Abstract
The c-Myc oncogenic transcription factor (Myc) is pathologically activated in many human malignancies. Myc is known to directly upregulate a pro-tumorigenic group of microRNAs (miRNAs) known as the miR-17-92 cluster. Through the analysis of human and mouse models of B cell lymphoma, we show here that Myc regulates a much broader set of miRNAs than previously anticipated. Unexpectedly, the predominant consequence of activation of Myc is widespread repression of miRNA expression. Chromatin immunoprecipitation reveals that much of this repression is likely to be a direct result of Myc binding to miRNA promoters. We further show that enforced expression of repressed miRNAs diminishes the tumorigenic potential of lymphoma cells. These results demonstrate that extensive reprogramming of the miRNA transcriptome by Myc contributes to tumorigenesis.
Dysregulated expression or function of the oncogenic transcription factor Myc occurs frequently in human malignancies. Through the positive and negative regulation of an expansive network of target genes, Myc globally reprograms cells to drive proliferation and, in some settings, to induce cell death1-4. Myc uses distinct mechanisms for activating and repressing gene expression. When inducing transcription, Myc dimerizes with its binding partner Max and binds to genomic DNA directly upstream or within the first introns of target genes5,6. When repressing transcription, Myc does not seem to contact DNA directly. Rather, Myc is recruited through protein-protein interactions to core promoters, where it antagonizes the activity of positive regulators of transcription7. For example, Myc can bind to and inhibit the activity of the transcription factor Myc-interacting zinc finger protein 1 (Miz1), thereby preventing Miz1 from activating transcription of the CDKN1A (also known as p21WAF1/CIP1) and CDKN2B (p15INK4b) cell cycle-inhibitory genes8,9. Repression of other Myc targets is probably mediated through the ability of Myc to interact with and to antagonize the activity of additional transcriptional regulators, including Sp1, Smad2 and NF-Y (refs. 10-12).
miRNAs are a diverse family of RNA molecules, typically ~18-24 nucleotides in length, that have emerged as a class of Myc-regulated transcripts13. miRNAs regulate the stability and translational efficiency of partially complementary target mRNAs. miRNAs are initially transcribed by RNA polymerase II as long primary transcripts (pri-miRNAs) that are capped, polyadenylated and frequently spliced14,15. The mature miRNA sequences are located in introns or exons of pri-miRNAs, within regions that fold into hairpin structures of ~60-80 nucleotides. Although most pri-miRNAs are noncoding transcripts, some miRNAs are located within introns of protein-coding genes16. miRNA maturation requires a series of endonuclease reactions in which miRNA hairpins are excised from pri-miRNAs, the terminal loop of the hairpin is removed, and one strand of the resulting duplex is selectively loaded into the RNA-induced silencing complex (RISC)17. This miRNA-programmed RISC is the effector complex that carries out regulation of target mRNAs.
A large body of evidence has documented nearly ubiquitous dysregulation of miRNA expression in cancer cells18. These changes in miRNA expression are highly informative for the classification and prognosis of cancer19,20. In addition, altered expression of specific miRNAs has been shown to promote tumorigenesis. For example, a group of six co-transcribed miRNAs known as the miR-17-92 cluster is amplified in lymphoma and solid tumors21. These miRNAs are frequently overexpressed in tumors, promote proliferation in cell lines, and accelerate angiogenesis and tumorigenesis in mouse models of Myc-induced colon cancer and lymphoma22-25. Although select miRNAs are upregulated in cancer cells, global miRNA abundance seems to be generally reduced in tumors20. Downregulation of miRNA probably contributes to neoplastic transformation by allowing an increased expression of proteins with oncogenic potential26. Evidence suggests that a block in the first step of miRNA processing may contribute to the reduced abundance of select miRNAs in cancer cells27. Additional mechanisms of miRNA downregulation, including direct transcriptional repression, have not been investigated.
We previously showed that Myc directly activates transcription of the miR-17-92 cluster13. Through the analysis of human and mouse models of Myc-mediated lymphomagenesis, we have now identified a large set of additional Myc-regulated miRNAs. Unexpectedly, induction of Myc results primarily in widespread downregulation of miRNA expression. Chromatin immunoprecipitation (ChIP) studies indicate that Myc binds directly to promoters or conserved regions upstream of the miRNAs that it represses. We also show that expression of Myc-repressed miRNAs markedly impedes lymphoma cell growth in vivo. These observations suggest that repression of tumor-suppressing miRNAs is a fundamental component of the Myc tumorigenic program.
RESULTS
Identification of Myc-repressed miRNAs
We previously described the use of a spotted oligonucleotide array to identify the miR-17-92 cluster as a direct transcriptional target of Myc13. To determine whether Myc regulates additional miRNAs, we produced custom microarrays with an expanded set of probes capable of assaying the expression of 313 human miRNAs and 233 mouse miRNAs. Two models of Myc-mediated tumorigenesis were chosen for analysis. As in our previous studies, we used P493-6 cells, which are Epstein-Barr virus-immortalized human B cells that carry a tetracycline (tet)-repressible allele of Myc28. These cells are tumorigenic in immunocompromised mice and represent a model of human B cell lymphoma29. miRNA expression profiles were examined in the high-Myc (-tet) and low-Myc (+tet) states. miRNA expression was also assayed in a mouse model of Myc-induced B cell lymphoma. In this system, bone marrow from p53-null (Trp53-/-) mice is infected with a retrovirus that produces a fusion protein of Myc and the estrogen receptor (MycER). Infected cells form polyclonal B cell lymphomas in the presence of 4-hydroxytamoxifen (4-OHT), which activates the MycER fusion protein30,31. RNA from subcutaneous tumors with high-Myc activity (mice treated continuously with 4-OHT) and low Myc activity (mice from which 4-OHT was withdrawn after tumor formation) was analyzed (see Supplementary Tables 1 and 2 online for complete expression profiling data for both models). All miRNAs showing a twofold or greater upregulation or downregulation in the high-Myc state in both human and mouse models were chosen for further analysis. We also selected miRNAs that showed a 1.5-fold or greater change in expression in both models if either the miRNA or a related family member was known to be deleted or mutated in cancer, or if the expression of a related family member changed twofold or greater in both models.
Unexpectedly, the predominant consequence of Myc induction in both model systems was widespread repression of miRNA expression. Very few upregulated miRNAs satisfied the criteria for inclusion in the study. Consistent with our previous findings, miRNAs derived from the miR-17-92 cluster were upregulated more than twofold by Myc in both models. miR-7 was the only other consistently upregulated miRNA identified by the microarray experiments. However, we were unable to detect this miRNA by RNA blotting (data not shown), so it was not studied further. At least 13 downregulated miRNAs, potentially representing 21 distinct transcription units, satisfied our criteria for inclusion in the study (Table 1). Of these downregulated miRNAs, miR-15a, miR-22, miR-26a, miR-29c, miR-34a, miR-195 and let-7 have been found to be mutated in some cancers or are located in genomic regions known to be deleted in cancer19,32.
Table 1.
Candidate Myc-repressed miRNAs identified by microarray
| Criteria | miRNA | Transcription unita |
|---|---|---|
| Repressed >2-fold in both models | miR-22 | miR-22 |
| miR-26a | miR-26a-1; miR-26a-2 | |
| miR-29c | (miR-29b-2/miR-29c) | |
| miR-30e | (miR-30e/miR-30c-1) | |
| miR-146a | miR-146a | |
| miR-150 | miR-150 | |
| Repressed > 1.5-fold in both models miRNA or family member deleted or mutated in cancers |
||
| let-7 | 8 clusters (see Fig. 4a) | |
| miR-15a | (miR-15a/miR-16-1) | |
| miR-29a | (miR-29b-1/miR-29a) | |
| miR-34a | miR-34a | |
| miR-195 | (miR-497/miR-195) | |
| Family member repressed twofold or more in both models |
miR-26b | miR-26b |
| miR-30c | (miR-30a/miR-30c-2); | |
| (miR-30e/miR-30c-1) |
Individual transcription units are separated by semicolons; clustered miRNAs are indicated in parentheses.
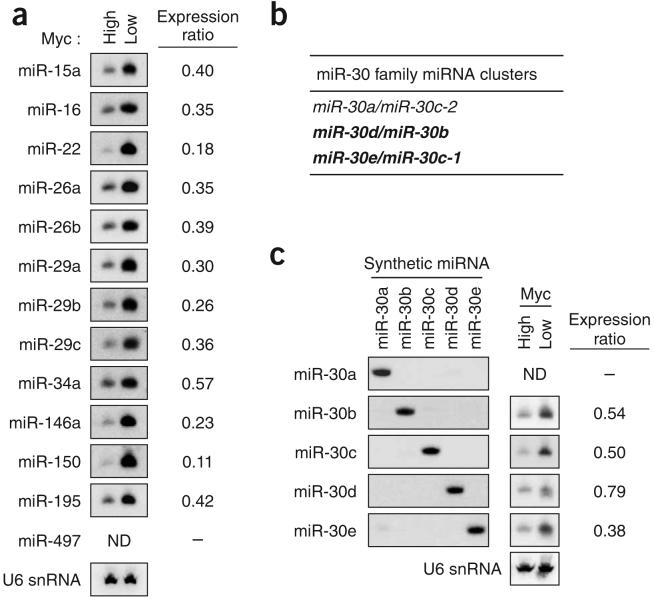
To confirm the expression changes detected by microarray analyses, RNA blotting was used to examine miRNA expression in P493-6 cells with high (-tet) and low (+tet) Myc expression (Fig. 1). Where more than one member of a miRNA family showed changes in expression (miR-26a and miR-26b, miR-29a and miR-29c, miR-30e and miR-30c, and members of the let-7 family), we considered the possibility that cross-hybridization contributed to the microarray signals. We previously established RNA blotting conditions that can assay specific members of the miR-29 family that differ by as few as two nucleotides33. We used these conditions to assay expression of miR-26a and miR-26b, which differ by three nucleotides, and of all other miRNAs with the exception of the more complex miR-30 and let-7 families (Fig. 1a). The results obtained by RNA blotting were highly concordant with those obtained by microarray. Three other miRNAs that are in clusters with the downregulated miRNAs were included in these RNA blotting studies (miR-16, miR-29b and miR-497). In most cases, clustered miRNAs behaved similarly (for example, miR-29a and miR-29b; miR-29b and miR-29c; and miR-15a and miR-16), with the exception of miR-497, which is clustered with miR-195 and was undetectable by microarray or RNA blotting.
Figure 1.
Repression of miRNA expression by Myc. (a) RNA blot analysis of miRNAs in P493-6 cells with high or low Myc expression. U6 small nuclear RNA (snRNA) was used as a loading control in this and all subsequent experiments (a representative blot is shown). ‘Expression ratio’ in this and subsequent figures indicates expression of the miRNA in the high-Myc state relative to the low-Myc state. ND, not detectable. (b) Organization of the human miR-30 clusters. miRNA clusters that are downregulated by Myc, as determined in c, are shown in bold. (c) RNA blots showing repression of miR-30 family members by Myc. Synthetic RNA oligonucleotides identical in sequence to each miR-30 family member and total RNA from P493-6 cells were hybridized with probes specific for each miRNA.
For the larger miR-30 and let-7 families, additional experiments were performed to establish specific hybridization conditions for each family member. Because of the considerable complexity of the let-7 family, analysis of this group of miRNAs is described separately below. The miR-30 family consists of five distinct mature miRNA sequences (miR-30a to miR-30e) organized in three clusters (Fig. 1b). Specific RNA blotting conditions were established by hybridizing probes to synthetic RNA oligonucleotides identical in sequence to each miR-30 family member (Fig. 1c). Endogenous miR-30a was not detected, suggesting that the miR-30a/miR-30c-2 cluster is not expressed in this cell line. The other two miR-30 clusters were expressed and down-regulated in the high-Myc state.
The expression of several miRNAs was examined further in MycER tumors, where the expected repression was also observed (Supplementary Fig. 1 online). We then determined whether human tumor cells associated with Myc overexpression show low expression of the repressed miRNAs. Analysis of a previously published miRNA expression profiling data set24 indicated that most Myc-repressed miRNAs are expressed at lower levels in Burkitt's lymphoma cells than in nontransformed B cells (Supplementary Fig. 2a online). In addition, inhibition of Myc expression using short hairpin RNA (shRNA) in a Burkitt's lymphoma cell line resulted in a modest but consistent upregulation of these miRNAs (Supplementary Fig. 2b,c).
Association of Myc with promoters of pri-miRNAs
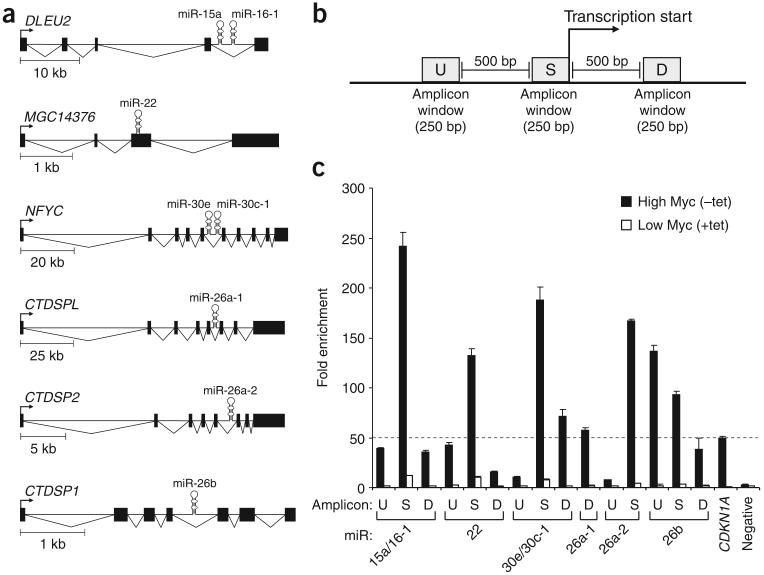
Studies have shown that Myc associates with the core promoters of the genes that it represses7. We therefore used ChIP to assay for the presence of Myc at the promoters of miRNAs downregulated in P493-6 cells. We first focused on miRNAs that are contained within pri-miRNAs with previously defined transcription start sites. Six such transcripts, encoding eight miRNAs (miR-15a, miR-16-1, miR-22, miR-30e, miR-30c-1, miR-26a-1, miR-26a-2 and miR-26b), were considered putative negative targets of Myc on the basis of our expression studies (Fig. 2a). Of note, a genome-wide analysis of Myc binding sites has shown association of Myc with the promoter of DLEU2, the primary transcript of the miR-15a/miR16-1 cluster34. Although expression of the miRNAs was not examined, expression of DLEU2 was shown to be reduced in the high-Myc state34.
Figure 2.
Myc associates with repressed pri-miRNA promoters. (a) Representations of repressed pri-miRNAs of known structure. (b) Real-time PCR amplicons for ChIP were designed within 250-bp windows immediately upstream of the transcription start site (amplicon S), 500-bp windows upstream of amplicon S (amplicon U) or 500-bp windows downstream of amplicon S (amplicon D). (c) Real-time PCR analysis of Myc chromatin immunoprecipitates. Fold enrichment in this and subsequent ChIP experiments represents the signal obtained after Myc immunoprecipitation relative to the signal obtained after immunoprecipitation with an irrelevant antibody. A validated Myc-bound amplicon in the promoter region of CDKN1A was used as a positive control. The 50-fold enrichment threshold for positive Myc binding is indicated as a broken line. Data are mean ± s.d. from three independent measurements.
To assay for Myc binding, we designed real-time PCR amplicons within three 250-bp windows near the transcription start sites of the miRNA transcripts: amplicon S, located immediately upstream of the transcription start site; amplicon U, located 500 bp upstream of amplicon S; and amplicon D, located 500 bp downstream of amplicon S (Fig. 2b). Owing to the high G+C content of the promoters of miR-26a-1 and miR-26a-2, only a subset of these amplicons could be designed for these miRNAs. As a positive control, an amplicon was designed within the promoter region of CDKN1A, a validated down-regulated target of Myc8. We observed 50-fold enrichment of the CDKN1A promoter amplicon in Myc ChIP samples as compared with ChIP samples generated with an irrelevant antibody (Fig. 2c). We therefore set 50-fold enrichment as the threshold for positive Myc binding in all subsequent studies. Signals above this threshold were obtained near the transcription start sites for each of the six pri-miRNAs assayed (Fig. 2c), providing strong evidence for the association of Myc with these promoters. These signals were greatly reduced when Myc expression was inhibited by treatment with tetracycline, demonstrating the specificity of these findings.
Myc association with conserved regions upstream of miRNAs
The remaining downregulated miRNAs, with the exception of some of the let-7 miRNA transcription units (see later), have unmapped transcription start sites, and therefore identification of the associated Myc binding sites required a different strategy. As illustrated by the pri-miRNAs in Figure 2a, miRNA promoters may be located a few kilobases to more than 50 kb upstream of the miRNAs. Because miRNAs are, in general, highly conserved, we reasoned that their promoters would tend to be conserved. We therefore selected conserved candidate regions upstream of miRNAs in which to assess Myc binding. As an initial test of this strategy, we examined the miR-29b-2/miR-29c cluster. Using Vista software, we identified a clear region of conservation ~20 kb upstream of these miRNAs (Fig. 3a, amplicon C). ChIP analysis in P493-6 cells revealed strong association of Myc specifically with this conserved region (Fig. 3b). Myc was not bound to nearby nonconserved regions (Fig. 3a, amplicon N), demonstrating the specificity of this finding.
Figure 3.
Myc associates with conserved regions upstream of repressed miRNAs. (a) Phylogenetic conservation of the intergenic region containing the miR-29b-2/miR-29c cluster. Vista was used to generate pairwise alignments between the genomic sequence from human (May 2004 assembly) and that from the indicated species. The graph is a plot of nucleotide identity for a 100-bp sliding window centered at a given position. Annotated transcripts produced from this locus are shown at the top. Note that the 5′ end of miR-29b-2/miR-29c is toward the right. Arrows indicate locations of the real-time PCR amplicons used for ChIP experiments. (b) Real-time PCR analysis of Myc chromatin immunoprecipitates (see Fig. 2c). The conserved amplicon that showed maximal Myc binding (C) and a representative negative control amplicon (N) are shown for each miRNA. Locations of these and additional amplicons for the miR-29b-1/miR-29a cluster, the miR-30d/miR-30b cluster, miR-34a, miR-146a, the miR-195/miR-497 cluster and miR-150 are shown in Supplementary Figures 3-8. (c) Conserved Myc binding sites correspond to pri-miRNA promoters. Shown are the structures of pri-miRNA transcripts, as defined by 5′ and 3′ RACE. For some transcripts, alternative splicing gives rise to major and minor isoforms. Plots representing evolutionary conservation, shown below each transcript, were taken from the UCSC Genome Browser (human genome May 2004 assembly). Arrows indicate the locations of ChIP amplicons that yielded the highest Myc binding signals.
We used the same strategy to assess Myc binding upstream of the remaining downregulated miRNAs. We found evidence for Myc binding to conserved regions upstream of the miR-29b-1/miR-29a cluster, the miR-30d/miR30b cluster, miR-34a and miR-146a (Fig. 3b and Supplementary Figs. 3-6 online). Strong Myc binding was also observed upstream of the miR-195/miR-497 cluster. Because this binding site is also near the transcription start site of BCL6B, however, we cannot rule out the possibility that Myc binding leads to the regulation of BCL6B transcripts, not the miRNAs (Supplementary Fig. 7 online). Despite assaying several amplicons, we obtained no evidence for Myc binding in the vicinity of miR-150 (Supplementary Fig. 8 online). As a further negative control, we assayed for Myc binding at six conserved sites upstream of the miR-30a/miR-30c-2 cluster, which is not expressed in P493-6 cells (Fig. 1c). As expected, none of these amplicons yielded positive ChIP signals (Supplementary Fig. 9 online).
Given that Myc bound in the vicinity of the transcription start sites of six out of six tested miRNA transcription units of known structure (Fig. 2), it seemed likely that the conserved Myc binding sites that we identified upstream of miR-29b-1/miR-29a, miR-29b-2/miR-29c, miR-30d/miR-30b, miR-34a, miR-146a and possibly miR-195/miR-497 were within miRNA promoters. To test this possibility directly, we used RACE to characterize completely a subset of these pri-miRNAs. For three of the transcripts, spliced ESTs were available to use as a starting point for RACE; in addition, we previously reported the complete structure of the primary transcript of miR-34a (ref. 35). For each of these pri-miRNAs, the experimentally determined 5′ end corresponded precisely to the conserved site that showed maximal Myc binding (Fig. 3c). Another study has also defined an identical transcription start site for miR-146a (ref. 36).
In summary, we identified sites bound by Myc upstream of 12 of 13 repressed miRNA transcription units of both known and unknown structure. In ten of these cases, the Myc binding site was determined to correspond precisely to the 5′ end of the pri-miRNA. These findings indicate that much of the repression of miRNAs observed in the high-Myc state is likely to be a direct consequence of Myc binding to miRNA promoters.
Dissection of regulatory control of let-7 miRNA clusters
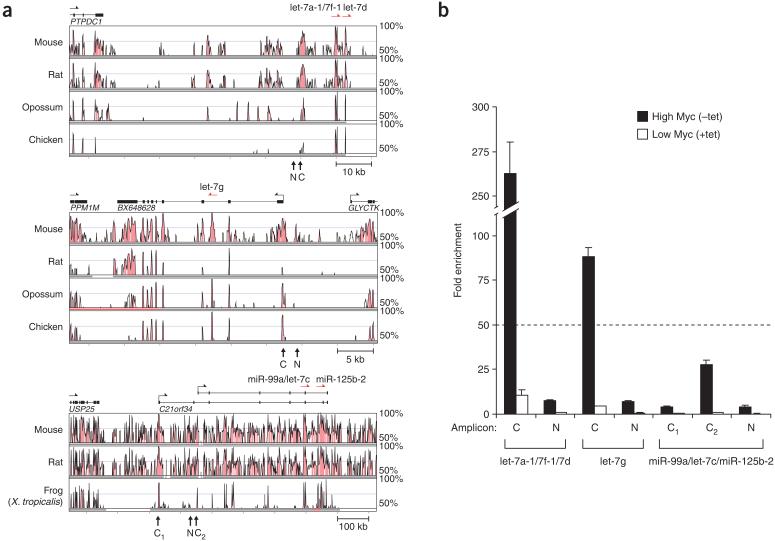
The miRNAs downregulated in the high-Myc state included members of the let-7 family, which comprises nine highly related mature miRNA sequences produced from eight different transcription units (Fig. 4a). let-7 miRNAs are known to be downregulated in lung tumors, and evidence suggests that they possess tumor suppressor activity37-39. We established hybridization conditions specific for nearly all of the human let-7 miRNAs by hybridizing DNA oligonucleotide probes against synthetic RNA oligonucleotides identical in sequence to each let-7 family member (Fig. 4b). Specific hybridization conditions were also identified for members of the miR-99/miR-100 family, which are clustered with a subset of let-7 miRNAs (Fig. 4c). Three let-7 clusters also include members of the miR-125 family that are sufficiently different to distinguish by standard RNA blotting conditions (seven nucleotides differ between miR-125a and miR-125b). We detected the expression of let-7a, let-7d, let-7g, miR-99a and miR-125b in P493-6 cells, and all of these miRNAs were downregulated in the high-Myc state (Fig. 4b-d). The remaining miRNAs assayed were not detectable. These data are most consistent with expression of only the let-7a-1/let-7f-1/let-7d cluster, the miR-99a/let-7c/miR-125b-2 cluster, and let-7g in this cell line.
Figure 4.
let-7 miRNAs are downregulated by Myc. (a) Organization of the human let-7 clusters. miRNA clusters downregulated by Myc, as determined in b-d, are shown in bold. (b-d) RNA blot analysis of synthetic RNA oligonucleotides or total RNA from P493-6 cells performed with probes specific for each member of the let-7 family (b), the miR-99/miR-100 family (c), and the miR-125 family (d). ND, not detectable.
We used ChIP to assess Myc binding to promoters or conserved sites upstream of these miRNA transcription units. Strong evidence was obtained for Myc binding to a conserved site upstream of the let-7a-1/let-7f-1/let-7d cluster, which is contained within a pri-miRNA that has not been characterized, and to the transcription start site of the let-7g pri-miRNA (Fig. 5). Signals above the 50-fold enrichment threshold were not obtained at either of two alternative transcription start sites for the miR-99a/let-7c/miR-125b-2 pri-miRNA, suggesting that this transcript is not a direct target of Myc.
Figure 5.
Myc binds to conserved regions upstream of let-7 miRNAs. (a) Vista analysis of phylogenetic conservation encompassing the let-7a-1/let-7f-1/let-7d cluster, let-7g and the miR-99a/let-7c/miR-125b-2 cluster (see Fig. 3a). (b) Real-time PCR analysis of Myc chromatin immunoprecipitates (see Fig. 2c).
Repressed miRNAs disadvantage lymphoma cell growth in vivo
To examine whether downregulation of specific miRNAs contributes to Myc-mediated tumorigenesis, we used an in vivo selection model of B cell lymphomagenesis40. Retroviral expression vectors were first generated by cloning individual human miRNAs or miRNA clusters into a derivative of the murine stem cell virus (MSCV-PIG) that also expresses green fluorescent protein (GFP; ref. 41 and Fig. 6a). Ten distinct miRNA expression constructs were generated (miR-15a/miR-16-1, miR-22, miR-26a-2, miR-29b-1/miR-29a, miR-30b, miR-34a, miR-146a, miR-150, miR-195/miR-497 and let-7a-1/let-7f-1). This set included all unique miRNAs that were downregulated in the high-Myc state and at least one member of each downregulated miRNA family. Each of the mature miRNA sequences is identical in human and mouse. Retroviral constructs were used to infect Myc3 cells, a B cell lymphoma line generated by expressing Myc in bone marrow from Trp53-/- mice42. To determine the consequences of expressing these miRNAs in the setting of transformation by other oncogenes, 38B9 cells, pro-B cells transformed by the v-Abl (Abl1) oncogene43, were used in a parallel series of experiments. Retroviral infection conditions were adjusted to achieve a level of ~50% GFP positivity in recipient cells, and these mixed cultures were injected subcutaneously into severe combined immunodeficient (SCID) mice. After ~3 weeks, the resulting tumors were removed and the percentage of remaining GFP-positive cells was measured. Expression of miRNAs that inhibit tumorigenesis will impart a selective disadvantage to retrovirally infected cells and will therefore result in a decrease in the percentage of GFP-positive cells in tumors.
Figure 6.
Expression of Myc-repressed miRNAs disadvantages lymphoma cell growth in vivo. (a) Myc3 or 38B9 lymphoma cells were infected with a retrovirus expressing a miRNA and GFP. The percentage of GFP-positive cells was measured before and after tumor formation. (b) Cells expressing select miRNAs are eliminated from tumors. Data are mean ± s.d. from three independent trials. At least 30% of cells were positive for GFP before injection into recipient mice.
To assess whether retroviral expression produced physiologically relevant levels of mature miRNAs, we compared the expression of miRNAs in retrovirally infected Myc3 and 38B9 cells to endogenous expression in the nontransformed pro-B cell line YS-PB11 (ref. 44 and Supplementary Fig. 10 online). Expression of miR-150, which is not expressed in YS-PB11, was compared to that in MycOFF tumors. For nearly all miRNAs, retroviral expression levels ranged from 0.6 to 6 times those observed in the physiologic setting. Higher expression was observed for miR-22 in both tumor cell lines and for miR-195 in 38B9 cells, and therefore the results obtained with these viruses in these settings must be interpreted with caution.
We could not establish stably infected cell populations with the let-7a-1/let-7f-1, miR-29b-1/29a and miR-146a viruses. This result may indicate that these miRNAs impose strong negative selection during in vitro cell growth, although this might also be a consequence of inefficient packaging of these viruses. For the remaining viruses, we attained 30-70% infection of recipient cells, as assessed by GFP positivity. The percentage of GFP-positive cells in Myc3 and 38B9 cell populations infected with empty, miR-18a or miR-30b viruses remained constant before and after tumor formation (Fig. 6b). In contrast, Myc3 and 38B9 cells infected with miR-34a, miR-150, miR-195/miR-497 and miR-15a/miR-16-1 viruses were nearly eliminated from tumors, indicating that these miRNAs possess tumor-suppressing properties in the setting of both Myc- and v-Abl-mediated transformation. miR-26a inhibited tumori-genesis specifically in Myc-transformed cells, whereas miR-22 expression affected tumori-genesis only in v-Abl-transformed cells. Notably, there was no correlation between the magnitude of miRNA expression and the phenotype observed, indicating that these results are unlikely to represent an artifact of retroviral overexpression. For example, miR-15a/miR-16-1, which had one of the strongest negative effects on tumorigenesis in both cell lines, showed the lowest retroviral expression levels (Supplementary Fig. 10). These data demonstrate that several of the miRNAs that Myc represses have tumor-suppressing activity in the setting of both Myc-mediated transformation and transformation by other oncogenes. Future work is necessary to determine whether the anti-tumorigenic properties of these miRNAs results from their ability to downregulate a housekeeping gene that is rate limiting for proliferation or from the specific inhibition of pro-oncogenic pathways.
To determine whether downregulation of anti-tumorigenic miRNAs correlates with enhanced cellular proliferation after Myc activation, we examined the kinetics of miRNA repression in P493-6 cells (Supplementary Fig. 11 online). These cells do not begin proliferating until 48 h after tetracycline removal and do not reach maximal growth rates until at least 72 h after Myc induction45. We observed substantial downregulation of miRNAs by these time points, consistent with a requirement for their repression to precede Myc-induced proliferation.
DISCUSSION
Pathologically activated expression of Myc is one of the most common oncogenic events in human cancers4. Here we have shown that a major consequence of Myc activation is extensive reprogramming of the miRNA expression pattern of tumor cells. Although the protumorigenic miR-17-92 cluster is directly upregulated by Myc13, our current findings unexpectedly show that the predominant influence of Myc on miRNA expression is widespread downregulation. Repression of miRNA expression by Myc is consistent with the previous observation that miRNA levels are globally reduced in tumors20. Although it has been demonstrated that a block in miRNA biogenesis contributes to the suppression of specific miRNAs in cancer27, our findings suggest that direct transcriptional repression is also likely to contribute to this reduction in miRNA abundance.
Much evidence supports the conclusion that miRNA repression favors Myc-mediated tumorigenesis. First, several of the miRNAs downregulated by Myc are mutated or located in regions known to be deleted in cancer, suggesting that they act as tumor suppressors19,32. Indeed, the anti-tumorigenic properties of several Myc-repressed miRNAs have been extensively documented. For example, miR-15a and miR-16-1 are deleted or downregulated in over two-thirds of individuals with chronic lymphocytic leukemia, and they target the anti-apoptotic gene BCL2 (refs. 46,47). Members of the let-7 miRNA family target the RAS oncogene and are frequently downregulated in lung cancer37-39. Evidence has implicated miR-34a as a crucial component of the p53 tumor suppressor network with potent anti-proliferative and pro-apoptotic activity35,48,49. Repression of these miRNAs by Myc is likely to be an important mechanism contributing to their reduced function in cancer cells. In addition, we have shown that several Myc-repressed miRNAs have marked anti-tumorigenic activity in a mouse model of B cell lymphoma. In particular, miR-26a, miR-150 and miR-195/miR-497 were previously unknown to have tumor-suppressing properties. Taken together, the available data support the idea that the control of miRNA expression has an important role in Myc-mediated tumorigenesis. In addition, given recent successes in the systemic delivery of small RNAs to animals, these results raise the possibility that delivery of Myc-repressed miRNAs might represent a therapeutic strategy for cancer50. Indeed, these findings suggest that re-expression of even a single crucial miRNA may be sufficient to block tumor formation.
Our study also highlights the importance of careful dissection of the regulatory control of related miRNAs both in cancer and in other biological processes. miRNAs frequently exist in multiple highly related or identical copies distributed throughout the genome of a given species. This organization is exemplified by the nine distinct miRNAs of the let-7 family, which are produced from eight individual transcription units in humans. Although previous studies have observed downregulation of let-7 miRNAs in cancer37-39, the expression of individual let-7 transcription units, and therefore the origin of let-7 miRNAs in a given tumor, has rarely been examined. We have shown here the feasibility of dissecting the complex regulatory control of these miRNAs. Because related miRNAs do not always have identical functions33,51, characterization of the specific miRNA family members that are dysregulated in a given tumor type is a necessary prerequisite for elucidating their roles in cancer pathogenesis.
Lastly, our data provide insight into the significance of the nearly ubiquitous dysregulation of miRNA expression that has been observed in diverse subtypes of cancer. Our results indicate that these abnormal miRNA expression patterns cannot be explained solely as an indirect consequence of the loss of cellular identity that accompanies malignant transformation. Rather, oncogenic events appear to directly reprogram the miRNA transcriptome to favor tumorigenesis. In light of the importance of miRNA regulation in Myc-mediated tumorigenesis, future studies are warranted to determine the extent to which other oncogenes have incorporated miRNA regulation into their tumorigenic programs.
METHODS
Cell culture
We cultured P493-6 cells (see Acknowledgments) in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin and streptomycin. To repress Myc expression, cells were grown in the presence of 0.1 μg/ml of tetracycline (Sigma) for 72 h. Mouse lymphoma cells with high and low Myc expression were obtained as described30,31.
miRNA microarray analysis
Custom microarrays containing oligonucleotide probes complementary to 313 human miRNAs or 233 mouse miRNAs were synthesized by Combimatrix. Probes containing two mismatches were included for all miRNAs. Array hybridization and data analysis were performed as described35. Signals that were less than twice the background were removed from subsequent analyses (listed as ‘zero’ in Supplementary Tables 1 and 2). For miRNA profiling of mouse B cell lymphomas, two tumors with high Myc and two tumors with low Myc expression were analyzed. miRNAs that were absent in three of the four tumors, or absent in one of each of the high-Myc and low-Myc tumors, were removed from subsequent analyses. Fold-change values were calculated for all four pairwise comparisons between the high-Myc and low-Myc tumors and then averaged to generate a mean fold-change value.
RNA blotting
For all miRNAs except those of the miR-30, miR-99/miR100 and let-7 families, RNA blotting was performed as described33 using Ultrahyb-Oligo (Ambion) and DNA oligonucleotide probes complementary to the mature miRNA sequences. To establish specific hybridization conditions for related miRNAs, 1 μl of 10 nM RNA oligonucleotide was separated on polyacrylamide gels and probed as above. Blots were washed once in 2× SSC plus 0.5% SDS at 42 °C, and a second time at a higher temperature such that less than 10% cross-hybridization was observed. Specific wash temperatures for each probe are listed in Supplementary Table 3 online.
Myc knockdown in Burkitt's lymphoma cells
We transfected 293T packaging cells with a pLKO.1-Puro lentivirus expressing a shRNA targeting Myc or a control shRNA (Sigma). EW36 cells were infected three times with lentiviral supernatant. At 48 h after the initial infection, cells were selected in puromycin for 48 h before total RNA and protein were collected.
ChIP and quantitative real-time PCR
We carried out ChIP as described13. Real-time PCR was performed using an ABI 7900 Sequence Detection System with the SYBR Green PCR core reagent kit (Applied Biosystems). Sequences of primers used to amplify ChIP samples are provided in Supplementary Table 4 online.
RACE mapping of miRNA primary transcripts
The GeneRacer kit (Invitrogen) was used to characterize the miR-29b-2/miR-29c, miR-29b-1/miR-29a and miR-146a primary transcripts. Before total RNA was isolated for use in these assays, expression of Drosha was inhibited by electroporating previously described short interfering RNAs33 into tetracycline-treated P493-6 cells. Electroporation was performed as described45. Primer sequences are provided in Supplementary Table 4.
Tumorigenesis assays
The miRNAs and at least 100 bp of flanking sequence were amplified from genomic DNA and cloned into the XhoI site of the retroviral vector MSCV-PIG41. Primer sequences are provided in Supplementary Table 4. Correct vector construction was verified by direct sequencing. Retroviral infection of Myc3 and 38B9 cells, flow cytometry and tumor formation were performed as described40. All mouse experiments were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Accession numbers
The sequences of miRNA primary transcripts have been deposited in GenBank under the following accession numbers: miR-29b-1/miR-29a cluster, EU154353; miR-29b-2/miR-29c cluster, EU154351 and EU154352; and miR-146a, EU147785. Microarray data have been deposited in the Gene Expression Omnibus database under accession number GSE9129.
URLs
UCSC Genome Browser, http://genome.ucsc.edu/; Vista software package, http://genome.lbl.gov/vista/index.shtml.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Eick (GSF Research Centre, Munich) for P493-6 cells; S. Hammond and M. Thomson for assisting with miRNA arrays and for providing Burkitt's lymphoma miRNA expression data; and S. McMahon for advice and reagents for Myc knockdown experiments; and K. O'Donnell and members of the Mendell lab for critical reading of the manuscript. J.T.M. is a Rita Allen Foundation Scholar and receives support from the Lustgarten Foundation for Pancreatic Cancer Research. This work was also supported by grants from the National Institutes of Health (R01CA120185 to J.T.M., and R01CA122334 and R01CA102709 to A.T.-T.).
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
References
- 1.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 2.Cole MD, McMahon SB. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 3.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 4.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 5.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 6.Zeller KI, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleine-Kohlbrecher D, Adhikary S, Eilers M. Mechanisms of transcriptional repression by Myc. Curr.Top.Microbiol. Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- 8.Seoane J, Le HV, Massague J. Myc suppression of the p21Cip1 Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 9.Staller P, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 10.Feng XH, Liang YY, Liang M, Zhai W, Lin X. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-β-mediated induction of the CDK inhibitor p15Ink4B. Mol. Cell. 2002;9:133–143. doi: 10.1016/s1097-2765(01)00430-0. [DOI] [PubMed] [Google Scholar]
- 11.Gartel AL, et al. Myc represses the p21WAF1/CIP1 promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. USA. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izumi H, et al. Mechanism for the transcriptional repression by c-Myc on PDGF β-receptor. J. Cell Sci. 2001;114:1533–1544. doi: 10.1242/jcs.114.8.1533. [DOI] [PubMed] [Google Scholar]
- 13.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 14.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 18.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 20.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 21.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 22.Dews M, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 24.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 27.Thomson JM, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pajic A, et al. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int. J. Cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 29.Gao P, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu D, Dews M, Park A, Tobias JW, Thomas-Tikhonenko A. Inactivation of Myc in murine two-hit B lymphomas causes dormancy with elevated levels of interleukin 10 receptor and CD20: implications for adjuvant therapies. Cancer Res. 2005;65:5454–5461. doi: 10.1158/0008-5472.CAN-04-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu D, Thomas-Tikhonenko A. A non-transgenic mouse model for B-cell lymphoma: in vivo infection of p53-null bone marrow progenitors by a Myc retrovirus is sufficient for tumorigenesis. Oncogene. 2002;21:1922–1927. doi: 10.1038/sj.onc.1205244. [DOI] [PubMed] [Google Scholar]
- 32.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 34.Mao DY, et al. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 2003;13:882–886. doi: 10.1016/s0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- 35.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 39.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 40.Yu D, Cozma D, Park A, Thomas-Tikhonenko A. Functional validation of genes implicated in lymphomagenesis: an in vivo selection assay using a Myc-induced B-cell tumor. Ann. NY Acad. Sci. 2005;1059:145–159. doi: 10.1196/annals.1339.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemann MT, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 42.Yu D, et al. Oscillation between B-lymphoid and myeloid lineages in Myc-induced hematopoietic tumors following spontaneous silencing/reactivation of the EBF/Pax5 pathway. Blood. 2003;101:1950–1955. doi: 10.1182/blood-2002-06-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MuLV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981;27:381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- 44.Lu LS, Auerbach R. Characterization and differentiation of an early murine yolk sac-derived IL-7-independent pre-pro-B cell line. J. Immunol. 1998;161:1284–1291. [PubMed] [Google Scholar]
- 45.O'Donnell KA, et al. Activation of transferrin receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis. Mol. Cell. Biol. 2006;26:2373–2386. doi: 10.1128/MCB.26.6.2373-2386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 50.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 51.Leaman D, et al. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.