-6 (red; bottom); DNA is shown in blue (bottom). Scale bar, 10 μm. Arrows point to centriolar HsSAS-6.
(B) HeLa cells were released from a double thymidine block, fixed at the indicated time points, and stained with antibodies against centrin (green) and HsSAS-6 (red). Scale bar, 500 nm. Cells were placed into three categories based on the distribution of these two proteins: (1) two centrin foci, no HsSAS-6 (top; empty circles); (2) two centrin foci, HsSAS-6 juxtaposed next to one (image not shown) or two (middle; filled squares) centrin foci; (3) three (image not shown) or four centrin foci with HsSAS-6 localized next to two of them (bottom; empty triangles). The relative representation of each category is shown in the graph (top; 100 cells counted for each time point). The DNA content of cells from each time point was analyzed by FACS (bottom).
(C and D) HeLa cell expressing centrin-GFP and stained with antibodies against GFP (green; shown in C and D), C-Nap1 (blue; shown in C and D), and HsSAS-6 (red; shown in D). The orientation of the centrioles (rectangles) is illustrated in gray. Scale bar, 500 nm. (E and F) HeLa cells transfected with control siRNAs or siRNAs directed against Plk4, fixed after 72 hr and stained with antibodies against centrin (green) and HsSAS-6 (red); DNA is shown in blue. Cells with a high cytoplasmic signal of HsSAS-6 were scored. Insets are magnified 5-fold. Scale bar, 10 μm.
HsSAS-6 Is Recruited to the Procentriole before Centrin
We set out to determine when HsSAS-6 is first detected on the procentriole during the centrosome duplication cycle. Because centriolar HsSAS-6 is present in essentially all S-phase cells (>99%; n = 109), the protein appears to be recruited very early during procentriole formation, as indicated also by immunoelectron microscopy (see Figure 2I). To investigate this further, we compared the timing of the appearance of centriolar HsSAS-6 to that of centrin, the earliest known marker of procentriole formation. We examined the distribution of HsSAS-6 and centrin in HeLa cells released from a double thymidine block and fixed at successive time points spanning the G1 to S transition (Figure 3B). Initially, most cells harbored only two foci of centrin, corresponding to the two centrioles (Figure 3B; 13 hr, open disks). Thereafter, many cells harbored two centrin foci with two juxtaposed HsSAS-6 foci (Figure 3B; 15 hr and 17 hr, filled squares). Interestingly, centrin recruitment to the two newly forming procentrioles was not detectable in this subset of cells. Later still, most cells harbored four centrin foci and two HsSAS-6 foci (Figure 3B; 21 hr, open triangles). Triple-labeling experiments with centrin-GFP and C-Nap1 established that HsSAS-6 could be found asymmetrically distributed on the side of the centriole in cells with two centrin foci and two juxtaposed HsSAS-6 foci (Figures 3C and 3D). Although we cannot exclude the possibility that undetectable amounts of centrin are present on the newly forming procentrioles in such cells, our findings taken together indicate that HsSAS-6 is recruited before centrin during procentriole formation. Compatible with this view, we found that depletion of centrin-2, which was reported to be required for procentriole formation (Salisbury et al., 2002), did not prevent centriolar HsSAS-6 recruitment (Figures S3A–S3D). By contrast, depletion of HsSAS-6 resulted in the absence of centrin from centrioles (see Figure 1). Overall, we conclude that HsSAS-6 is recruited before and independently of centrin during procentriole formation.
We then investigated whether HsSAS-6 recruitment requires Plk4 function, which is crucial for procentriole formation (Habedanck et al., 2005). Importantly, we found that the vast majority (94%; n = 100) of HsSAS-6-positive interphase cells depleted of Plk4 harbored some HsSAS-6 on their single centriole (Figure 3F; compare with Figure 3E), although the amount of protein present at centrioles was lower than that in control cells (data not shown). We noted also that most (51%; n = 100) mitotic cells depleted of Plk4 no longer had detectable centriolar HsSAS-6 (Figures S3E–S3I). Overall, we conclude that the initial recruitment of HsSAS-6 to the procentriole appears to be Plk4 independent. In addition, our results indicate that Plk4 plays a role in the maintenance of centriolar HsSAS-6. This role may be direct or indirect, reflecting for instance the requirement of Plk4 for proper procentriole formation (Habedanck et al., 2005).
HsSAS-6 Is Degraded by the 26S Proteasome Starting in Anaphase
Next, we investigated the mechanisms that enable HsSAS-6 levels to fluctuate during the cell cycle. First, to verify that protein levels are cell-cycle regulated, we analyzed synchronized cells by western blot and fluorescence activated cell sorter (FACS). As shown in Figure 4A, HsSAS-6 levels increased gradually from early S phase until mitosis, when the protein disappeared abruptly. Given the minute size of procentrioles, this fluctuation in total HsSAS-6 levels represents primarily that of the cytoplasmic pool, although centriolar HsSAS-6 follows analogous kinetics (see Figure 3A). We found that the fluctuation in overall protein levels is not due to significant variations in mRNA levels, as demonstrated by quantitative RT-PCR experiments (Figure 4B).
Figure 4. HsSAS-6 Protein Levels Are Cell-Cycle Regulated.
(A) HeLa cells released from a double thymi-dine block and analyzed at the indicated time points by FACS for DNA content (bottom) and by western blot using HsSAS-6 antibodies (top). The membrane was also probed with antibodies against α-tubulin as a loading control (middle). Unsynchronized cells (Uns) were processed in parallel.
(B) HeLa cells were released from a double thymidine block and RNA was isolated at the indicated time points. HsSAS-6 mRNA levels were analyzed by quantitative RT-PCR, with values being expressed relative to the total RNA amount (top). Error bars, 95% confidence intervals. Cells from each time point were also analyzed by FACS for DNA content (bottom). (C) HeLa cells at the indicated stages were fixed and stained with antibodies against Cyclin B1 (green) and HsSAS-6 (red). DNA is shown in blue. Scale bar, 10 μm.
(D) Cells were processed as in (C) and the average cytoplasmic signal intensity of metaphase, anaphase, telophase, and G1 cells was quantified (n = 10; error bars, 95% confidence intervals). Note that while Cyclin B1 levels drop already in anaphase, those of HsSAS-6 start dropping during telophase.
Next, we addressed whether HsSAS-6 protein degradation is mediated by the 26S proteasome. To this end, we synchronized cells in mitosis, subjected them to the 26S proteasome inhibitor MG132, and processed them 30 min later for analysis by immunofluorescence with HsSAS-6 antibodies. We focused our analysis on the subset of cells that were past the metaphase to anaphase transition at the time of drug exposure, and that were thus able to proceed to telophase. Importantly, we found that HsSAS-6 is present at centrioles and in the cytoplasm of such telophase cells (Figure 5B), in contrast to control telophase cells (Figure 5A). We conclude that HsSAS-6 degradation is mediated by the 26S proteasome.
Figure 5. HsSAS-6 Degradation Is Mediated by the 26S Proteasome and Depends on a KEN Box.
(A and B) HeLa cells 10 hr after release from a thymidine block were treated with MG132 or DMSO alone (Control) for 30 min, fixed, and stained with antibodies against HsSAS-6 (red). DNA is shown in blue. Telophase cells were picked based on DNA staining and imaged, and the average cytoplasmic HsSAS-6 signal was quantified (n = 10; arbitrary units; errors represent 95% confidence intervals). Scale bar, 10 μm.
(C) Schematic representation of HsSAS-6 fragments: N-ter (aa 1–173), C-ter (aa 148–657), and C-ter-ΔKEN (aa 148–657; K589A, E590A, N591A). The putative D boxes are indicated in black; the predicted coiled-coil domain, in gray; and the KEN box, in red.
(D–F) Dual GFP fluorescence and phase-contrast time lapse microscopy of HeLa cells expressing GFP-HsSAS-6N-ter (D), GFPHsSAS-6C-ter (E), or GFP-HsSAS-6C-ter-ΔKEN (F). Time 0 min corresponds to the metaphase to anaphase transition. See also Movies S1–S3. Scale bar, 10 μm.
(G) HeLa cells were transfected with Myc-Cdh1 and treated with aphidicolin (for 48 hr) starting 24 hr after transfection. The average cytoplasmic HsSAS-6 signal was quantified for randomly picked transfected cells and their nontransfected neighbors (n = 10; arbitrary units; errors represent 95% confidence intervals). Scale bar, 10 μm. Note that we performed staining with PCNA antibodies (data not shown) to verify that cells transfected with Myc-Cdh1 are arrested in S phase, as previously reported (Sorensen et al., 2000). Note also that the arbitrary units in (G) cannot be compared to those in (A) and (B), because the imaging was performed under different conditions in the two experiments.
Proteins are degraded by the 26S proteasome after being subjected to polyubiquitination through the action of an E3 ubiquitin ligase (reviewed by Peters, 2006). The E3 ubiquitin ligase APC/C is active during mitosis and in G1 (reviewed by Peters, 2006), raising the possibility that it may be involved in HsSAS-6 degradation. APC/C functions with two adaptor proteins at different times during the cell cycle: Cdc20 during metaphase and early anaphase, and Cdh1 during late anaphase and in G1 (reviewed by Peters, 2006).
We investigated in more detail the timing of HsSAS-6 degradation to address whether HsSAS-6 may be a substrate of APCCdc20 or APCCdh1. To this end, we performed quantitative immunofluorescence analysis of mitotic cells stained with antibodies directed against HsSAS-6 and against Cyclin B1, a known APCCdc20 substrate. Importantly, we found that the levels of Cyclin B1 diminish during anaphase already, as anticipated, while those of HsSAS-6 diminish only starting in telophase (Figures 4C and 4D). This timing of disappearances strongly suggests that HsSAS-6 is not a target of APCCdc20, and is compatible with the notion that HsSAS-6 is a target of APCCdh1.
A KEN Box Is Important for HsSAS-6 Degradation
Cdc20 usually binds substrates containing a D box, whereas Cdh1 binds substrates with a KEN box (Glotzer et al., 1991; Pfleger and Kirschner, 2000). We identified three putative D boxes in the N terminus of HsSAS-6 and one putative KEN box in the C terminus of the protein (Figure 5C). To address whether fragments containing these putative recognition motifs can mediate protein degradation, we fused GFP to the N- and C-terminal portions of HsSAS-6 (Figure 5C) and conducted time-lapse microscopy of HeLa cells expressing these fusion proteins. We found that the signal of GFPHsSAS-6N-ter, which contains two of the three putative D boxes, remained constant as cells progressed through and exited from mitosis (Figure 5D; see Movie S1 in theSupplemental Data), indicating that these two putative D boxes are not sufficient to mediate degradation, at the least in the context of such a fusion protein. By contrast, the signal of GFP-HsSAS-6C-ter, which was high at metaphase, dropped abruptly during mitosis and was not detectable in early G1, much like endogenous HsSAS-6 (Figure 5E; Movie S2). We conclude that the C-terminal portion of HsSAS-6 contains a degradation signal.
To test whether this signal corresponds to the putative KEN box, we mutated this motif in GFP-HsSAS-6C-ter. Strikingly, we found that this alteration prevented degradation of the fusion protein (Figure 5F; Movie S3). To address whether the KEN box is also functional in the context of full-length HsSAS-6, we conducted immunofluorescence analysis of cells overexpressing HsSAS-6-δKEN. As shown in Figures S4A and S4B, we found that HsSAS-6-δKEN is present in G1 cells, indicating that it is not efficiently degraded, in contrast to the endogenous protein (see Figure 3A).
To further address whether APCCdh1 is important for degradation of endogenous HsSAS-6, we examined the consequence of overexpressing Myc-tagged Cdh1, which results in constitutive activation of APC/CCdh1 (Sorensen et al., 2000). Cells transfected with Myc-Cdh1 were arrested in S phase by aphidicolin and their levels of cytoplasmic HsSAS-6 were compared with those in neighboring nontransfected cells using quantitative immunofluorescence analysis (see Experimental Procedures). Importantly, we found that levels of HsSAS-6 were substantially lower in cells expressing Myc-Cdh1 (Figure 5G). Therefore, constitutive APC/CCdh1 activation results in diminution of HsSAS-6 levels.
Although other mechanisms may additionally contribute to degradation, these results taken together suggest that HsSAS-6 levels oscillate during the cell cycle because the protein is ubiquitinated by APC/CCdh1 starting in anaphase and is thus targeted for degradation by the 26S proteasome.
HsSAS-6 Levels Must Be Limited to Restrict Procentriole Formation
To investigate the biological significance of having limited levels of HsSAS-6, we analyzed cells overexpressing HsSAS-6-δKEN. Strikingly, we found that ∼60% of cells overexpressing HsSAS-6-δKEN have more than four centrin foci 72 hr after transfection, compared with only ∼4% of control cells (Figures 6A–6C). The presence of excess centrin foci was not due to an arrest in cell cycle progression, which can result in centriole overduplication (Meraldi et al., 1999), because we found a similar cell cycle distribution when comparing cells overexpressing either GFP or HsSAS-6-δKEN (Figures S4A and S4B). Because centrin localizes to the distal end of procentrioles and centrioles (Paoletti et al., 1996; Piel et al., 2000), these findings suggest that higher levels of HsSAS-6 promote formation of excess procentrioles or centrioles.
Figure 6. HsSAS-6 Levels Must Be Limited to Restrict Procentriole Formation.
(A and B) U2OS cells transfected with HsSAS-6 or HsSAS-6-ΔKEN, fixed after 72 hr and stained with antibodies against centrin (green) and HsSAS-6 (red). Representative images of cells without (A) or with (B) excess centrin foci are shown.
(C) Cells transfected with GFP, HsSAS-6, or HsSAS-6-ΔKEN fixed after 72 hr and stained with antibodies against centrin and GFP or centrin and HsSAS-6. The number of centrin foci in GFP-positive cells (for GFP) or in cells with elevated HsSAS-6 levels (for HsSAS-6 and HsSAS-6-ΔKEN) was quantified (n ≥ 100 for each of three independent experiments). Error bars, 95% confidence intervals.
(D) HeLa cells were cotransfected with GFP and HsSAS-6 or GFP and HsSAS-6-ΔKEN, and HsSAS-6 levels were analyzed after 48 hr by western blot. GFP was used as a transfection and loading control. The images are from one membrane and a single exposure. Note that there is significantly more HsSAS-6 accumulating in cells expressing HsSAS-6-ΔKEN, as compared with HsSAS-6.
(E and F) U2OS cells transfected with HsSAS-6-ΔKEN, fixed after 24 hr and stained with antibodies against centrin (green, shown in E and F) and HsSAS-6 (red, shown in E) or Odf2 (red, shown in F). Arrows point to a centriole that seeds formation of three (E) or two (F) centrin foci. Scale bar, 500 nm.
(G) U2OS cells expressing centrin-GFP were transfected with HsSAS-6-ΔKEN. Seventy-two hours later, cells were screened using fluorescence microscopy (top panel; arrow points to the area shown on the EM images below; note also the other centrin-GFP focus), fixed, and analyzed by serial section EM (1–4: selected sections spanning the entire centriole and associated procentrioles; scale bar, 500 nm). Arrows point to the centriole; arrowheads, to procentrioles, three of which are present in this particular case.
We then set out to address whether a more moderate increase in HsSAS-6 levels causes a more modest increase in centrin foci number. To this end, we analyzed cells over-expressing wild-type HsSAS-6, which accumulated at lower overall levels than HsSAS-6-δKEN (Figure 6D,Figure S5), as expected from the fact that the KEN box is intact in wild-type HsSAS-6. Interestingly, we found that only ∼30% cells overexpressing wild-type HsSAS-6 had more than four centrin foci 72 hr after transfection, compared with ∼60% in the case of HsSAS-6-δKEN (Figure 6C). Therefore, HsSAS-6 levels appear to dictate the number of centrin foci that are formed.
Excess HsSAS-6 Results in Multiple Procentrioles Forming Next to One Centriole
To gain further insight into the mechanisms by which increased HsSAS-6 levels result in excess centrin foci, we analyzed the distribution of HsSAS-6-δKEN at centrioles. We examined cells 24 hr after transfection, when overduplication events were rarer than at later time points but when the spatial distribution of centrin foci was less complex and thus could be more readily determined. Importantly, we observed cells with one strongly staining centrin focus, probably corresponding to the centriole, surrounded by a cloud of HsSAS-6 from which several weakly staining centrin foci emanated, presumably corresponding to procentrioles (Figure 6E). Staining with the centriole-specific marker Odf2 (Ishikawa et al., 2005; Nakagawa et al., 2001) confirmed that one centriole could seed the formation of several weakly staining centrin foci following HsSAS-6-δKEN overexpression (Figure 6F).
To address whether these centrin foci correspond to bona fide procentrioles, U2OS cells expressing centrin-GFP and overexpressing HsSAS-6-δKEN were filmed; cells with three or more centrin foci moving coordinately were fixed and analyzed by serial section EM. Strikingly, we found that such cells contained a single centriole surrounded by several procentrioles (Figure 6G), which were usually morphologically normal (16/19 procentrioles). Moreover, we observed that the centriole in such cells often had a slightly altered morphology with a seemingly expanded lumen, for reasons that remain to be determined (Figure 6G; 4/6 centrioles). Overall, we conclude that elevated levels of HsSAS-6 result in the seeding of more than one procentriole per centriole.
DISCUSSION
How a single procentriole forms next to each centriole once per cell cycle is a long-standing question in cell and developmental biology. Our findings provide evidence that HsSAS-6 is a key element in answering this question, since the protein is crucial for ensuring procentriole formation, as is the case for its counterparts in C. elegans and Drosophila (Dammermann et al., 2004; Leidel et al., 2005; Peel et al., 2007), and since regulation of its levels allows the occurrence of this event to be restricted to one per centriole.
HsSAS-6 Asymmetric Distribution May Dictate Procentriole Formation
How does HsSAS-6 ensure procentriole formation? The protein is recruited asymmetrically next to the centriole before any other known marker of procentriole formation, supporting the hypothesis that HsSAS-6 defines the position where the procentriole forms. Interestingly, time-resolved electron tomography of C. elegans embryos revealed that centriole formation begins with the assembly of a central tube, whose assembly notably requires SAS-6 function (Pelletier et al., 2006). Perhaps an analogous structure exists in human cells and is similarly dependent on the presence of HsSAS-6. Regardless, the asymmetric localization of HsSAS-6 suggests a model in which the protein recognizes a structural or molecular feature associated with the proximal end of the centriole. Interestingly, in addition, ultrastructural analysis of cells with excess HsSAS-6 demonstrates that several procentrioles can be formed per centriole, indicating that more than one site on the centriole can act as a seed for procentriole formation. Perhaps HsSAS-6 provides a seed for procentriole formation in a manner analogous to the satellite that foreshadows spindle pole body formation in S. cerevisiae (Adams and Kilmartin, 1999; Byers and Goetsch, 1974). In the case of human cells, recruitment of other proteins, including centrin, would follow and contribute to further procentriole growth.
The impact of HsSAS-6 on the centrosome duplication cycle bears resemblance to that of Plk4, since this kinase is also required for procentriole formation and since Plk4 overexpression leads to excess centrioles (Habedanck et al., 2005). In Drosophila, overexpression of either DSas-6 or SAK/Plk4 can similarly induce centriole amplification (Peel et al., 2007; Rodrigues-Martins et al., 2007). Therefore, SAS-6-related proteins and Plk4 kinases appear necessary and sufficient for dictating procentriole formation. Although Plk4 seems dispensable for HsSAS-6 centriolar recruitment, it remains possible that Plk4 kinase activity is needed for proper HsSAS-6 function.
HsSAS-6 Levels Must Be Regulated during the Cell Cycle
How are HsSAS-6 levels regulated in normal proliferating cells so that a single procentriole forms next to each centriole? Our findings suggest that HsSAS-6 is a substrate for the E3 ligase APCCdh1 and that the protein is degraded by the 26S proteasome starting in anaphase and throughout G1. Importantly, our results additionally demonstrate that HsSAS-6 levels must be limited to restrict procentriole formation to a single event per cell cycle.
Experiments with Xenopus extracts have shed light in a different manner on how procentriole formation is restricted to once per cell cycle. It was shown that separase-mediated disorientation must occur at the end of mitosis so that the two centrioles can seed formation of one procentriole each at the next G1 to S transition (Tsou and Stearns, 2006). Our findings lead us to conclude that dis-orientation is not sufficient for restricting this event to one per centriole, but that concomitant degradation of HsSAS-6 is essential for this to be the case.
It will be interesting to determine when during the cell cycle HsSAS-6 levels matter for ensuring formation of a single procentriole per centriole. Given that procentriole formation begins at the G1 to S transition, when HsSAS-6 also begins to accumulate, HsSAS-6 levels may be critical at this juncture for determining procentriole number. Perhaps HsSAS-6 levels are sufficiently low at this moment to enable just a single site to act as a seed for procentriole formation. This would direct further growth exclusively to this site despite HsSAS-6 levels subsequently increasing in the cell cycle.
On the Importance of HsSAS-6 Oscillation
Progression through the cell cycle is notably driven by the periodic accumulation and degradation of cyclins (Murray, 2004). For instance, accumulation of B type cyclins is necessary for Cdk1 activation and mitotic entry, whereas their degradation is required for Cdk1 inactivation and mitotic exit. Oscillation of HsSAS-6 protein levels during the centrosome duplication cycle plays a role reminiscent of that of cyclins during the cell cycle. The presence of HsSAS-6 is necessary for procentriole formation, whereas its degradation is required for the number of procentrioles seeded by each centriole at each cell cycle to be limited to one (Figure 7).
Figure 7. HsSAS-6 Level Oscillations Are Crucial for the Centrosome Duplication Cycle.

Schematic representation of HsSAS-6 distribution during the cell cycle. Red disks represent HsSAS-6; green disks, centrin; blue disks, C-Nap1; and gray diamonds, active APC/C; one centriole/procentriole pair is shown in opaque in M phase. HsSAS-6 is absent in G1 and is first clearly detected in S phase when it is recruited before centrin in an asymmetric position next to the proximal end of the centriole. As cells progress through S phase and G2, HsSAS-6 accumulates further until it is degraded after ubiquitination by APC/CCdh1. Our findings indicate that the presence of HsSAS-6 in S phase is essential for procentriole formation, whereas its degradation during M phase is essential for limiting procentriole formation to one event per centriole at each cell cycle.
In conclusion, we discovered that HsSAS-6 is essential for procentriole formation and that its levels must be tightly regulated to restrict the number of procentrioles formed per centriole to one, thus providing important novel insight into the mechanisms driving the centrosome duplication cycle.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa and U2OS cells, as well as HeLa cells stably expressing centrin-GFP (Piel et al., 2000) (gift from Michel Bornens), were grown at 37°C and 5% CO2 in high-glucose DMEM supplemented with GlutaMAX (Invitrogen, Carlsbad, CA) and 10% fetal calf serum (Invitrogen). Cells were split to 60%–90% confluency and transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) using 1 μg and 2 μl (U2OS) and 4 μg and 8 μl (HeLa) of DNA and Lipofectamine, respectively, per well in 6-well plates. When the phenotype was assayed >24 hr after transfection, cells were split at that time to prevent overgrowth.
For HsSAS-6, Plk4, and centrin-2 depletion, siRNAs with the sequences GCACGUUAAUCAGCUACAAUU, GAGCAAAAGCAGGA GAUCCUU, and GAAAUGAACAGGUAUCUAAUU, respectively, were ordered from the siGENOME collection (Dharmacon, Lafayette, CO) and used essentially as described (Leidel et al., 2005), with a cell density of ∼5 × 104 cells/well in 6-well plates. We used siRNAs directed against GFP as a negative control.
Molecular Biology and Antibody Production
A full length HsSAS-6 cDNA was cloned into pET30a+ (EMD Biosciences, Novagen, San Diego, CA) to express His(6)-HsSAS-6. The protein was affinity purified and injected into a rabbit according to standard procedures (Eurogentec, Seraing, Belgium). Full-length HsSAS-6 was also cloned into pGEX-6P-3 (Amersham Pharmacia Biotech, Freiburg, Germany) to express GST-HsSAS-6, which was blotted from a 7.5% SDS-PAGE gel onto a PVDF (Millipore, Billerica, MA) membrane. Antibodies were affinity purified using a piece of membrane with immobilized GST-HsSAS-6, eluted with 100 mM glycine (pH 2.5), neutralized with 1M Tris-HCl buffer (pH 8.8), and stored in 50% glycerol at -20°C.
For overexpression experiments, full-length HsSAS-6 was cloned into pcDNA5/FRT/TO (Invitrogen), which drives expression from a CMV promoter. The relevant mutations in the KEN box (K589A, E590A, N591A) were generated using the QuickChange Kit (Strata-gene, La Jolla, CA) and introduced into full-length HsSAS-6 to generate HsSAS-6-δKEN. HsSAS-6 fragments (Figure 5C) were cloned into pEGFP-C1 (Clontech, Mountain View, CA), and pcDNA3.1-6xMyc-Cdh1 (Sorensen et al., 2000) (gift from Jiri Lukas) was used to overex-press Myc-Cdh1.
Indirect Immunofluorescence
Cells grown on glass coverslips were fixed rapidly after removal from the culture medium for ∼10 min in -20°C methanol, washed in PBS, and blocked for 20 min in antibody dilution solution (1% bovine serum albumin, 0.05% Triton X-100 in PBS). Cells were incubated overnight at 4°C with primary antibodies, washed four times for 5 min in PBST (0.05% Triton X-100 in PBS), incubated for 1 hr at room temperature with secondary antibodies, washed four times for 5 min in PBST (with ∼1 μg/ml Hoechst 33258 included in the third wash), and mounted. Primary antibodies were 1:400 mouse anti-tubulin (DM1A, Sigma), 1:2000 mouse anti-centrin (Sanders and Salisbury, 1994) (20H5; gift from Jeffrey L. Salisbury), 1:2000 rabbit anti-centrin-3 (Middendorp et al., 2000) (gift from Michel Bornens), 1:60 rabbit anti-HsSAS-6, 1:400 mouse anti-C-Nap1 (BD Biosciences, Franklin Lakes, NJ), 1:400 rabbit anti-Odf2 (Ishikawa et al., 2005; Nakagawa et al., 2001) (gift from Hiroaki Ishikawa), 1:400 mouse anti-GFP (Chemikon International, Temecula, CA), 1:200 rabbit anti-GFP (gift from Viesturs Simanis), 1:500 mouse anti-PCNA, and 1:500 mouse anti-Cyclin B1 (both from Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies were 1:1000 goat anti-mouse coupled to Alexa 488, 1:1000 goat anti-rabbit coupled to Alexa 568 (both from Molecular Probes, Eugene, OR), and 1:400 goat anti-mouse coupled to Cy5 (gift from Joachim Lingner). For triple-color labeling, 1:400 goat anti-GFP coupled to FITC (Abcam, UK) was included with the secondary antibodies.
Microscopy, Image Processing, and Analysis
Indirect immunofluorescence was imaged on a Leica TCS SP2 (Leica Microsystems, Germany) inverted confocal microscope. Images were taken as z-stacks with distances between planes of 0.2 μm for zoom 15, and 0.4 μm for zoom 6; projections of relevant planes are shown. Live HeLa cells grown on glass-bottom dishes (MatTek, Ashland, MA) were imaged using dual time-lapse fluorescent and phase-contrast microscopy on a Zeiss Axiovert 100 equipped with a 633 oil-immersion objective (Zeiss, Germany) and a cooled CCD camera (Photometrics, Tucson, AZ) controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA). Images were processed in Adobe Photoshop or ImageJ. To determine the position of the signal centers (see Figures 2F and 2G), the background was subtracted and the center of mass of a region of interest spanning the vast majority of relevant pixels was measured in ImageJ.
To measure cytoplasmic signal intensity by quantitative immunofluorescence microscopy, cells were imaged in one session under identical laser and detector settings. The average signal intensity in the cytoplasm (excluding centriole and chromosomes) was quantified using Leica confocal software (Leica Microsystems, Germany). In addition, the relative intensities between images were preserved whenever appropriate.
After transfection with HsSAS-6 or HsSAS-6-δKEN, cells with elevated levels of HsSAS-6 or HsSAS-6-δKEN were scored for the number of centrin foci. Care was taken to exclude satellites (Kubo et al., 1999), noted for their weak centrin staining intensity and their position away from stronger centrin foci and centriolar HsSAS-6. Cells with bulky aggregates of HsSAS-6 or HsSAS-6-δKEN, formed probably as a consequence of expression levels that were too high, were excluded from the analysis. Binucleated cells were also not considered, as they may have arisen from cytokinesis failure.
Cell Synchronization and FACS Analysis
To analyze endogenous HsSAS-6 during the cell cycle, HeLa cells were synchronized by a double thymidine block. Briefly, 2 mM thymi-dine was added to the culture medium for 19 hr, cells were incubated in fresh medium for 9 hr, and were incubated again in 2 mM thymidine for 16 hr. After release into the fresh medium, samples of synchronized cells were taken at successive time points. The DNA content of cells was analyzed after propidium iodide staining using a FACS instrument (FACScan, BD Biosciences).
To investigate the role of the 26S proteasome, cells were synchronized by a single thymidine block for 19 hr, then released for 10 hr to enrich for mitotic cells, after which MG132 (Sigma) (20 mM in DMSO) or DMSO alone as a control was added at a dilution of 1:1000 to the culture medium; cells were fixed and stained 30 min later.
Quantitative RT-PCR
RNA was isolated from synchronized HeLa cells using the RNeasy mini kit (QIAGEN, Chatsworth, CA) and cDNA was generated using oligo dT primers and the Omniscript RT kit (QIAGEN, Chatsworth, CA) according to the manufacturer’s instructions. Quantitative RT-PCR was performed on a LightCycler system (Roche, Mannheim, Germany) with FastStart DNA MasterPLUS SYBR Green I kit (Roche, Mannheim, Germany) and the following primers: CCAGCAGCAACACAATCAGAA and CCACAGGTTGGGTAAGTCAGTCT. The HsSAS-6 mRNA level was normalized with respect to both total RNA level and GAPDH level (not shown), with both sets of values being similar.
Immunoelectron Microscopy and Serial Section EM
Serial section EM was conducted as described (La Terra et al., 2005). In brief, cells depleted of HsSAS-6 or overexpressing HsSAS-6-δKEN were chosen, fixed in 1% glutaraldehyde, postfixed in 1% OsO4, and embedded using conventional protocols. Mitotic cells containing a single centrin dot or interphase cells with several centrin dots moving coordinately were chosen under the fluorescence microscope, and full 3D differential interference contrast (DIC) and fluorescence datasets (0.2 μm z-steps) were recorded immediately before fixation. The positions of cells were marked on the coverslip with a diamond-tipped objective scribe. After embedding, cells were relocated using phase-contrast microscopy and serially sectioned (10 nm sections). Sections were imaged in a Zeiss 910 electron microscope operated at 100KV. Images were recorded on film and then scanned using a flatbed scanner.
For immunoelectron microscopy analysis, HeLa cells were grown on plastic coverslips (Thermanox, Nunc, Naperville) for 24 hr, then lysed for 60 s with 1% Triton X-100 in PHEM buffer followed by fixation with cold methanol (-20°C) for 15 min. Dehydration, embedding in Lowicryl, labeling, and staining were carried out as described before (Euteneuer et al., 1998).
Supplementary Material
ACKNOWLEDGMENTS
We thank Michel Bornens, Hiroaki Ishikawa, Joachim Lingner, Jiri Lukas, Jeffrey L. Salisbury, and Viesturs Simanis for reagents. We are grateful to Peter Beard, Marie Delattre, Virginie Hachet, Daiju Kitagawa, Gregor Kohlmaier, and Viesturs Simanis for critical reading of the manuscript, as well as José Artacho, Claude Bonnard, and Nathalie Garin for microscopy support. We acknowledge financial support from Oncosuisse (OCS-01495-02-2004) to P.G. and from National Institutes of Health (GM59363) to A.K.
Footnotes
Note Added in Proof While this manuscript was under review, it was reported that HsSAS-6 is also required for centriole assembly in ciliated epithelial cells; Vladar and Stearns, (2007). J. Cell. Biol. 178, 31-42.
REFERENCES
- Adams IR, Kilmartin JV. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Chretien D, Buendia B, Fuller SD, Karsenti E. Reconstruction of the centrosome cycle from cryoelectron micrographs. J. Struct. Biol. 1997;120:117–133. doi: 10.1006/jsbi.1997.3928. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Euteneuer U, Graf R, Kube-Granderath E, Schliwa M. Dictyostelium gamma-tubulin: molecular characterization and ultra-structural localization. J. Cell Sci. 1998;111:405–412. doi: 10.1242/jcs.111.3.405. [DOI] [PubMed] [Google Scholar]
- Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yub-Kuboa A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Borisy GG. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 2005;168:713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gönczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J. Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Middendorp S, Kuntziger T, Abraham Y, Holmes S, Bordes N, Paintrand M, Paoletti A, Bornens M. A role for centrin 3 in centrosome reproduction. J. Cell Biol. 2000;148:405–416. doi: 10.1083/jcb.148.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Yamane Y, Okanoue T, Tsukita S, Tsukita S. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol. Biol. Cell. 2001;12:1687–1697. doi: 10.1091/mbc.12.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 2002;12:1287–1292. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- Sanders MA, Salisbury JL. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 1994;124:795–805. doi: 10.1083/jcb.124.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Lukas C, Kramer ER, Peters JM, Bartek J, Lukas J. Nonperiodic activity of the human anaphase-promoting complex-Cdh1 ubiquitin ligase results in continuous DNA synthesis uncoupled from mitosis. Mol. Cell. Biol. 2000;20:7613–7623. doi: 10.1128/mcb.20.20.7613-7623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YS. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 1982;98:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.