Abstract
The genetic structures of past human populations are obscured by recent migrations and expansions, and can been observed only indirectly by inference from modern samples. However, the unique link between a heritable cultural marker, the patrilineal surname, and a genetic marker, the Y chromosome, provides a means to target sets of modern individuals that might resemble populations at the time of surname establishment. As a test case, we studied samples from the Wirral peninsula and West Lancashire, in northwest England. Place names and archaeology show clear evidence of a past Viking presence, but heavy immigration and population growth since the Industrial Revolution are likely to have weakened the genetic signal of a thousand-year-old Scandinavian contribution. Samples ascertained on the basis of two generations of residence were compared with independent samples based on known ancestry in the region, plus the possession of a surname known from historical records to have been present there in medieval times. The Y-chromosomal haplotypes of these two sets of samples are significantly different, and in admixture analyses the surname-ascertained samples show markedly greater Scandinavian ancestry proportions, supporting the idea that northwest England was once heavily populated by Scandinavian settlers. The method of historical surname-based ascertainment promises to allow investigation of the influence of migration and drift over the last few centuries in changing the population structure of Britain, and will have general utility in other regions where surnames are patrilineal and suitable historical records survive.
Keywords: Human, Y chromosome, surnames, population, Vikings, admixture
Introduction
Studies of the human past draw on lines of evidence from many fields, including paleoclimatology, archaeology, paleontology, linguistics, and history (Jobling, Hurles, and Tyler-Smith 2004), and recent years have seen an increasing influence of genetics. Unlike most other sources of evidence, genetic studies generally rely upon the collection of data from contemporary sources - modern, living populations - and use various statistical methods of inference to make deductions about events many generations ago. However, modern populations have been influenced by recent migration and expansion, associated with events such as the Industrial Revolution, and may provide a blurred and misleading picture of the past. Ancient DNA methods have promised direct access to the genetic diversity of past populations, but unfortunately the practical difficulties of retrieving verifiable and contamination-free DNA data mean that sample sizes are inconveniently small for population genetic analysis (Topf et al. 2006), except in rare cases of exceptional preservation (Keyser-Tracqui, Crubézy, and Ludes 2003).
A possible alternative approach is suggested by the link between a heritable cultural marker, the patrilineal surname, and an informative genetic marker, the non-recombining region of the Y chromosome, demonstrated through studies of men sharing surnames (Sykes and Irven 2000; King et al. 2006; McEvoy and Bradley 2006). Although the link between surname and Y-chromosomal haplotype is imperfect, due to multiple founders for names, and historical nonpaternities and adoptions (Jobling 2001), unrelated men sharing surnames are significantly more likely to share haplotypes than are men carrying different names. This demonstrates that surnames have been associated with specific haplotypes for many generations, and suggests that access to the Y-chromosomal diversity of past populations might be possible through the selection of modern samples based on surnames known to exist in a particular region during the medieval period. A similar approach has been proposed independently (Manni et al. 2005), based on a spatial analysis of ∼200-year old Netherlands surnames.
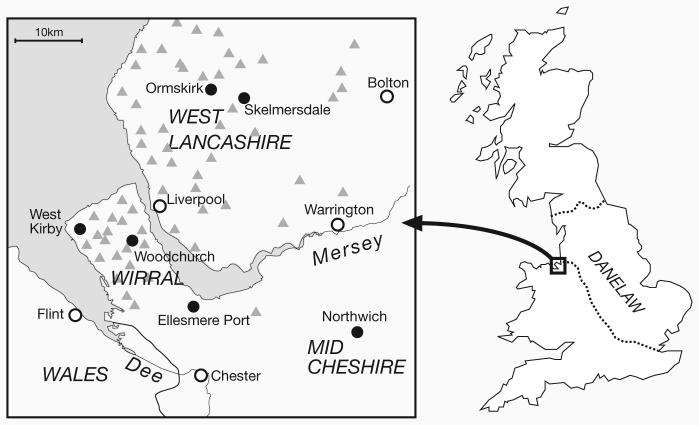
A useful test case to evaluate the potential of surname-based ascertainment is provided by the thousand-year-old Viking settlement of the British Isles. The Viking age of raiding, exploration, trading and colonization began in the late eighth century, with a series of attacks on the coasts of Britain, Ireland and France. In England, a shift from raiding to permanent settlement began after 865AD. The modern consensus has moved from the earlier view of a mass migration of Viking settlers, to a more geographically variable and gradual process, with much assimilation of local culture, under the administration of a Scandinavian elite (Richards 2004). Even though Viking rule in England came to an end nearly a thousand years ago, and the settlers were soon integrated linguistically and culturally, abundant evidence of Scandinavian influence remains today. Important archaeological findings exist, but the most striking evidence is linguistic and onomastic – many words in standard English and in local dialects, and many place-names, are of Scandinavian origin. In some areas of English counties that formed part of the Danelaw (Figure 1), the region under the administrative control of the Vikings from the late 9th century, up to 70% of major place-names are in this category, with endings such as –by and –thorp(e).
Figure 1. Location of the Wirral peninsula and West Lancashire.
Sampling ocations for the ‘modern’ samples are shown by filled circles, and unfilled circles show major towns for orientation. Gray triangles show the locations of Scandinavian major place-names (Harding 2002).
Attempts to identify a corresponding genetic legacy of Viking settlement in modern populations of the British Isles have focused on North Atlantic islands with archaeologically and culturally defined histories of Viking contact (Helgason et al. 2000a; Helgason et al. 2000b; Helgason et al. 2001; Wilson et al. 2001; Capelli et al. 2003; Goodacre et al. 2005). These studies have exploited the phylogeographic resolution of the paternally inherited Y chromosome, and the maternally inherited mitochondrial DNA to reveal strong signals of Scandinavian admixture. For example, the proportion of Scandinavian patrilineal ancestry in Iceland has recently been estimated at 75%, and this proportion has been described as declining through Shetland (44.5%), Orkney (31%), the Western Isles and Skye (22.5%), to the coastal regions of northern and western Scotland (15%) (Goodacre et al. 2005).
In mainland Britain, abundant evidence exists for a past Viking presence in the Wirral peninsula and West Lancashire, in the northwest of England. Scandinavian major place-names are common (Cavill, Harding, and Jesch 2000; Harding 2002) (Figure 1), and these two regions possess the only definite examples of the place-name ‘Thingwall’ (from Old Norse þing-völlr meaning ‘Assembly Field’) in England, indicating settlements of sufficient density and autonomy to warrant their own parliaments. Moreover, in the case of Wirral, the intensity and distribution of minor place name elements attests to the persistence of a Scandinavian-influenced dialect through the centuries that may reflect the intensity of the original settlement (Wainwright 1943; Cavill, Harding, and Jesch 2000). This onomastic evidence is supported by archaeological discoveries such as jewelry, weaponry or treasure hoards at Meols, Crosby, and Cuerdale (Graham-Campbell 1992; Harding 2002) or Hiberno-Norse ring-headed crosses (Bu'Lock 1958) and hogback tombstones (Collingwood 1928; Bailey et al. 2006) which reflect Scandinavian presence.
In one version of events, Vikings of Norwegian origin, under their leader Ingimund, arrived in the region in 902AD after having been expelled from Dublin. Æthelflæd, Lady of the Mercians, granted them land in north Wirral, where they settled (Wainwright 1942; Wainwright 1948; Wainwright 1975). More complex models have also been proposed, based on place-names analysis, including elements of migration from the Isle of Man as well as Dublin (Fellows-Jensen 1992). Specific historical evidence for the migration of Vikings into West Lancashire is lacking, although place-names and archaeological evidence clearly demonstrate that this occurred.
Widespread migration and population expansion in recent centuries presents a particular problem in genetic analyses of mainland regions of Britain. The Wirral experienced a major influx of people since the medieval period, its population growing over seventy-fold between 1545 and 1921 (Roberts 2002) – almost ten times the national average increase. In conventional sampling strategies, where two generations of residence in an area are sufficient to qualify a DNA donor for participation in a study, the signal of Viking influence is likely to be weakened by the noise of more recent population movement.
Here we exploit the relationship between patrilinearly inherited surnames and Y-chromosomal haplotype, using surname-based ascertainment to ask if targeted sub-sampling of modern populations can retrieve samples that more accurately represent the structures of populations in the past. Historical documents can provide lists of relevant surnames, and in our study we have used such sources to provide a set of names present in the Wirral and West Lancashire prior to 1572. We show that samples taken under these criteria are significantly different from samples taken on the two generations of residence criterion. They preserve a markedly stronger signal of Scandinavian admixture than their modern counterparts, support the idea the northwest England was once heavily populated by Scandinavian settlers, and validate the surname-ascertainment approach.
Materials and Methods
DNA samples
Buccal samples were taken with informed consent, and DNA extracted as described (King et al. 2006).
Samples were collected from Mid-Cheshire, the Wirral and West Lancashire under the two generations of residence criterion.
Independent samples were also collected from the Wirral and West Lancashire based upon a patrilineal residence criterion and possession of a surname known from historical documentary sources to have been present in the region prior to 1572. For the Wirral, we compiled a list of 236 surnames from the 1545 subsidy rolls (Irvine and Sanders 1894), listing all households in the Wirral recorded during the reign of Henry VIII, supplemented with two surnames from records of court proceedings dating from 1353 (Booth 1983), one from ale-house licensing records from 1572 (Bennett and Dewhurst 1940) and three based on specific old local place-names. For West Lancashire, we used a list of 232 surnames based on a list of inhabitants of Ormskirk, Scarisbrick-with-Hurlton, Bickerstaffe, Burscough-with-Marton, Westhead-with-Lathom and Skelmersdale who promised to contribute to the stipend of the priest of the altar of Our Lady at Ormskirk in 1366 (Peet 1991); this was supplemented with 37 surnames based on specific old local place-names (Ekwall 1922; McKinley 1981), and allowing additional surnames containing particular old place-name elements of Scandinavian origin. Some surnames were converted into their current equivalents to allow matching to modern donors' names. Within each sample, duplicate surnames (including known spelling variants) were avoided, though some surnames are found in both Wirral and West Lancashire samples, carried by different and, to our knowledge, unrelated individuals.
Comparative data were also taken from the literature (Capelli et al. 2003).
Y-chromosomal haplotyping
Binary markers (Y Chromosome Consortium 2002) shown in Figure 2 were typed hierarchically using the SNaPshot minisequencing procedure (Applied Biosystems) and an ABI3100 Genetic Analyzer (Applied Biosystems). Initially all chromosomes were typed with Multiplex I of Bosch et al. (Bosch et al. 2006) containing markers M9, M69, M89, M145, M170, M172, M201 and 12f2. Chromosomes belonging to haplogroup DE were further typed with Multiplex III (Bosch et al. 2006), containing markers M33, M35, M75, M78, M81, M96, M123 and P2, while chromosomes in Haplogroup K*(xR) were typed with the markers M17 and M173 (Bosch et al. 2006), and M269 (Adams et al. 2006).
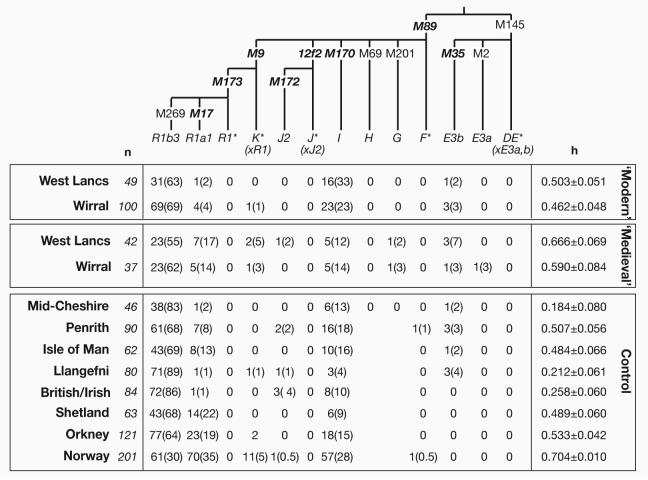
Figure 2. Y-chromosomal haplogroups in Wirral and West Lancashire samples, and controls.
Binary marker phylogeny of the Y chromosome, showing mutations on the branches of the tree, and shorthand haplogroup names (Jobling and Tyler-Smith 2003) beneath. Mutations in bold italic type were also typed in the study of Capelli et al. (Capelli et al. 2003). Below the phylogeny are given the number of chromosomes carrying the haplogroup, with percentages in parentheses. n: sample size. h: Nei's unbiased estimator of gene diversity. With the exception of Mid-Cheshire, data on the control populations are from Capelli et al. (Capelli et al. 2003), and in these samples haplogroups G and H are subsumed under F*, since the markers M69 and M201 were not typed. Lancs: Lancashire.
For the purposes of comparison with published data, haplogroups were combined in some cases; in particular, chromosomes defined by us as hgG were placed into the superhaplogroup F*(xIJK). The haplogroup we defined as R1b3 was taken to be equivalent to R1(xR1a1) (Capelli et al. 2003).
Six Y-specific microsatellites (DYS19, DYS388, DYS390, DYS391, DYS392, DYS393), chosen to be compatible with published control data (Capelli et al. 2003), were typed in a single multiplex as described (Thomas, Bradman, and Flinn 1999).
Analysis
Population differentiation tests and the generation of FST and RST matrices were carried out using Arlequin (Schneider, Roessli, and Excoffier 2000). Multidimensional scaling was done using PROXSCAL within SPSS 14.0.
Admixture analysis was carried out using the mρ estimator (Helgason et al. 2001). The probability that a particular haplotype from the admixed population originated from the Norwegian parental population is determined by its relative frequency in Norwegian and British/Irish samples. If the haplotype is not present in the parental populations, this probability is derived from the relative frequency of the most closely related haplotypes in terms of mutational differences. The average proportion of haplotypes assigned a Norwegian origin, plus standard deviation, was derived from 5000 runs of a Monte Carlo simulation. Note that the assumptions underlying the admixture estimation do not permit significance testing of differences between proportions. Binary markers and microsatellites were used together, with the former given a relative weight of 100 to reflect their lower mutation rates. The Norwegian parental sample (n=201) was from a published dataset (Capelli et al. 2003), and the British/Irish parental sample was assembled from the same source. We followed precedent (Goodacre et al. 2005), by pooling central Scottish and central Irish haplotypes (Pitlochry and Castlerea samples (Capelli et al. 2003) combined, n=84). To reduce bias, we equalized the sizes of the parental population samples by random sub-sampling of 84 chromosomes from the larger Norwegian set prior to analysis. Comparison of ten random sub-sets (Fisher exact test) showed no significant differences.
Results
Two independent samples of unrelated males representing the Wirral, and two representing West Lancashire, were collected using different criteria. The first type of sample (which we refer to here as ‘modern’) is based simply on two generations of residence, and ignores surname. Subjects for the second sample (‘medieval’) were also required to have at least two generations of residence, and to have their earliest recorded patrilineal ancestor born in the relevant area. However, in addition they were required to carry surnames (Supplementary Information) that were present in the relevant region prior to 1572 as judged by documentary sources (see Materials and Methods). Note that hereditary surnames were not established until long after the end of Viking rule, so our ascertainment does not require names themselves to be of Scandinavian origin; the fact that many of them do contain Scandinavian elements reflects the use of local place-names in the locative surnames. The approach of surname-based ascertainment yielded samples of 37 and 40 males respectively for Wirral and West Lancashire.
The Y chromosomes of the ‘modern’ and ‘medieval’ Wirral and West Lancashire samples were analyzed using six microsatellites (Supplementary Table), and a total of 13 binary markers defining 13 haplogroups (Figure 2), of which eight were observed. These haplogroups are typical of western European Y chromosomes, with one exception: a single example of hgE3a in the Wirral ‘medieval’ sample. This haplogroup is typical of sub-Saharan African populations, comprising 48% of a continent-wide sample of 1122 African chromosomes (Wood et al. 2005), and is not reported in a composite sample of 2193 British Isles males (Capelli et al. 2003; King et al. 2007). It probably represents an African migrant, but not a recent one, since the individual carrying the chromosome reports at least four generations of residence in the Wirral. It may, like a hgA1 chromosome found previously in a man with ancestry in Yorkshire (King et al. 2007), represent African presence (Fryer 1984) via the Roman occupation, or the Atlantic slave trade, and the proximity of the Wirral to the port of Liverpool might suggest the latter.
To ask if our two sampling methods led to a significant difference in haplogroup profiles, the ‘modern’ and ‘medieval’ samples were compared in population differentiation tests. Both Wirral and West Lancashire showed significant modern-medieval differences, with p-values of 0.032 and 0.006 respectively. In both cases, the ‘medieval’ samples contain a greater number of haplogroups than do the ‘modern’ samples. Gene diversity in both ‘medieval’ samples is increased compared to their ‘modern’ counterparts (Figure 2), suggesting that some degree of recent homogenization has occurred, possibly through immigration from relatively low-diversity populations.
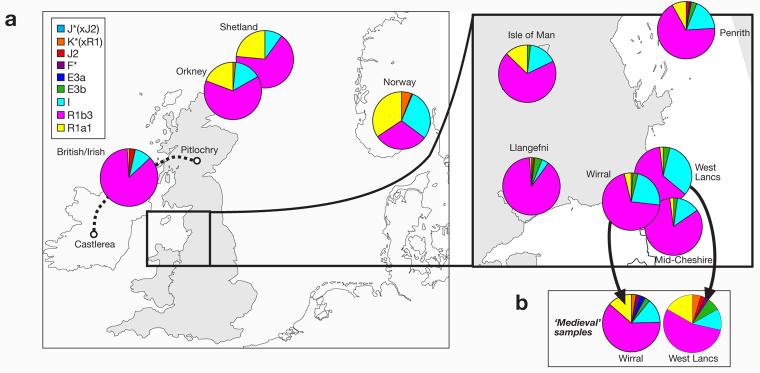
To illuminate the basis for this modern-medieval difference, we compared all four samples with a ‘modern’ sample from Mid-Cheshire (a nearby region lacking Scandinavian place-names) collected and analyzed as part of this study, and with other ‘modern’ data from the literature (Capelli et al. 2003). From Britain we chose nearby western British populations (the Isle of Man, Llangefni in Anglesey, and Penrith), and a sample (‘British/Irish’) representing the putative population of the western British Isles prior to the Viking incursions, made by pooling central Scottish and central Irish samples (see Materials & Methods). Samples from Orkney and Shetland were included since these are known to show substantial Scandinavian admixture (Goodacre et al. 2005), and a sample from Norway was used to represent a possible Viking source population (Figure 3).
Figure 3. Haplogroup distributions in Wirral, West Lancashire and control populations.
a) Haplogroup profiles of ‘modern’ samples from the Wirral and West Lancashire, and from control populations. Sectors in pie charts are colored according to haplogroup, and sector areas are proportional to haplogroup frequency. Lancs: Lancashire.
b) Haplogroup profiles of ‘medieval’ samples from the Wirral and West Lancashire, ascertained by surname.
HgR1a1 is a relatively frequent lineage in Norway, Shetland, Orkney and the Isle of Man, but rare in most mainland English and Welsh samples (Capelli et al. 2003) (Figure 3). The ‘medieval’ sample from West Lancashire shows a significant increase in the proportion of hgR1a1 with respect to its ‘modern’ counterpart (p=0.044, Fisher exact test), and for the Wirral samples the increase is close to significance (p=0.051). These observations seem compatible with a higher proportion of Viking lineages in the ‘medieval’ than in the ‘modern’ Wirral and West Lancashire samples. Comparison of the microsatellite haplotypes of these ‘modern’ and ‘medieval’ examples of hgR1a1 chromosomes (Supplementary Table) shows that they form a coherent haplotype cluster, suggesting that the use of the surnames-based sampling is not altering the nature of the hgR1a1 haplotypes sampled, but simply increases their proportions. However, this conclusion might change if the resolution of microsatellite typing were higher.
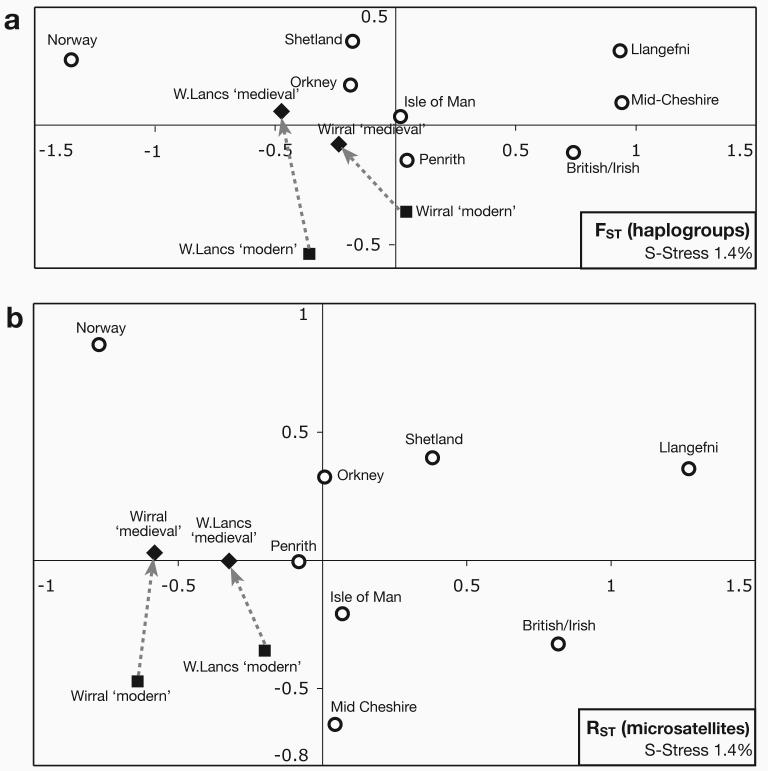
Figure 4a shows the result of a multidimensional scaling analysis based on population pairwise FST values, calculated from haplogroup frequencies. The ‘medieval’ Wirral and West Lancashire samples are differentiated from their ‘modern’ counterparts primarily in dimension 2 of the plot, and lie closer to samples from the Isle of Man and Orkney, known to have experienced significant Scandinavian input (Capelli et al. 2003; Goodacre et al. 2005). In a parallel analysis based on population pairwise RST values calculated from microsatellite data, the trend is similar (Figure 4b).
Figure 4. Multidimensional scaling (MDS) plots illustrating the relationships between Wirral, West Lancashire and control populations.
a) MDS based on population pairwise FST values from haplogroup data. Dotted arrows highlight the differences between ‘modern’ and ‘medieval’ Wirral and West Lancashire samples. Lancs: Lancashire.
b) MDS based on population pairwise RST values from microsatellite data.
Admixture analysis, in which individual samples are treated as hybrid populations made up of contributions from two parental populations, provides a means of assessing statistically the relative degree of putative Viking genetic input into the ‘modern’ and ‘medieval’ samples. Following published precedent (Goodacre et al. 2005), we assessed admixture between the Norwegian and the ‘British/Irish’ samples in the two Wirral and two West Lancashire samples. As controls, we also considered admixture within the samples from Llangefni in Anglesey (Wales), Mid-Cheshire, Penrith, the Isle of Man, and Shetland and Orkney. We employed the mρ estimator of Helgason et al. (Helgason et al. 2001), analysing admixture using a combination of binary and microsatellite marker data (Table 1).
Table 1. Admixture proportions in British Isles populations.
| Admixed population | na | Proportion Scandinavian ancestry ± standard deviation |
Proportion British ancestry ± standard deviation |
|---|---|---|---|
| Llangefni | 80 | 0.10 ± 0.02 | 0.90 ± 0.02 |
| Mid-Cheshire | 46 | 0.21 ± 0.03 | 0.79 ± 0.03 |
| Isle of Man | 62 | 0.39 ± 0.04 | 0.61 ± 0.04 |
| Penrith | 90 | 0.37 ± 0.03 | 0.63 ± 0.03 |
| Orkney | 121 | 0.50 ± 0.03 | 0.50 ± 0.03 |
| Shetland | 63 | 0.41 ± 0.04 | 0.59 ± 0.04 |
|
| |||
| Wirral ‘modern’ | 100 | 0.38 ± 0.03 | 0.62 ± 0.03 |
| Wirral ‘medieval’ | 37 | 0.47 ± 0.05 | 0.53 ± 0.05 |
| Wirral ‘medieval’ rarer surnames | 26 | 0.51 ± 0.06 | 0.49 ± 0.06 |
|
| |||
| West Lancsb ‘modern’ | 49 | 0.38 ± 0.04 | 0.62 ± 0.04 |
| West Lancsb ‘medieval’ | 40 | 0.51 ± 0.04 | 0.49 ± 0.04 |
| West Lancsb ‘medieval’ rarer | 30 | 0.53 ± 0.05 | 0.47 ± 0.05 |
| surnames | |||
sample size
Lancashire.
Where comparisons can be made, the proportions of Scandinavian admixture that we estimate differ somewhat from those seen in a previous study (Goodacre et al. 2005): for Shetland we observe 41% Scandinavian ancestry compared to the previously published figure of 44%, and the corresponding figures for Orkney are 50% compared to 31%. Such differences may reflect sampling variance or differences in the compositions of the parental and hybrid samples and in the marker resolution – we used greater numbers of both binary markers and microsatellites. Note that direct comparisons with the study from which we draw our comparative data (Capelli et al. 2003) are problematic, since it used a different admixture method, and considered admixture between a Norwegian sample and a sample chosen to represent Anglo-Saxons. The lowest proportions of Scandinavian admixture among our samples are seen in Llangefni and Mid-Cheshire, at 10% and 21% respectively. In agreement with previous results (Capelli et al. 2003), a higher proportion (39%) is seen in the Isle of Man, where there is a known history of Viking presence, and in Penrith (37%), which shows Scandinavian dialect influence (Reaney 1927). The ‘modern’ samples from Wirral and West Lancashire both show 38% Scandinavian admixture, markedly higher than the nearby sample from Mid-Cheshire (21%). This is consistent with the historical and place-name evidence for greater Viking presence in Wirral and West Lancashire than in Mid-Cheshire.
The ‘medieval’ sample from West Lancashire shows an increased proportion (51±4%) of Scandinavian ancestry compared to its ‘modern’ counterpart (38±4%); the equivalent values for Wirral are 47±5% and 38±3%. These differences, revealed by our different sampling strategies, are likely to reflect a change in haplotype frequencies due to post-medieval immigration, and are supported by genetic distances (FST) between the Norwegian sample and the Wirral and West Lancashire samples. FST between Norwegians and the West Lancashire ‘modern’ sample is 0.130, while the value for the ‘medieval’ sample is only 0.069; corresponding values for the Wirral samples are 0.162 and 0.096.
The surnames that we used to ascertain the ‘medieval’ samples belong to a wide range of frequencies – from Otty, with only 146 bearers in 1998 (www.spatial-literacy.org/UCLnames/) to Brown, with 242,765 (Supplementary Table). The more frequent names are likely to have had multiple founders and are relatively widespread in Britain, and so may provide less reliable links to medieval presence in the specific regions under study. To address this, we sub-sampled from the two ‘medieval’ samples by removing surnames with frequencies of greater than 20,000 (Supplementary Table), resulting in reduced sample sizes of 26 and 30 for Wirral and West Lancashire, respectively. We then repeated the admixture analysis. In both cases, the proportions of Scandinavian admixture in the sub-sampled ‘medieval’ populations increased further, compared to the original ‘medieval’ samples: admixture proportions in the ‘modern’ and ‘medieval’ sub-sampled Wirral populations are now 38±3% compared to 51±6%, and the corresponding values for West Lancashire are 38±4% compared to 53±5%.
Discussion
Since the discovery of the first genetic polymorphisms over a century ago, genetics has contributed much to our understanding of the human past. However, despite advances in ancient DNA technologies, genetic studies remain limited by the requirement to study extant populations, drawing conclusions about the compositions of past populations using various methods of inference. The tempo and mode of change in populations through time is difficult to discern, though we know that migration, drift and alterations in social structure through changes in lifestyle can all influence population structure markedly, and, in some cases, can cause dramatic change in only a few generations (Thomas, Stumpf, and Harke 2006; Chaix et al. 2007).
The association of Y chromosomes with patrilineal surnames, which were established in England some 700 years ago (McKinley 1990), provides a potential tool to investigate changes in population structure over the last few centuries (Manni et al. 2005) – an era in which population growth and migration have been considerable. Here, we have focused on a region in which historical and place-names evidence suggests a past presence of Viking incomers, and demonstrated that samples ascertained using surnames present in medieval times show a markedly greater proportion of Scandinavian ancestry. Our genetic analysis appears to confirm a belief, based on archaeological and place-names evidence, that the northwest of England was once heavily settled by Scandinavians, many of whom were refugees expelled from Ireland in 902AD (Wainwright 1942; Wainwright 1945; Wainwright 1948; Wainwright 1975).
Nearby Mid-Cheshire, with its absence of Viking place-names, shows a much weaker signal of Viking presence than West Lancashire or the Wirral (Table 1). Prior to arriving in Wirral, Ingimund's Scandinavian refugees expelled from Ireland are reported to have made an attempt to settle in Anglesey (Wainwright 1942; Wainwright 1975; Cavill, Harding, and Jesch 2000; Harding 2002); recent archaeological evidence (Redknap 2000) for some Scandinavian activity supports this, but we observe a low level of Scandinavian admixture there. In contrast, significant and lasting settlements appear to have been made in the northwest of England and the Isle of Man, and there is genetic evidence (Capelli et al. 2003) (Table 1) for a substantial contribution in Cumbria (Penrith). The detailed relationship of the settlements described in this paper to other Viking activities in the Irish Sea region still remains to be unravelled, but the combination of linguistic, archaeological, historical, literary and, where possible, genetic evidence will continue to provide valuable clues.
Our findings suggest that the approach of surname-based ascertainment is promising, and may provide a means to address the issue of population change through time, as well as some specific questions in the histories of regions having patrilineal surnames. The robustness of conclusions will be improved by the inclusion of both ‘modern’ and ‘medieval’ samples from places not thought to have experienced particular population incursions, such as Mid-Cheshire in our study (surname-based sampling from this region will form part of a future project). The approach relies on the existence of appropriate historical surname lists, which are plentiful, and, as we have shown, can also be supplemented by lists based on place-names. However, regions in which the time-depth of patrilineal surnames is shallow, such as Wales, where patronymic systems persisted to some extent until the 19th century (McKinley 1990), will be more difficult to study in this way.
To maximize the benefits of surname-based ascertainment, care needs to be taken in sampling. The effects on the Scandinavian admixture proportions of sub-sampling from our ‘medieval’ samples suggest that common surnames should be avoided where possible. In this study we sampled one male per surname for each of the two regions studied. However, previous studies of the relationships between surnames and Y haplotypes have shown the effects of nonpaternities in breaking the link between the two (Sykes and Irven 2000; King et al. 2006; McEvoy and Bradley 2006), and a recent nonpaternity could introduce the Y haplotype of an ‘outsider’ into a surname which has medieval roots in a study area. To obviate this problem, it may be desirable to sample more than one male per surname, and to treat the majority haplotype as a founder. In fact, the chance of sampling recently introgressed Y chromosomes increases as sample size per surname increases, but the power to distinguish such chromosomes from the earlier founding types also increases, since a consensus Y chromosome type can more readily be derived.
Genetic drift is a powerful force patterning the diversity of Y haplotypes, and computer simulations (Sturges and Haggett 1987), as well as the experience of genealogists (McKinley 1990), shows that the effect of drift upon the survival of surnames and on the number of their bearers is correspondingly strong. The degree of drift since the period of surname adoption my have a large impact on the diversity of haplotypes and surnames, and the value of using our approach to compare different populations may be compromised if these populations have experienced different magnitudes of drift. In comparing populations it will be important to take into account historical information on their demographic histories. Furthermore, drift during the 300-400 years between the Viking age and the era in which surname adoption took place may mean that estimates based on surnames may not be an accurate reflection of haplotype frequencies at the earlier time of the introduction of Viking Y chromosomes.
The admixture approach we have employed has been useful in providing evidence that surname-based ascertainment reveals some aspects of the past compositions of populations. However, it is clearly not ideal to use ‘modern’ parental populations to assess admixture in ‘medieval’ hybrids, and the results are likely to be more reliable if the parental samples themselves could be ascertained in a similar way to the hybrids. A further shortcoming of our admixture analysis is that we have assumed that the non-Celtic contributions are all due to Scandinavians. A more sophisticated approach would also consider an Anglo-Saxon contribution, though distinguishing this from a Danish Viking contribution to more easterly British populations would be difficult, since modern proxies for these two different source populations are so similar in their Y-chromosomal haplotype compositions (Capelli et al. 2003).
The use of patrilineal surnames in ascertainment provides both the power of our approach, and its major limitation, since any analysis of the structure or history of populations based on a single genetic locus, the Y chromosome, has inherently low power, and population statistics calculated from Y data have high variance (Wilson et al. 2001). Furthermore, we can learn nothing about changes in the maternal genetic landscape, which is unfortunate, since the role of women in the Viking colonizations was as important as that of men (Jesch 1991). Nonetheless, in the absence of a time-machine, the link between a haplotype and a cultural marker, the surname, may provide the only practical means to access the genetic composition of populations in the past. As well as allowing us to investigate the influence of migration and drift over the last few centuries in changing the population structure of Britain, the method should be applicable to other regions where surnames are patrilineal and suitable historical records survive.
Supplementary Material
Acknowledgements
We thank all DNA donors, and BBC Radio Merseyside, BBC Radio Lancashire, the Daily Post, Wirral News, Wirral Globe and Ormskirk Advertiser for assistance in the recruitment of volunteers. We thank Chris Tyler-Smith and Agnar Helgason for help with admixture analysis, Neil Bradman for contributions to haplotyping, and two anonymous referees for useful comments on the manuscript. M.A.J. was supported by a Wellcome Trust Senior Fellowship in Basic Biomedical Science (grant no. 057559), G.R.B., P.L.B. and A.C.L. by the Wellcome Trust, and T.E.K. by a Wellcome Prize Studentship (grant no. 061129). The project was also supported by a 2002 DNA anniversary award of the Biotechnology and Biological Sciences Research Council, and by the Melford Charitable Trust.
Literature cited
- Adams SM, King TE, Bosch E, Jobling MA. The case of the unreliable SNP: Recurrent back-mutation of Y-chromosomal marker P25 through gene conversion. Forens. Sci. Int. 2006;159:14–20. doi: 10.1016/j.forsciint.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Bailey R, Whalley J, Bowden A, Tresise G. A miniature Viking-Age hogback from the Wirral. Antiquaries J. 2006;86:345–356. [Google Scholar]
- Bennett JHE, Dewhurst JC. Quarter sessions records with other records of the Justices of the Peace for the County Palatine of Chester 1559-1760 together with a few earlier miscellaneous records deposited with the Cheshire County Council. Records Soc. Lancs. Ches. 1940;94:37–39. [Google Scholar]
- Booth P. Calendar of Cheshire Trailbaston Proceedings 1353. Ches. History. 1983;12 [Google Scholar]
- Bosch E, Calafell F, González-Neira A, Flaiz C, Mateu E, Scheil H-G, Huckenbeck W, Efremovska L, Mikerezi I, Xirotiris N, Grasa C, Schmidt H, Comas D. Male and female lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann. Hum. Genet. 2006;70:459–487. doi: 10.1111/j.1469-1809.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- Bu'Lock JD. Pre-Norman crosses of West Cheshire and the Norse settlements around the Irish Sea. Trans. Lancs. Ches. Antiquarian Soc. 1958;68:1–11. [Google Scholar]
- Capelli C, Redhead N, Abernethy JK, Gratrix F, Wilson JF, Moen T, Hervig T, Richards M, Stumpf MPH, Underhill PA, Bradshaw P, Shaha A, Thomas MG, Bradman N, Goldstein DB. A Y chromosome census of the British Isles. Curr. Biol. 2003;13:979–984. doi: 10.1016/s0960-9822(03)00373-7. [DOI] [PubMed] [Google Scholar]
- Cavill P, Harding SE, Jesch J. Wirral and its Viking Heritage. English Place-Name Society; Nottingham: 2000. [Google Scholar]
- Chaix R, Quintana-Murci L, Hegay T, Hammer MF, Mobasher Z, Austerlitz F, Heyer E. From social to genetic structures in central Asia. Curr. Biol. 2007;17:43–48. doi: 10.1016/j.cub.2006.10.058. [DOI] [PubMed] [Google Scholar]
- Collingwood WG. Early monuments of West Kirby. In: Brownbill J, Kirby West, Hilbre, editors. A Parochial History. Henry Young & Sons Ltd; Liverpool: 1928. pp. 14–26. [Google Scholar]
- Ekwall E. The Place-names of Lancashire. Manchester University Press; Manchester: 1922. [Google Scholar]
- Fellows-Jensen G. Scandinavian place-names of the Irish Sea Province. In: Graham-Campbell JA, editor. Viking treasure from the north-west: the Cuerdale hoard in its context. National Museums and Galleries on Merseyside Occasional Papers; Liverpool: 1992. pp. 31–42. [Google Scholar]
- Fryer P. Staying Power: The History of Black People in Britain. Pluto Press; London: 1984. [Google Scholar]
- Goodacre S, Helgason A, Nicholson J, Southam L, Ferguson L, Hickey E, Vega E, Stefansson K, Ward R, Sykes B. Genetic evidence for a family-based Scandinavian settlement of Shetland and Orkney during the Viking periods. Heredity. 2005;95:129–135. doi: 10.1038/sj.hdy.6800661. [DOI] [PubMed] [Google Scholar]
- Graham-Campbell JA. Viking treasure from the north-west: the Cuerdale hoard in its context. National Museums and Galleries on Merseyside Occasional Papers; Liverpool: 1992. [Google Scholar]
- Harding S. Viking Mersey: Scandinavian Wirral. Countyvise, Birkenhead; West Lancashire and Chester: 2002. [Google Scholar]
- Helgason A, Hickey E, Goodacre S, Bosnes V, Stefansson K, Ward R, Sykes B. mtDNA and the islands of the North Atlantic: estimating the proportions of Norse and Gaelic ancestry. Am. J. Hum. Genet. 2001;68:723–737. doi: 10.1086/318785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigurdardóttir S, Gulcher JR, Ward R, Stefansson K. mtDNA and the origin of the Icelanders: deciphering signals of recent population history. Am J Hum Genet. 2000a;66:999–1016. doi: 10.1086/302816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigurdardóttir S, Nicholson J, Sykes B, Hill EW, Bradley DG, Bosnes V, Gulcher JR, Ward R, Stefánsson K. Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am. J. Hum. Genet. 2000b;67:697–717. doi: 10.1086/303046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine WF, Sanders F. Wirral Notes and Queries, Being Gleanings Historical and Antiquarian. Vol. 2. Willmer Bros., Birkenhead; 1894. [Google Scholar]
- Jesch J. Women in the Viking Age. Boydell; Woodbridge: 1991. [Google Scholar]
- Jobling MA. In the name of the father: surnames and genetics. Trends Genet. 2001;17:353–357. doi: 10.1016/s0168-9525(01)02284-3. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Hurles ME, Tyler-Smith C. Human Evolutionary Genetics: origins, peoples and disease. Garland Science; New York/Abingdon: 2004. [Google Scholar]
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat. Rev. Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Keyser-Tracqui C, Crubézy E, Ludes B. Nuclear and mitochondrial DNA analysis of a 2,000-year-old necropolis in the Egyin Gol Valley of Mongolia. Am. J. Hum. Genet. 2003;73:247–260. doi: 10.1086/377005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TE, Ballereau SJ, Schürer K, Jobling MA. Genetic signatures of coancestry within surnames. Curr. Biol. 2006;16:384–388. doi: 10.1016/j.cub.2005.12.048. [DOI] [PubMed] [Google Scholar]
- King TE, Parkin EJ, Swinfield G, Cruciani F, Scozzari R, Rosa A, Lim SK, Xue Y, Tyler-Smith C, Jobling MA. Africans in Yorkshire? The deepest-rooting clade of the Y phylogeny within an English genealogy. Eur. J. Hum. Genet. 2007;15:288–293. doi: 10.1038/sj.ejhg.5201771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni F, Toupance B, Sabbagh A, Heyer E. New method for surname studies of ancient patrilineal population structures, and possible application to improvement of Y-chromosome sampling. Am. J. Phys. Anthropol. 2005;126:214–228. doi: 10.1002/ajpa.10429. [DOI] [PubMed] [Google Scholar]
- McEvoy B, Bradley DG. Y-chromosomes and the extent of patrilineal ancestry in Irish surnames. Hum. Genet. 2006;119:212–219. doi: 10.1007/s00439-005-0131-8. [DOI] [PubMed] [Google Scholar]
- McKinley R. The Surnames of Lancashire. Vol. 4. Leopard's Head Press; London: 1981. (Surnames Series). [Google Scholar]
- McKinley RA. A history of British surnames. Longman; London: 1990. [Google Scholar]
- Peet G. Inhabitants of Ormskirk, Scarisbrick with Hurlton, Bickerstaffe, Burscough with Marton, Westhead with Lathom and Skelmersdale who promised to contribute to the stipend of the priest of the altar of Our Lady at Ormskirk, 1366. Ormskirk Distr. Fam. Historian. 1991;1:2–6. [Google Scholar]
- Reaney PH. A Grammar of the Dialect of Penrith (Cumberland): Descriptive and Historical, with Specimens and Glossary. Manchester University Press; Manchester: 1927. [Google Scholar]
- Redknap M. Vikings in Wales. National Museums and Galleries of Wales; Cardiff: 2000. [Google Scholar]
- Richards JD. Viking age England. Tempus, Stroud; 2004. [Google Scholar]
- Roberts SJ. A History of Wirral. Philimore; Chichester: 2002. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin ver. 2.0: A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva; Geneva, Switzerland: 2000. [Google Scholar]
- Sturges CM, Haggett BC. Inheritance of English surnames. Hawgood Computing; London: 1987. [Google Scholar]
- Sykes B, Irven C. Surnames and the Y chromosome. Am. J. Hum. Genet. 2000;66:1417–1419. doi: 10.1086/302850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Bradman N, Flinn HM. High throughput analysis of 10 microsatellite and 11 diallelic polymorphisms on the human Y chromosome. Hum. Genet. 1999;105:577–581. doi: 10.1007/s004399900181. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Stumpf MP, Harke H. Evidence for an apartheid-like social structure in early Anglo-Saxon England. Proc. Biol. Sci. 2006;273:2651–2657. doi: 10.1098/rspb.2006.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf AL, Gilbert MT, Dumbacher JP, Hoelzel AR. Tracing the phylogeography of human populations in Britain based on 4th-11th century mtDNA genotypes. Mol. Biol. Evol. 2006;23:152–161. doi: 10.1093/molbev/msj013. [DOI] [PubMed] [Google Scholar]
- Wainwright FT. North-west Mercia AD 871-924. Trans. Hist. Soc. Lancs. Ches. 1942;94:3–55. [Google Scholar]
- Wainwright FT. Wirral Field Names. Antiquity. 1943;27:57–66. [Google Scholar]
- Wainwright FT. The Scandinavians in Lancashire. Trans. Lancs. Ches. Antiquarian Soc. 1945;58:71–116. [Google Scholar]
- Wainwright FT. Ingimund's Invasion. Engl. Hist. Rev. 1948;247:145–167. [Google Scholar]
- Wainwright FT. In: Scandinavian England: Collected Papers. Finberg HPR, editor. Phillimore; Chichester: 1975. [Google Scholar]
- Wilson JF, Weiss DA, Richards M, Thomas MG, Bradman N, Goldstein DB. Genetic evidence for different male and female roles during cultural transitions in the British Isles. Proc. Natl. Acad. Sci. USA. 2001;98:5078–5083. doi: 10.1073/pnas.071036898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ET, Stover DA, Ehret C, Destro-Bisol G, Spedini G, McLeod H, Louie L, Bamshad M, Strassmann BI, Soodyall H, Hammer MF. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur. J. Hum. Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.