The major hazelnut allergen Cor a 9 was purified from the natural source and crystallized. Diffraction data were collected to 1.9 Å resolution using a synchrotron-radiation source.
Keywords: hazelnuts, 11S seed-storage proteins, food allergies, tree-nut allergens
Abstract
Hazelnut (Corylus avellana) is one of the food sources that induce allergic reaction in a subpopulation of people with food allergy. The 11S legumin-like seed-storage protein from hazelnut has been identified as one of the major hazelnut allergens and named Cor a 9. In this study, Cor a 9 was extracted from hazelnut kernels using a high-salt solution and was purified by desalting out and FPLC to a highly purified state. Diffraction-quality single crystals were obtained using the hanging-drop vapour-diffusion method. Diffraction data were collected and a structure solution has been obtained by molecular-replacement calculations. Further refinement of the structure is currently in progress.
1. Introduction
Hazelnuts are widely consumed, especially in Europe. The nut kernel is rich in plant oils (>50%), protein (∼15%) and dietary fiber (∼10%) (Savage & McNeil, 1998 ▶), as well as minerals, vitamins and other flavor components (Alasalvar et al., 2003 ▶). Recent studies have suggested that hazelnuts are a source of phenolic antioxidants (Alasalvar et al., 2006 ▶; Shahidi et al., 2007 ▶) and a hazelnut supplement may reduce the risk of coronary heart disease by enhancing the plasma antioxidant potential and lowering the plasma cholesterol level (Mercanligil et al., 2007 ▶). However, many proteins in the nut kernel have been found to be capable of inducing anaphylactic responses in certain populations (Pastorello et al., 2002 ▶).
Allergic reactions to hazelnuts range from oral allergy syndrome to severe anaphylactic reactions (Beyer et al., 2002 ▶; Birmingham et al., 2007 ▶; Ortolani et al., 2000 ▶; Schocker et al., 2000 ▶). Allergy to hazelnuts is a classical example of a birch-pollen-related food allergy, as certain hazelnut allergens are known to cross-react with IgE of patients sensitized to homologous allergens in birch pollen (Bohle et al., 2005 ▶). In addition, hazelnuts give rise to cross-reactions in patients with allergies to other tree nuts. The sera of patients with anaphylaxis induced by macadamia nut (Sutherland et al., 1999 ▶), coconut (Nguyen et al., 2004 ▶) and walnut (Asero et al., 2004 ▶) have been reported to cross-react with hazelnut allergens.
Cor a 9, also known as corylin, is a major allergen in hazelnuts (Pastorello et al., 2002 ▶). Similar to Ara h 3 (a peanut allergen) and Ber e 2 (a brazil nut allergen), Cor a 9 belongs to the 11S legumin-like seed-storage protein family. It is expressed by a gene that encodes a protein containing 515 amino acids with a theoretical molecular mass of 59 kDa; however, the 11S proteins are known to be post-translationally processed into more than one peptide (Albillos et al., 2008 ▶; Dickinson et al., 1989 ▶; Guo et al., 2007 ▶; Jin et al., 2007 ▶). The IgE-binding epitope(s) of Cor a 9 has not been defined, but one known linear IgE-binding epitope of Ara h 3 has 67% sequence identity to the corresponding region in Cor a 9 (Beyer et al., 2002 ▶). To date, the structural basis of the allergenicity of tree-nut allergens and their cross-reactivity are poorly understood and no structure has been reported for any of the hazelnut allergens. Here, we report the crystallization and X-ray data collection of Cor a 9, with the aim of determining its structure.
2. Materials and methods
2.1. Protein purification
Bagged raw shelled hazelnuts were purchased from a local supermarket. 30 g of the hazelnut kernels was ground in 35 ml 2 M NaCl solution using a KitchenAid blender. After a 20 min incubation at 333 K with stirring at 2 min intervals, the sample was centrifuged at 2000g for 15 min at room temperature (298 K) and the supernatant was collected as a crude protein extract. The crude extract was defatted by mixing 20 ml of the sample with 20 ml hexane and incubating for 2 h at room temperature after vigorous vortexing. After centrifugation at 2000g for 15 min at room temperature, the aqueous portion was collected and subjected to the same defatting process four more times. The salt concentration of the sample was reduced to 0.4 M by diluting it with water at 333 K. Cor a 9 was precipitated by incubating the diluted sample at 273 K for 2 h. After the sample had been centrifuged at 300g for 5 min at 298 K, the pellet was redissolved in 2 M NaCl solution to a final volume of 5 ml. 5 ml hexane was added to further remove the fat component from the protein sample. The defatted sample was collected by centrifugation at 2000g and diluted with water to bring the salt concentration to 0.4 M as described above. This precipitation and defatting procedure was repeated two more times. The above procedures were carried out in the presence of protease inhibitors (100 nM aprotinin, 50 µM antipain, 50 µM leupeptin and 0.5 µg ml−1 pepstatin) and antibiotics (50 µg ml−1 ampicillin and 50 µg ml−1 kanamycin) in the sample.
Cor a 9 was then further purified by size-exclusion chromatography (SEC) and hydrophobic interaction chromatography (HIC) at room temperature. For SEC, the precipitated Cor a 9 was dissolved in buffer C (1 M NaCl, 10 mM Tris pH 8.0) to a final concentration of 4 mg ml−1. The sample was filtered with a 0.2 µm syringe filter before application onto an XK 26/70 Superdex-200 column (GE Healthcare, Piscataway, New Jersey, USA), which was equilibrated and eluted with buffer C. Column calibration was carried out using thyroglobulin (669 kDa), ferritin (440 kDa), catalase (242 kDa), aldolase (158 kDa) and bovine serum albumin (67 kDa) obtained as a protein standard kit (GE Healthcare) and the flow rate was 1 ml min−1 for all purification and calibration runs.
Fractions containing Cor a 9 eluted from the Superdex 200 column were pooled and adjusted to 2 M ammonium sulfate by quick mixing with an equal volume of buffer AS (4 M ammonium sulfate, 50 mM Tris–HCl pH 8). The diluted sample was then injected onto two 5 ml HiTrap Phenyl HP columns (GE Healthcare) in tandem that were pre-equilibrated with buffer AS mixed with 50 mM Tris–HCl pH 8 in a 1:1(v:v) ratio. The columns were washed with 20 ml 50% buffer AS and eluted with a linear ammonium sulfate gradient from 2 to 0 M over 100 ml in 50 mM Tris–HCl pH 8. The Cor a 9 peak fractions were pooled and the buffer was changed to buffer C using an Amicon Ultra centrifugal filter device (Millipore, Billerica, Massachusetts, USA) by repeated dilution and concentration. The concentration of purified Cor a 9 was calculated using its molecular weight and a theoretical extinction coefficient of ∊280 = 47 900 cm−1 mol−1.
2.2. SDS–PAGE analysis
SDS–PAGE was carried out with 4–20% polyacrylamide gels in Tris–HEPES–SDS running buffer (100 mM Tris, 100 mM HEPES, 3 mM SDS pH 8). Precision Plus Protein Standards (Bio-Rad, Hercules, California, USA) were used as markers. The reducing SDS–PAGE sample buffer was prepared by adding 288 mM 2-mercaptoethanol to the twofold-concentrated nonreducing sample buffer [20%(v/v) glycerol, 150 mM Tris–HCl pH 6.8, 4 mM EDTA, 4%(w/v) SDS, 0.4%(w/v) bromophenol blue dye]. The protein was dissolved in 8 M urea and mixed with the reducing or nonreducing sample buffer in a 1:1 ratio. It was boiled for 5 min and then cooled on ice before being loaded onto the gel. The separated proteins in the gel were stained with a solution containing 0.1%(w/v) Coomassie Brilliant Blue R-250, 40%(v/v) ethanol and 10%(v/v) acetic acid for 1 h and destained in a 40%(v/v) ethanol and 10%(v/v) acetic acid solution. The gel image was taken with a Nikon (Coolpix-995) digital camera.
2.3. Crystallization of Cor a 9
For crystallization trials, a 170 mg ml−1 Cor a 9 sample in buffer C was quickly diluted in a 1:1 ratio with water. The diluted sample was used to prepare samples with concentrations of 64, 43 and 21 mg ml−1 by mixing with different amounts of 0.5 M NaCl solution at 333 K. For each Cor a 9 sample (170, 85, 64, 43 or 21 mg ml−1), 11 2 µl aliquots were spotted onto EasyXtal CrystalSupports (Qiagen, Valencia, California, USA) and screw-sealed over 0.5 ml 0–0.5 M NaCl reservoir solutions with 0.05 M intervals; the crystallization trials were set up both at room temperature and at 277 K.
2.4. N-terminal amino-acid sequencing
After successful crystallization of Cor a 9, a sample was prepared by redissolving a number of the single crystals in 8 M urea and was mixed with the reducing sample buffer in a 1:1 ratio. This sample was subjected to SDS–PAGE using 8–25% gradient gels in a PhastSystem (GE Healthcare) and was electrophoretically transferred to a Problott membrane (Applied Biosystems, Foster City, California, USA) in transfer buffer [10 mM 3-(cyclohexylamino)-1-propanesulfonic acid containing 10%(v/v) methanol pH 11.0]. The transfer was carried out at 298 K at a constant current (0.35 A). The blot was stained with Coomassie Brilliant Blue and the main peptide bands were excised and subjected to N-terminal amino-acid sequencing by Edman degradation using a Procise Model 491 Protein Sequencer (Applied Biosystems). No attempts were made to derivatize cysteine residues to the PE-Cys form so that they could be analyzed. The obtained sequences were aligned with the published Cor a 9 sequence using the BLAST program.
2.5. X-ray diffraction experiments and crystal characterization
Sucrose, glycerol, PEG 400 and ethylene glycol were screened for cryoprotection of the Cor a 9 crystals. Single crystals were picked up with Mounted CryoLoops (Hampton Research, Aliso Viejo, California, USA) staked to CrystalCap Copper Magnetic (Hampton Research) pins, briefly immersed in cryoprotectant solution and flash-cooled in liquid nitrogen before X-ray diffraction. Data collection was performed using the SER-CAT 22-ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory. A complete data set was collected from crystals obtained using 30%(v/v) glycerol as a cryoprotectant after determining a data-collection strategy with HKL-2000 (Otwinowski & Minor, 1997 ▶): 120 1° frames were collected with 2.5 s exposure time and a crystal-to-detector (MAR CCD 300) distance of 200 mm. To solve the phase problem by molecular replacement (MR), one protomer of proglycinin (PDB code 1fxz, chain A; Adachi et al., 2001 ▶) was used as the starting model in MR calculations using the programs CHAINSAW (Stein, 2008 ▶) and Phaser (McCoy et al., 2005 ▶; Storoni et al., 2004 ▶).
3. Results and discussion
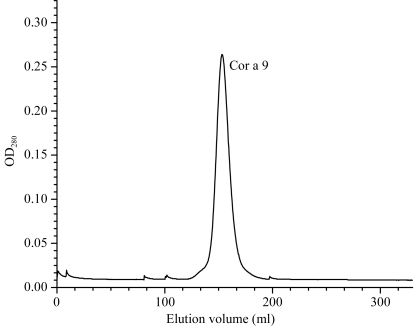
Cor a 9 is the hazelnut 11S seed-storage protein. Its physical and biological properties are similar to those of the 11S protein from brazil nut, Ber e 2 (Guo et al., 2007 ▶). Its solubility increases rapidly with increasing salt concentration and it can easily be desalted out as microcrystals. After threefold desalting-out, the Cor a 9 sample was very pure and the SEC purification profile showed only one peak at a position consistent with Cor a 9 being a hexamer (Fig. 1 ▶) based on the calibration curve obtained for the column. To ensure high purity for crystallization, Cor a 9 was further purified by HIC, but no noticeable improvement in the purity was observed (data not shown).
Figure 1.
Elution profile of Superdex-200 size-exclusion chromatography of Cor a 9.
As shown in Fig. 2 ▶, purified Cor a 9 gives multiple bands on SDS–PAGE, indicating that it consists of more than one type of subunit. This is consistent with the electrophoresis band patterns of 11S proteins from other species analyzed by SDS–PAGE (Albillos et al., 2008 ▶; Dickinson et al., 1989 ▶; Guo et al., 2007 ▶). It is known that 11S proteins are expressed as single peptide chains and are post-translationally cleaved to an N-terminal and a C-terminal domain by protease processing. The two domains are connected by a disulfide bond, which results in different patterns on SDS–PAGE under reducing and nonreducing conditions (Fig. 2 ▶). Interchain disulfide-bond link(s) between the cleaved peptides have also been observed in 11S proteins from other species (Kitamura et al., 1976 ▶). 11S proteins are rarely glycosylated. The origins of the split of the major bands into two and the nature of the low-molecular-weight bands present in the SDS gel are still not clear and require further investigations.
Figure 2.

Purification of Cor a 9. Purified Cor a 9 was analyzed by SDS–PAGE under both nonreducing (lane NR) and reducing (lane R) conditions. The molecular weights (in kDa) of the protein standards contained in the marker (lane M) are shown on the left of the gel image.
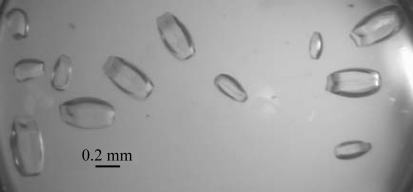
The solubility of Cor a 9 is greatly affected by salt concentration, which is useful for crystallization of the protein. At room temperature, Cor a 9 remains soluble at concentrations as high as 170 mg ml−1 in buffer C, while it saturates at ∼3 mg ml−1 in 0.1 M NaCl. Thus, crystallization trials were set up such that the reservoir solutions used were at a lower salt concentration than that in the starting crystallization drops and water molecules would diffuse from the reservoir to the drops during the equilibration process. The desalting-out effect was sufficient to drive both the crystal-nucleation process and subsequent crystal growth. About a week after crystallization setup, small crystals appeared in most of the trials containing 43 mg ml−1 protein. Only microcrystals grew in the trials containing 21 mg ml−1 protein, while a thick protein shell formed on the surface of drops containing 170 mg ml−1 Cor a 9. Diffraction-quality crystals were obtained at room temperature in drops containing 85 mg ml−1 Cor a 9 (in 0.5 M NaCl, 5 mM Tris pH 8) sealed over 0.2 M NaCl reservoir solution (Fig. 3 ▶). Further optimization did not result in significant improvement of the crystallization results.
Figure 3.
Single crystals of Cor a 9 obtained using the vapour-diffusion method.
11S proteins are known to be post-translationally cleaved by an asparaginyl endopeptidase after the formation of an interchain disulfide bond between the acidic N-terminal and basic C-terminal subunits (Badley et al., 1975 ▶; Kitamura et al., 1976 ▶). To confirm that the crystallized protein was Cor a 9, both the N-terminal subunit (which gives double bands in the SDS gel at ∼35 kDa) and the C-terminal subunit (which gives double bands at ∼24 kDa) were subjected to N-terminal amino-acid sequencing by Edman degradation. 20 cycles of N-terminal sequencing of the upper band of the N-terminal domain resulted in a peptide (IDVGLRRQQQRHFGEXNLDR) that has 85% sequence identity to residues 23–42 of the published Cor a 9 sequence (Beyer et al., 2002 ▶). 27 cycles of N-terminal sequencing of the upper band of the C-terminal domain resulted in a peptide (GFEETIXSLRLMENIRTRSRADIYTEQ) that has 89% sequence identity to residues 321–347 of the Cor a 9 sequence (Beyer et al., 2002 ▶). An X in the N-terminal sequencing result indicates a possible Cys residue that was not derivatized. If these residues were derivatized and determined to be Cys, the sequence identities would be 90% and 93% for the N- and C-terminal domains, respectively. This is consistent with the crystallized protein being Cor a 9. However, the data also indicated that there were significant sequence variations between the Cor a 9 crystallized in this study and the Cor a 9 isolated previously (Beyer et al., 2002 ▶), possibly because the sources of the hazelnuts used in this and the previous study were different and heterogeneous. Isolation of the Cor a 9 genes from the single source of hazelnuts that was used for protein purification is under way in order to deduce the entire sequence of the purified protein.
The cryoprotection tests gave 30% glycerol in 0.2 M NaCl as the best cryoprotectant for Cor a 9 crystals. The best Cor a 9 crystal diffracted to 1.89 Å resolution and a complete data set was collected. Data processing with HKL-2000 (Otwinowski & Minor, 1997 ▶) revealed that the crystal belonged to space group P21 (Table 1 ▶). Although the theoretical molecular mass of Cor a 9 deduced from its cDNA sequence is 59 kDa, removal of the signal peptide and post-translational peptidase processing of the 11S protein may give rise to a smaller protein. Owing to the very low solubility of Cor a 9 in low-salt buffer, determination of its molecular mass by mass spectrometry was not successful. Because of the presence of urea, estimation of its mass by the distance the protein migrated in the SDS gel may not be accurate. Using a molecular mass of 50 kDa for Cor a 9 and assuming the presence of six monomers in an asymmetric unit, the Matthews coefficient was estimated to be 1.69 Å3 Da−1, corresponding to protein crystals containing ∼27.4% solvent given an average partial specific volume of 0.74 cm3 g−1 for proteins. Assuming the presence of three Cor a 9 monomers in an asymmetric unit, the Matthews coefficient was estimated to be 3.39 Å3 Da−1, corresponding to protein crystals containing ∼63.7% solvent.
Table 1. X-ray data-collection statistics.
Values in parentheses are for the outer shell.
| Resolution (Å) | 50–1.89 (1.96–1.89) |
| Wavelength (Å) | 1.0 |
| Data-collection temperature (K) | 110 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 74.9, b = 91.1, c = 149.2, β = 90.32 |
| No. of observed reflections | 393136 |
| No. of unique reflections | 151061 |
| Completeness (%) | 94.1 (90.4) |
| Mean I/σ(I) | 10.8 (2.0) |
| Rmerge† (%) | 8.4 (47.4) |
R
merge = 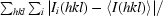
 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
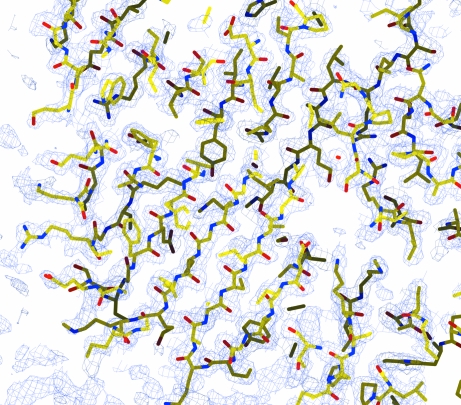
Searching the PDB database using BLAST showed that Cor a 9 and soybean proglycinin (PDB code 1fxz) have 46% sequence identity over 473 residues, with three gaps in Cor a 9 covering ten residues and three gaps in glycinin covering 13 residues. Proglycinin is the 11S seed-storage protein of soybean (Glycine max) and one of its isoforms has been purified and crystallized from a recombinant source (Adachi et al., 2001 ▶). A molecular-replacement calculation was carried out using CHAINSAW and Phaser (McCoy et al., 2005 ▶; Storoni et al., 2004 ▶) with monomeric proglycinin (PDB code 1fxz, chain A) as the initial structural model. This resulted in a structure solution with six Cor a 9 molecules in the asymmetric unit and a log-likelihood gain of 1829. The R factor of the solution after one round of ten cycles of restrained refinement with the program REFMAC (Murshudov et al., 1997 ▶) was 32.4%, with an R free of 40.5% using a 5% test set of reflections selected from thin shells. A preliminary inspection of the map calculation and refinement led to an experimental electron-density map at 1.89 Å resolution with a clear protein–solvent boundary (Fig. 4 ▶). The electron-density map allowed modeling of most of the main-chain atoms for all six monomers. However, as the Cor a 9 crystallized here consisted of multimeric hetero isoforms, full determination of this crystal structure will require further determination of the sequences of the different isoforms of this protein. Currently, model building and refinement of the structure and isolation of the Cor a 9 genes from a single hazelnut source are under way. Obtaining structural information on tree-nut allergens should result in a better understanding of the structural basis of the allergenicity and the cross-reactivity of nut allergens.
Figure 4.
A section of the 2F o − F c electron-density map. The map was calculated after a quick ten-cycle restrained refinement starting from a Phaser molecular-replacement solution. The map was contoured at 1.0σ, with the structure solution shown in stick representation using the CPK coloring scheme.
Acknowledgments
This work was supported by a cooperative agreement between the US Food and Drug Administration and the National Center for Food Safety and Technology (FD-0004331) and a fund from Illinois Institute of Technology. X-ray diffraction data were collected on Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the APS, Argonne National Laboratory. Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract W-31-109-Eng-38.
References
- Adachi, M., Takenaka, Y., Gidamis, A. B., Mikami, B. & Utsumi, S. (2001). J. Mol. Biol.305, 291–305. [DOI] [PubMed]
- Alasalvar, C., Karamac, M., Amarowicz, R. & Shahidi, F. (2006). J. Agric. Food Chem.54, 4826–4832. [DOI] [PubMed]
- Alasalvar, C., Shahidi, F., Liyanapathirana, C. M. & Ohshima, T. (2003). J. Agric. Food Chem.51, 3790–3796. [DOI] [PubMed]
- Albillos, S. M., Jin, T., Howard, A., Zhang, Y., Kothary, M. H. & Fu, T. J. (2008). J. Agric. Food Chem.56, 5352–5358. [DOI] [PubMed]
- Asero, R., Mistrello, G., Roncarolo, D. & Amato, S. (2004). J. Allergy Clin. Immunol.113, 358–360. [DOI] [PubMed]
- Badley, R. A., Atkinson, D., Hauser, H., Oldani, D., Green, J. P. & Stubb, J. M. (1975). Biochim. Biophys. Acta, 412, 214–228. [DOI] [PubMed]
- Beyer, K., Grishina, G., Bardina, L., Grishin, A. & Sampson, H. A. (2002). J. Allergy Clin. Immunol.110, 517–523. [DOI] [PubMed]
- Birmingham, N. P., Parvataneni, S., Hassan, H. M., Harkema, J., Samineni, S., Navuluri, L., Kelly, C. J. & Gangur, V. (2007). Int. Arch. Allergy Immunol.144, 203–210. [DOI] [PubMed]
- Bohle, B., Radakovics, A., Luttkopf, D., Jahn-Schmid, B., Vieths, S. & Ebner, C. (2005). Clin. Exp. Allergy, 35, 1392–1399. [DOI] [PubMed]
- Dickinson, C. D., Hussein, E. H. & Nielsen, N. C. (1989). Plant Cell, 1, 459–469. [DOI] [PMC free article] [PubMed]
- Guo, F., Jin, T., Howard, A. & Zhang, Y.-Z. (2007). Acta Cryst. F63, 976–979. [DOI] [PMC free article] [PubMed]
- Jin, T., Howard, A. & Zhang, Y.-Z. (2007). Acta Cryst. F63, 848–851. [DOI] [PMC free article] [PubMed]
- Kitamura, K., Takagi, T. & Shibasaki, K. (1976). Agric. Biol. Chem.40, 1837–1844.
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Mercanligil, S. M., Arslan, P., Alasalvar, C., Okut, E., Akgul, E., Pinar, A., Geyik, P. O., Tokgozoglu, L. & Shahidi, F. (2007). Eur. J. Clin. Nutr.61, 212–220. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Nguyen, S. A., More, D. R., Whisman, B. A. & Hagan, L. L. (2004). Ann. Allergy Asthma Immunol.92, 281–284. [DOI] [PubMed]
- Ortolani, C., Ballmer-Weber, B. K., Hansen, K. S., Ispano, M., Wuthrich, B., Bindslev-Jensen, C., Ansaloni, R., Vannucci, L., Pravettoni, V., Scibilia, J., Poulsen, L. K. & Pastorello, E. A. (2000). J. Allergy Clin. Immunol.105, 577–581. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pastorello, E. A. et al. (2002). J. Allergy Clin. Immunol.109, 563–570. [DOI] [PubMed]
- Savage, G. P. & McNeil, D. L. (1998). Int. J. Food Sci. Nutr.49, 199–203. [DOI] [PubMed]
- Schocker, F., Luttkopf, D., Muller, U., Thomas, P., Vieths, S. & Becker, W. M. (2000). Eur. J. Nutr.39, 172–180. [DOI] [PubMed]
- Shahidi, F., Alasalvar, C. & Liyana-Pathirana, C. M. (2007). J. Agric. Food Chem.55, 1212–1220. [DOI] [PubMed]
- Stein, N. (2008). J. Appl. Cryst.41, 641–643.
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Sutherland, M. F., O’Hehir, R. E., Czarny, D. & Suphioglu, C. (1999). J. Allergy Clin. Immunol.104, 889–890. [DOI] [PubMed]