FliJ is one of the essential cytoplasmic components of the flagellar type III protein-export apparatus; it has been expressed, produced and crystallized and the crystals have been characterized by X-ray diffraction.
Keywords: FliJ, type III protein-export apparatus, bacterial flagella
Abstract
The axial component proteins of the bacterial flagellum are synthesized in the cytoplasm and then translocated into the central channel of the flagellum by the flagellar type III protein-export apparatus for self-assembly at the distal growing end of the flagellum. FliJ is an essential cytoplasmic component of the export apparatus. In this study, Salmonella FliJ with an extra three residues (glycine, serine and histidine) attached to the N-terminus as the remainder of a His tag (GSH-FliJ) was purified and crystallized. Crystals were obtained by the sitting-drop vapour-diffusion technique using PEG 300 as a precipitant. GSH-FliJ crystals grew in the hexagonal space group P6122 or P6522. While the native crystals diffracted to 3.3 Å resolution, the diffraction resolution limit of mercury derivatives was extended to 2.1 Å. Anomalous and isomorphous difference Patterson maps of the mercury-derivative crystal showed significant peaks in their Harker sections, indicating the usefulness of the derivative data for structure determination.
1. Introduction
Bacteria swim in liquid environments by rotating a long helical filamentous organelle called the flagellum, which is composed of basal body rings and a tubular axial structure. The basal body rings act as the rotor of the rotary motor, which is embedded in the cell surface (Berg & Anderson, 1973 ▶; Silverman & Simon, 1974 ▶). The tubular axial structure can be divided into three parts: the filament, the hook and the rod (DePamphilis & Adler, 1971a ▶,b ▶). The filament, which has a typical length of 10–15 µm, is a helical assembly made up of a few tens of thousands of flagellin molecules and propels the cell by its rotation, which is driven by the rotary motor. The ∼55 nm long hook acts as a universal joint that smoothly transmits the torque generated by the motor to the filament regardless of its orientation. The hook is a helical assembly of about 120 copies of a single protein, FlgE. The rod is a drive shaft that connects the rotor rings and the hook. The rod, which has an approximate length of 30 nm, is a rather complex structure that is composed of at least four proteins (FlgB, FlgC, FlgF and FlgG). The axial structure contains three further proteins: HAP1 and HAP3, which form a joint connecting the filament and the hook, and HAP2, which forms a pentameric assembly at the distal end of the filament that assists in filament growth (for reviews, see Macnab, 2003 ▶; Berg, 2003 ▶).
All the component proteins of the flagellar axial structure are synthesized in the cytoplasm and are translocated into the central channel of the growing flagellum by the flagellar type III protein-export apparatus; they then travel through the channel to the growing end for self-assembly (Macnab, 2004 ▶; Minamino et al., 2008 ▶). The export apparatus consists of six integral membrane proteins, FlhA, FlhB, FliO, FliP, FliQ and FliR, and three cytoplasmic proteins, FliH, FliI and FliJ (Minamino & Macnab, 1999 ▶, 2000a ▶). The membrane components are believed to be located within the annular pore of the basal body MS ring to form the export gate. FliI is an ATPase (Fan & Macnab, 1996 ▶) and its entire molecular structure highly resembles those of the α and β subunits of F1-ATPase (Imada et al., 2007 ▶). FliH binds to the extreme N-terminal region of FliI to form the FliH2–FliI complex in the cytoplasm and suppresses the ATPase activity of FliI (Auvray et al., 2002 ▶; González-Pedrajo et al., 2002 ▶; Minamino & Macnab, 2000b ▶). FliI forms a hexameric ring to fully exert its ATPase activity when the FliH2–FliI complexes dock with the export gate formed by the membrane-protein components of the export apparatus (Minamino et al., 2006 ▶). FliI only facilitates initial entry of export substrates into the gate, with the energy of ATP hydrolysis being used to disassemble and release the FliI complex from the protein about to be exported (Minamino & Namba, 2008 ▶).
FliJ consists of 147 amino-acid residues with a molecular mass of 17 kDa. FliJ is an essential component for protein export and as it interacts with various flagellar axial proteins, it has been postulated to be a general chaperone for the export substrate proteins (Minamino et al., 2000 ▶). FliJ interacts not only with the FliH2–FliI complex and the export-gate proteins FlhA and FlhB (Minamino & Macnab, 2000a ▶; Fraser et al., 2003 ▶), but also with FliM (which is a component protein of the basal body C ring; González-Pedrajo et al., 2006 ▶) and flagellar specific chaperones such as FliT and FlgN (Evans et al., 2006 ▶). FliJ is therefore considered to regulate the whole export process through complex interactions, but its detailed function is still obscure. Therefore, the three-dimensional structure of FliJ is essential in order to understand the mechanism of flagellar protein export and the role of FliJ in this process. Here, we report the expression, purification and preliminary X-ray crystallographic study of FliJ.
2. Materials and methods
2.1. Protein expression and purification
A NdeI–BamHI fragment encoding the fliJ gene of Salmonella enterica serovar Typhimurium (GenBank accession No. M62408) from pMM405 (Minamino & Macnab, 2000a ▶) was inserted into the NdeI–BamHI site of pET15b (Novagen) to create the plasmid pMMIJ001. A 30 ml overnight culture of BL21(DE3)pLysS carrying pMMIJ001, which encodes FliJ with an N-terminal hexahistidine tag followed by a thrombin site (His-FliJ; MGSS-6×His-SSGLVPRGSH-FliJ), was inoculated into 3 l LB medium (10 g Bacto tryptone, 5 g yeast extract, 10 g NaCl per litre) containing 50 µg ml−1 ampicillin and 30 µg ml−1 chloramphenicol. Cells were grown at 303 K until the culture density reached an OD600 of 0.6. Expression of His-FliJ was induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 1 mM and the culture was continued for 4 h. The cells were harvested by centrifugation (6400g, 10 min, 277 K) and stored at 193 K. The cells were thawed, suspended in binding buffer (50 mM Tris–HCl pH 8.0, 500 mM NaCl, 50 mM imidazole) with a tablet of Complete protease-inhibitor cocktail (Boehringer Mannheim) and sonicated (ASTRASON model XL2020 sonicator, Misonix Inc.). The cell lysate was centrifuged (19 000g, 20 min, 277 K) to remove cell debris. The supernatant was loaded onto a HiTrap chelating column (GE Healthcare) equilibrated with binding buffer. Proteins were eluted using a linear gradient of 50–500 mM imidazole and fractions containing His-FliJ were collected. The His tag was removed using proteolytic cleavage by adding 50 units of thrombin (GE Healthcare) to the His-FliJ solution. The protease cleavage produced FliJ with extra three residues, glycine, serine and histidine, attached to its N-terminus as the remainder of the His tag (GSH-FliJ). The reaction was carried out at 277 K in a solution dialyzed overnight against binding buffer and the reactant was purified on a HiTrap chelating column followed by a HiTrap Benzamidine FF column (GE Healthcare) to remove the N-terminally His-tagged peptide, noncleaved His-FliJ and thrombin. The eluate was dialyzed against 50 mM MES–NaOH pH 6.0, 5 mM EDTA for 3 h at 277 K and GSH-FliJ was then further purified by cation-exchange chromatography on a HiTrap SP HP column (GE Healthcare). The peak fractions were applied onto a HiLoad Superdex 75 (26/60) column (GE Healthcare) in 50 mM Tris–HCl pH 8.0, 2 mM EDTA, 500 mM NaCl. The purity of the product was examined by SDS–PAGE and MALDI–TOF mass spectrometry (Voyager DE/PRO). The purified protein sample was concentrated to 5.0–6.0 mg ml−1 for further use.
2.2. Crystallization
Initial crystallization screening of GSH-FliJ was performed at 293 K by the sitting-drop vapour-diffusion technique using the following screening kits: Wizard I and II, Cryo I and II (Emerald Biostructures) and Crystal Screens I and II (Hampton Research). Each drop was prepared by mixing 1 µl protein solution (4–8 mg ml−1 GSH-FliJ, 50 mM Tris–HCl pH 8.0, 2 mM EDTA, 500 mM NaCl) with 1 µl reservoir solution and was equilibrated against 150 µl reservoir solution.
2.3. Preparation and crystallization of mercury derivative
FliJ contains a single cysteine residue in its sequence and hence mercury derivatives were prepared by adding methylmercury chloride to the purified GSH-FliJ solution to a final concentration of 1 mM for 15 h at 277 K. Prior to treatment with the mercury compound, the purified GSH-FliJ solution was stored with 1 mM DTT for 3 h at 277 K for complete reduction of the cysteine side chain and was dialyzed against 50 mM Tris–HCl pH 8.0, 500 mM NaCl to remove DTT. After treatment with mercury, the protein solution was dialyzed against 50 mM Tris–HCl pH 8.0, 500 mM NaCl to remove unbound methylmercury chloride.
2.4. Data collection and processing
All X-ray diffraction data were collected on SPring-8 beamline BL41XU (Harima, Japan). Since the concentration of PEG 300 in the crystallization drops was sufficiently high for cryoprotection, the crystals were directly transferred into liquid nitrogen for vitrification and mounted in a cryo-gas flow. The diffraction data were recorded on an ADSC Quantum 315 CCD detector (Area Detector Systems Corporation) at 35 K using a helium-cryocooling device (Rigaku). The diffraction data were indexed, integrated and scaled using the programs MOSFLM (Leslie, 1992 ▶) and SCALA from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). The statistics of data collection are summarized in Table 1 ▶.
Table 1. Summary of the data statistics.
Values in parentheses are for the highest resolution shell.
| Native | Mercury derivative | |
|---|---|---|
| Space group | P6122 or P6522 | P6122 or P6522 |
| Unit-cell parameters (Å, °) | a = b = 52.9, c = 193.6, γ = 120 | a = b = 52.9, c = 192.6, γ = 120 |
| Wavelength (Å) | 1.000 | 1.008 |
| Resolution (Å) | 48.4–3.3 (3.48–3.3) | 38.5–2.1 (2.21–2.1) |
| Observations | 38500 (5686) | 109081 (15888) |
| Unique reflections | 2825 (378) | 10037 (1426) |
| Completeness (%) | 100 (100) | 99.7 (100) |
| Redundancy | 13.6 (15.0) | 10.9 (11.1) |
| 〈I/σ(I)〉 | 3.4 (1.9) | 4.7 (2.2) |
| Rmerge† (%) | 8.1 (39.7) | 8.0 (31.7) |
| Ranom‡ (%) | 5.6 (11.2) |
R
merge = 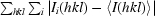 /
/ , where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
R
anom = 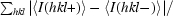
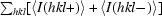 , where 〈I(hkl
, where 〈I(hkl
 )〉 and 〈I(hkl−)〉 correspond to the average intensities of each Friedel pair for reflection hkl.
)〉 and 〈I(hkl−)〉 correspond to the average intensities of each Friedel pair for reflection hkl.
3. Results and discussion
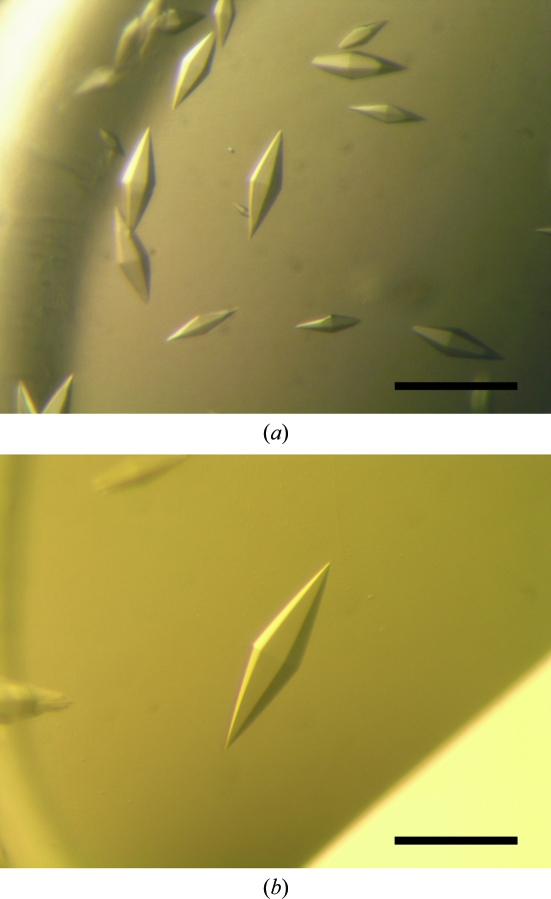
Within a week, small hexagonal bipyramidal crystals of GSH-FliJ appeared in several drops containing PEG 300 or PEG monomethyl ether 2000 in the pH range 4.2–5.2. We optimized the conditions by varying the precipitant concentration, pH and additives using the sitting-drop method. Finally, crystals suitable for X-ray analysis were obtained from drops prepared by mixing 1 µl protein solution (4.5 mg ml−1) with 1 µl reservoir solution containing 0.1 M phosphate–citrate pH 3.8, 20–30%(v/v) PEG 300 and 200–300 mM NaCl at 293 K. The crystals appeared within one week and grew to typical dimensions of 0.05 × 0.05 × 0.3 mm (Fig. 1 ▶ a).
Figure 1.
Hexagonal bipyramidal crystals of (a) GSH-FliJ and (b) its mercury derivative. The scale bar is 0.2 mm in length.
Although crystals of the mercury derivative appeared under the same conditions as used for the native GSH-FliJ crystals, only a limited number of small and highly stacked crystals were obtained. Therefore, silicone oil was added onto the reservoir solution to reduce nucleation and the speed of crystal growth by slowing the vapour-exchange rate (Chayen, 1997 ▶). This procedure improved the crystal growth and single crystals with typical dimensions of 0.1 × 0.1 × 0.4 mm were grown from a solution containing 0.1 M phosphate–citrate pH 3.8, 25–35%(v/v) PEG 300 and 200–300 mM NaCl using a protein concentration of 5.7 mg ml−1 (Fig. 1 ▶ b). We also tried to prepare SeMet-labelled FliJ crystals, but no crystals were obtained.
While both FliJ and His-FliJ tended to form insoluble aggregates, the cleavage of His-FliJ to GSH-FliJ resulted in highly soluble protein that was successfully crystallized. However, the detailed molecular nature of the dramatic increase in solubility of GSH-FliJ (which only has three additional N-terminal residues) remains unclear. The structure of GSH-FliJ should shed light on this question. Interestingly, MGSH-FliJ protein (GSH-FliJ with an additional N-terminal methionine residue) restored the swarming motility of a Salmonella ΔfliJ mutant to the wild-type level (data not shown), indicating that these extra amino-acid residues do not interfere with the function of FliJ.
Since the crystals of both GSH-FliJ and its mercury derivative were highly sensitive to X-ray irradiation, cryocooling to around 35 K was essential for data collection. In addition, we shifted the irradiating position on the crystal during measurement in order to minimize radiation damage.
The GSH-FliJ crystals diffracted to 3.3 Å resolution. The native crystals belonged to the hexagonal space group P6122 or P6522. The Matthews coefficient (V M; Matthews, 1968 ▶) suggested the presence of a single GSH-FliJ molecule in the asymmetric unit, with a solvent content of 53%. The GSH-FliJ mercury-derivative crystals, which were isomorphous to the wild-type protein crystals, grew larger than the native crystals and diffracted to 2.1 Å resolution. The dramatic improvement in the diffraction quality is probably a consequence of enhancement of the molecular-packing interactions in the crystal through the bound Hg atoms.
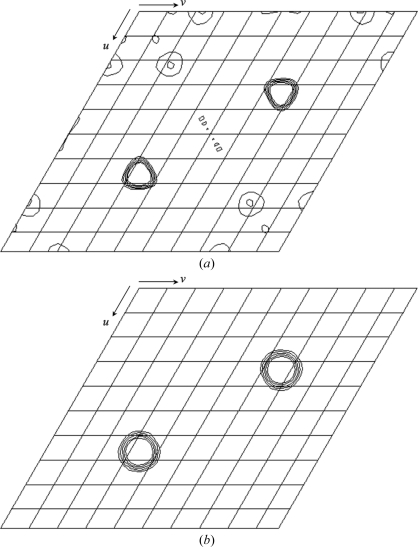
MALDI–TOF mass spectrometry of the mercury-derivative crystals indicated that about a third of the GSH-FliJ molecules did not bind mercury (data not shown). However, Bijvoet and isomorphous difference Patterson maps of the derivative showed common significant clear peaks on the Harker sections (Fig. 2 ▶), suggesting the usefulness of these data for phasing using the SAD or the SIRAS method.
Figure 2.
(a) Bijvoet and (b) isomorphous difference Patterson maps (w = 0.33 Harker section) calculated using data from the mercury derivative at 3.5 Å resolution. The contour lines are drawn from 2.0σ to 6.0σ with an increment of 1.0σ.
Acknowledgments
We thank N. Shimizu, M. Kawamoto and K. Hasegawa at SPring-8 for technical help in the use of beamline BL41XU. This work was supported in part by Grants-in-Aid for Scientific Research (18074006 to KI and 16087207 to KN) and the Targeted Proteins Research Program (TPRP) from the Ministry of Education, Science and Culture of Japan.
References
- Auvray, F., Ozin, A. J., Claret, L. & Hughes, C. (2002). J. Mol. Biol.318, 941–950. [DOI] [PMC free article] [PubMed]
- Berg, H. C. (2003). Annu. Rev. Biochem.72, 19–54. [DOI] [PubMed]
- Berg, H. C. & Anderson, R. A. (1973). Nature (London), 245, 380–382. [DOI] [PubMed]
- Chayen, N. E. (1997). Structure, 5, 1269–1274. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- DePamphilis, M. L. & Adler, J. (1971a). J. Bacteriol.105, 376–383. [DOI] [PMC free article] [PubMed]
- DePamphilis, M. L. & Adler, J. (1971b). J. Bacteriol.105, 384–395. [DOI] [PMC free article] [PubMed]
- Evans, L. D. B., Stafford, G. P., Ahmed, S., Fraser, G. M. & Hughes, C. (2006). Proc. Natl Acad. Sci. USA, 103, 17474–17479. [DOI] [PMC free article] [PubMed]
- Fan, F. & Macnab, R. M. (1996). J. Biol. Chem.271, 31981–31988. [DOI] [PubMed]
- Fraser, G. M., Gonzalez-Pedrajo, B., Tame, J. R. H. & Macnab, R. M. (2003). J. Bacteriol.185, 5546–5554. [DOI] [PMC free article] [PubMed]
- González-Pedrajo, B., Fraser, G. M., Minamino, T. & Macnab, R. M. (2002). Mol. Microbiol.45, 967–982. [DOI] [PubMed]
- González-Pedrajo, B., Minamino, T., Kihara, M. & Macnab, R. M. (2006). Mol. Microbiol.60, 984–998. [DOI] [PubMed]
- Imada, K., Minamino, T., Tahara, A. & Namba, K. (2007). Proc. Natl Acad. Sci. USA, 104, 131–137. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Macnab, R. M. (2003). Annu. Rev. Microbiol.57, 77–100. [DOI] [PubMed]
- Macnab, R. M. (2004). Biochim. Biophys. Acta, 1694, 207–217. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Minamino, T., Imada, K. & Namba, K. (2008). Mol. Biosyst.4, 1105–1115. [DOI] [PubMed]
- Minamino, T., Kazetani, K., Tahara, A., Suzuki, H., Furukawa, Y., Kihara, M. & Namba, K. (2006). J. Mol. Biol.360, 510–519. [DOI] [PubMed]
- Minamino, T. & Macnab, R. M. (1999). J. Bacteriol.181, 1388–1394. [DOI] [PMC free article] [PubMed]
- Minamino, T. & Macnab, R. M. (2000a). Mol. Microbiol.35, 1052–1064. [DOI] [PubMed]
- Minamino, T. & Macnab, R. M. (2000b). Mol. Microbiol.37, 1494–1503. [DOI] [PubMed]
- Minamino, T. & Namba, K. (2008). Nature (London), 451, 485–488. [DOI] [PubMed]
- Minamino, T., Ryan, C., Yamaguchi, S. & Macnab, R. M. (2000). J. Bacteriol.182, 4207–4215. [DOI] [PMC free article] [PubMed]
- Silverman, M. & Simon, M. (1974). Nature (London), 249, 73–74. [DOI] [PubMed]