Abstract
Localization and targeting of depth electrodes in specific regions of the human brain is critical for accurate clinical diagnoses and treatment as well as for neuroscientific electrophysiological research. By using high-resolution magnetic resonance imaging combined with 2D computational unfolding, the authors present a method that improves electrode localization in the medial temporal lobe. This method permits visualization of electrode placements in subregions of the hippocampus and parahippocampal gyrus, allowing for greater specificity in relating electrophysiological and anatomical features in the human medial temporal lobe. Such methods may be extended to therapeutic procedures targeting specific neuronal circuitry in subfields of structures deep in the human brain.
Keywords: coregistration, depth electrode, electrode localization, epilepsy, hippocampus
A hallmark of electrophysiological recordings in mammals is the ability to localize electrodes to specific cell-body layers in the brain by using histological reconstructions. Micrometer imaging of electrode placements permits statements about the interactions of different cell types in specific subregions of the mammalian brain.12 Correlations between electrophysiological findings in humans and anatomical locations of the electrodes used in gathering the data are gaining increasing importance in functional neurosurgery and are likely to be critical for directing therapeutic procedures such as DBS to precise neuronal circuitry. Such correlations are currently limited by the resolution of imaging modalities, including structural MR images or coregistration of CT scans and MR images.6
When identifying electrode locations following implantation, as in patients who undergo implantation of depth electrodes for delineation of seizure foci, current computational alignment methods permit registration of postim-plant CT scans and preimplant MR images with millimeter accuracy.7–9,11 Using high-resolution imaging and computational unfolding of the hippocampus, Zeineh et al.13,14 reported hippocampal voxel resolutions of 0.391 × 0.391 × 0.429 mm, providing clear visualization of hippocampal subfields in 2 dimensions. Because the hippocampus and surrounding tissue may be particularly impacted by recurrent seizures,5 we apply this method to improve localization of microelectrode recording sites in patients with epilepsy who are undergoing seizure monitoring with intracranial depth electrodes.
Materials and Methods
Prior to implantation with depth electrodes, 3-T high-resolution T2-weighted structural MR images of the hippocampus were obtained in all patients (TR 5.2 seconds, TE 105 msec, 19 slices, voxel size 0.4 × 0.4 × 3 mm). These parameters optimize signal strength in the hippocampal area, particularly in the anterior portion.13,14 Slices were obtained perpendicular to the long axis of the hippocampus. Patients also underwent 3-T whole-brain T1-weighted magnetization-prepared rapid acquisition gradient echo MR imaging (TR 1800 msec, TE 2.93 seconds, voxel size 0.9 × 0.9 × 0.8 mm) as part of depth electrode placement planning. Patients then underwent implantation of depth electrodes for surgical monitoring. At the tip of each electrode, a set of eight 40-μm platinum–iridium microwires provided possible cellular and electroencephalographic signals.6 After implantation with depth electrodes, patients underwent a spiral CT scan (1 second rotation, high-quality [also called HQ] mode, helical pitch 1.5, 1-mm slice collimation, and a 0.5-mm reconstruction interval delineated using axial slices) to localize electrodes. All patients who participated provided informed consent. All studies conformed to the guidelines of the Medical Institutional Review Board at our institution.
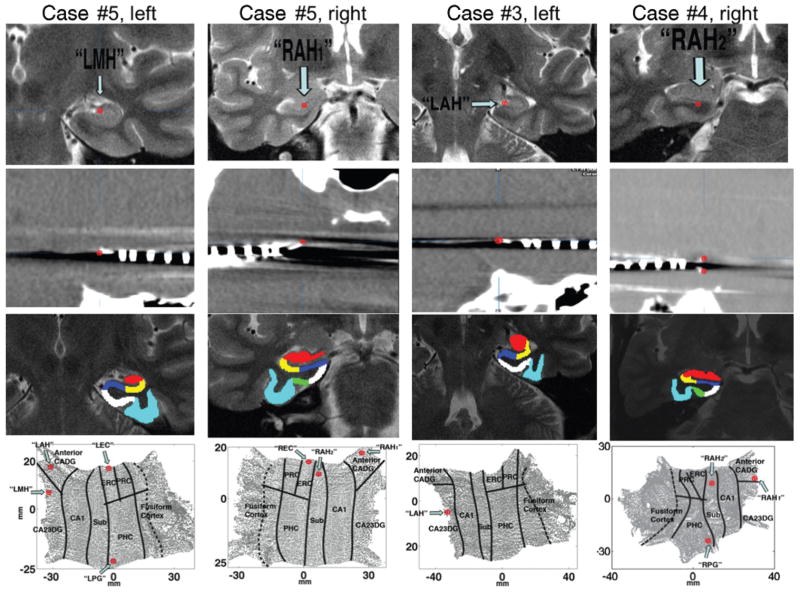
We performed a 3-way registration of the patients’ CT, whole-brain MR imaging, and high-resolution MR imaging studies to maximize our localization accuracy. This was done because we occasionally obtained unsatisfactory registrations between the high-resolution MR images and CT scans (> 1-mm translations). Three-way registrations were performed using BrainLab stereotactic and localization software (www.brainlab.com).7,9 The CT scan was first registered to the whole-brain MR image, followed by registration of the whole-brain image to the high-resolution MR imaging study. In most cases, microwire tracks were visible as artifacts in the coregistered CT–MR images (Fig. 1, Row 2). By localizing microwires on the CT scan, we could then visualize these locations on the high-resolution and whole-brain MR image simultaneously. In all cases, anatomical locations targeted by microwire tips on the high-resolution MR images corresponded with those on the whole-brain imaging studies.
Fig. 1.
Neuroimaging studies and maps used in localization of microelectrodes to hippocampal subregions. Row 1: High-resolution coronal–oblique MR images (perpendicular to the long axis of the hippocampus) obtained in 3 different patients. “Left” and “right” indicate the cerebral hemispheres. Red target location (arrows) shows location defined by the microtip identified on postimplant CT scans (see Row 2) coregistered to the preimplant MR images. Row 2: Postimplant axial CT scans coregistered to the high-resolution MR images shown in Row 1. In most cases, microelectrodes were clearly visible as artifacts on the CT scan, as indicated with red targets. Microwires occasionally bifurcated within a structure, leading to placement in 2 different hippocampal subfields (Case 4, Row 2). Row 3: Subregion definitions in 3D space based on back-projecting regions from the 2D maps shown in Row 4. Red area, CA2/CA3/dentate gyrus (CA23DG); yellow area, CA1; dark blue area, subiculum (Sub); green area, PRC; white area, ERC; and light blue area, fusiform cortex. The PHC is shown in white when slices are posterior to the ERC-PRC-PHC boundary (for example, Case 5 left and Case 3 left). Row 4: Two-dimensional hippocampal flat maps of each MR imaging sequence shown partially in Row 1. Top of map shows the most anterior aspect of the hippocampus, and bottom of map shows the most posterior end. Electrode locations in flat space, defined by coregistration from 3D neuroimages shown in Rows 1 and 2, are indicated with red targets. Black lines indicate subregion boundaries, and dotted black line indicates location of medial fusiform vertex. Electrode names indicate putative placement. Thus, “RAH” and “LAH” indicate putative placement in the right and left anterior hippocampus, respectively, and “LMH” indicates putative placement in the left medial hippocampus. The label “RAH1” indicates a bifurcated electrode placed in the superior portion of the right anterior hippocampus, and “RAH2” denotes a bifurcated electrode placed in the inferior portion of the right anterior hippocampus. Also visible in some cases are electrodes placed in the right and left ERC (designated “REC” and “LEC”) and in the right and left PHC (labeled “RPG” and “LPG”). CADG = CA/dentate gyrus.
High-resolution structural scans were computationally unfolded and flattened from 3 dimensions to 2 dimensions, producing 2D maps oriented from bottom to top (posterior to anterior) along the long axis of the hippocampus. Computational unfolding involved several steps. We first segmented hippocampal gray matter by outlining white matter and cerebrospinal fluid along the hippocampus proper and extending through the fusiform cortex. To improve the quality of the segmentation and our overall voxel resolution within the hippocampus, slices were computationally and then manually interpolated by a factor of 7 along the long axis of the hippocampus, producing a final voxel size of 0.391 × 0.391 × 0.429 mm. Gray matter voxels were computationally connected using the region-expansion algorithm “MrGray.”10 This 3D gray matter strip was then computationally flattened into 2 dimensions by using “mrUnfold” software.3 Using landmarks on the 3D hippocampal scan, we defined anatomical boundaries of the following structures: CA2/CA3/dentate gyrus (labeled CA23DG); CA1–subiculum; subiculum–ERC; ERC–PRC; subiculum–PHC; and PHC–fusiform cortex (for example, the collateral sulcus).1,2 We also demarcated the beginning of the hippocampal head (anterior CA/dentate gyrus) and the ERC-PRC-PHC boundary. These boundaries were then projected into 2D hippocampal space, thus outlining the anatomical locations defined by these boundaries in 3D space. A more complete explanation of the methods involved in computational unfolding, including validation and precision of the method, can be found in Zeineh et al.14
Results
Five patients underwent high-resolution hippocampal MR imaging studies prior to depth electrode placement. In all 5 cases, electrode locations were visible in hippocampal subnuclei (Table 1, Fig. 1) by using the coregistered high-resolution MR image (Fig. 1 Row 1) and CT scan (Fig. 1 Row 2), with microelectrode tracks visible as artifacts on the CT scan. On both the high-resolution 3D MR image (Fig. 1 Row 1) and the 2D hippocampal flat maps (Fig. 1 Row 4), microelectrodes could therefore be localized to specific hippocampal subregions. In cases in which electrodes were placed in areas from which the MR imaging slices we obtained allowed segmentation of the hippocampus, we were further able to localize placement of entorhinal and parahippocampal electrodes on 2D hippocampal maps (Fig. 1 Row 4, Cases 4 and 5). To validate our boundary demarcations in 2 dimensions (see Materials and Methods), gray matter regions defined in 2D space were back-projected into 3D space; the subregions defined in 2D space corresponded with the anatomical subregions on the 3D MR image (Fig. 1 Row 3).
TABLE 1.
Localization of electrodes in 5 patients with pharmacologically intractable epilepsy*
| Case No. | Electrode Placement | Intrahippocampal Localization |
|---|---|---|
| 1 | rt AH | CA2/CA3/DG |
| lt AH | subiculum | |
| lt PH | medial CA1 | |
| 2 | rt AH | CA2/CA3/DG |
| lt AH | anterior CA fields | |
| 3 | rt AH | CA2/CA3/DG |
| lt AH | medial CA2/CA3/DG | |
| 4 | rt AH | RAH1: anterior CA fields |
| RAH2: subiculum | ||
| lt AH | CA1/subiculum border | |
| 5 | rt AH | RAH1: anterior CA fields |
| RAH2: subiculum | ||
| lt AH | anterior CA fields | |
| lt MH | CA2/CA3/DG |
Column 2 indicates putative electrode placement, and column 3 indicates intrahippocampal localization of electrodes by using 2D patient flat maps. “Anterior” denotes the first third of the hippocampus, “medial” the middle third of the hippocampus, and “posterior” the posterior third of the hippocampus. Thus, “rt AH” indicates an electrode putatively placed in the right anterior hippocampus; “RAH1” indicates the superior portion of a bifurcated electrode placed in the right anterior hippocampus; and “RAH2” indicates the inferior portion of a bifurcated electrode placed in the right anterior hippocampus. In addition to localizing electrodes to hippocampal subregions, we further corroborated placement in the anterior–posterior plane. Unless otherwise indicated in column 3, putative anterior–posterior placements and actual intrahippocampal localizations were consistent. Abbreviations: AH = anterior hippocampus; DG = dentate gyrus; MH = medial hippocampus; PH = posterior hippocampus.
Discussion
The method we present here, which was previously used to improve visualization of the functional MR imaging activations to subregions of the hippocampus, localizes microelectrodes in 2D maps at high resolution to hippocampal subregions. In the 5 patients who underwent imaging and implantation with depth electrodes, we were able to localize microelectrode placements to subregions of the hippocampus in all cases. Previous methods similarly co-registered preoperative whole-brain MR images to postim-plant CT scans to localize electrodes.6–9,11 Our method introduces several new steps that improve localization and visualization of electrodes. First, we use a high-resolution hippocampal structural scan, providing imaging of hippocampal substructures. Second, we perform a 3-way registration between the high-resolution MR, whole-brain MR, and whole-brain CT imaging studies to ensure that no additional registration errors are introduced when aligning a partial-volume MR image to a whole-brain CT scan. Third, computational unfolding of the hippocampus augments voxel resolution in the 2D plane because the 3D hippocampal manifold is computationally and then manually interpolated to a final voxel resolution of ~ 0.4 × 0.4 × 0.4 mm. This step thus provides superior visualization of the hippocampal area. Fourth, our method permits clear visualization of electrodes in subregions that would otherwise not be as readily visible in 3D space because of variability in 3D subfield boundary locations.
The coupling of electrophysiological recordings with precise anatomical localization is gaining increasing importance in stereotactic and functional neurosurgery. In DBS for movement and other neurological disorders, electrodes are targeted to minute functional locations deep in the brain, where the electrophysiological signature of cell layers and cell aggregates are often critical for the identification of the relevant functional zones. Anatomical targeting of these regions is limited by the resolution of imaging methods. As we demonstrate here, computational methods such as unfolding offer better visualization of the cellular circuitry of relevant complex brain structures such as the hippocampus, and therefore present the potential for more accurate targeting as well. Because high-resolution imaging and cortical unfolding techniques have been applied successfully to the whole brain as well,4 our method could also provide information about extrahippocampally placed electrodes. Such methods could be extended to other brain regions such as thalamus, subthalamic nucleus, and globus pallidus, which are important therapeutic targets for DBS. Furthermore, as our understanding of the pathophysiological origins of neurological disorders increases, future procedures for diagnosis or stimulation may be aimed at much more refined brain circuitry than are current methods, which are limited to the several-millimeter accuracy range.
Conclusions
Our results demonstrate that electrode localization in the medial temporal lobe can be improved using registration of high-resolution MR imaging sequences and CT scans coupled with computational unfolding methods.
Acknowledgments
This study was supported by grants from the National Institutes for Neurological Disorders and Stroke (Grant No. F32 NS50067-01A1 to Dr. Ekstrom and No. NS033221 to Dr. Fried).
We thank the patients who participated in this study for the generous contribution of their time, and we thank Brooke Salaz for administrative assistance.
Abbreviations in this paper
- CT
computed tomography
- DBS
deep brain stimulation
- ERC
entorhinal cortex
- MR
magnetic resonance
- PHC
parahippocampal cortex
- PRC
perirhinal cortex
References
- 1.Amaral DG, Insausti R. The Hippocampal formation. In: Paxinos G, editor. The Human Nervous System. San Diego: Academic Press; 1990. pp. 711–755. [Google Scholar]
- 2.Duvernoy HM. The Human Hippocampus: Functional Anatomy, Vascularization, and Serial Sections with MRI. New York: Springer-Verlag; 1997. [Google Scholar]
- 3.Engel SA, Glover GH, Wandell BA. Retinotopic organization in human visual cortex and the spatial precision of functional MRI. Cereb Cortex. 1997;7:181–192. doi: 10.1093/cercor/7.2.181. [DOI] [PubMed] [Google Scholar]
- 4.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 5.Foldvary N, Lee N, Hanson MW, Coleman RE, Hullette CM, Friedman AH, et al. Correlation of hippocampal neuronal density and FDG-PET in mesial temporal lobe epilepsy. Epilepsia. 1999;40:26–29. doi: 10.1111/j.1528-1157.1999.tb01984.x. [DOI] [PubMed] [Google Scholar]
- 6.Fried I, Wilson CL, Maidment NT, Engel J, Jr, Behnke E, Fields TA, et al. Cerebral microdialysis combined with single-neuron and electroencephalographic recording in neurosurgical patients. Technical note J Neurosurg. 1999;91:697–705. doi: 10.3171/jns.1999.91.4.0697. [DOI] [PubMed] [Google Scholar]
- 7.Gumprecht HK, Widenka DC, Lumenta CB. BrainLab VectorVision Neuronavigation System: technology and clinical experiences in 131 cases. Neurosurgery. 1999;44:97–105. doi: 10.1097/00006123-199901000-00056. [DOI] [PubMed] [Google Scholar]
- 8.Nelles M, Koenig R, Kandyba J, Schaller C, Urbach H. Fusion of MRI and CT with subdural grid electrodes. Zentralbl Neurochir. 2004;65:174–179. doi: 10.1055/s-2004-820354. [DOI] [PubMed] [Google Scholar]
- 9.Schlaier JR, Warnat J, Dorenbeck U, Proescholdt M, Schebesch KM, Brawanski A. Image fusion of MR images and real-time ultrasonography: evaluation of fusion accuracy combining two commercial instruments, a neuronavigation system and an ultrasound system. Acta Neurochir (Wien) 2004;146:271–277. doi: 10.1007/s00701-003-0155-6. [DOI] [PubMed] [Google Scholar]
- 10.Teo PC, Sapiro G, Wandell BA. Creating connected representations of cortical gray matter for functional MRI visualization. IEEE Trans Med Imaging. 1997;16:852–863. doi: 10.1109/42.650881. [DOI] [PubMed] [Google Scholar]
- 11.West J, Fitzpatrick JM, Wang MY, Dawant BM, Maurer CR, Jr, Kessler RM, et al. Comparison and evaluation of retrospective intermodality brain image registration techniques. J Comput Assist Tomogr. 1997;21:554–566. doi: 10.1097/00004728-199707000-00007. [DOI] [PubMed] [Google Scholar]
- 12.Wittner L, Henze DA, Záborszky L, Buzsáki G. Hippocampal CA3 pyramidal cells selectively innervate aspiny interneurons. Eur J Neurosci. 2006;24:1286–1298. doi: 10.1111/j.1460-9568.2006.04992.x. [DOI] [PubMed] [Google Scholar]
- 13.Zeineh MM, Engel SA, Thompson PM, Bookheimer SY. Dynamics of the hippocampus during encoding and retrieval of face-name pairs. Science. 2003;299:577–580. doi: 10.1126/science.1077775. [DOI] [PubMed] [Google Scholar]
- 14.Zeineh MM, Engel SA, Thompson PM, Bookheimer SY. Unfolding the human hippocampus with high resolution structural and functional MRI. Anat Rec. 2001;265:111–120. doi: 10.1002/ar.1061. [DOI] [PubMed] [Google Scholar]