The enzyme cgHle from C. glutamicum, which has acetyl ester hydrolase activity, was crystallized in four different crystal forms. X-ray diffraction data have been collected to a resolution of 1.2 Å.
Keywords: CgHle, Corynebacterium glutamicum, homoserine acetyltransferases
Abstract
CgHle is an enzyme that is encoded by gene cg0961 from Corynebacterium glutamicum. The physiological function of cgHle is so far unclear. Bioinformatic annotations based on sequence homology indicated that cgHle may be an acetyl-CoA:homoserine acetyl transferase and as such may be involved in methionine biosynthesis, but recent evidence has shown that it is an esterase that catalyzes the hydrolysis of acetyl esters. Here, the crystallization of cgHle in two orthorhombic crystal forms, a trigonal crystal form and a monoclinic crystal form is described. The trigonal crystals have a solvent content of 83.7%, which is one of the highest solvent contents ever found for protein crystals. One of the orthorhombic crystals diffracted X-rays to at least 1.2 Å resolution.
1. Introduction
Corynebacterium glutamicum is a Gram-positive prokaryote that is of great biotechnological importance owing to its ability to produce and release glutamate and other amino acids (Hermann, 2003 ▶). The genome of C. glutamicum has been sequenced twice (Kalinowski et al., 2003 ▶; Ikeda & Nakagawa, 2003 ▶) and many of its genes have been functionally annotated on the basis of sequence homology.
One of these genes is called cg0961 in the KEGG database (Kanehisa et al., 2008 ▶). The corresponding accession codes of the cg0961 gene product are Q8NS43 in the UniProt knowledge base (The UniProt Consortium, 2008 ▶) and YP_225131 in the NCBI protein database, which is available via the NCBI’s Entrez database system (http://www.ncbi.nlm.nih.gov/Entrez). Basic information about the protein encoded by cg0961 extracted from these databases is that it comprises 349 amino acids with a calculated molecular weight of 38 836 Da and a predicted α/β-hydrolase fold (Ollis et al., 1992 ▶).
Here, we describe the crystallization of this protein, which is designated as a homoserine acetyltransferase (HAT) in all three databases (KEGG, UniProt and the NCBI protein database). This annotation is based on the protein’s sequence identity of nearly 30% to HAT enzymes. HAT activity is required in the methionine-biosynthesis pathway; therefore, genes that encode genuine HAT enzymes are typically called metX (sometimes metA). However, many bacteria, including C. glutamicum, also contain paralogues of metX; these paralogues are assumed to be involved in an alternative route of the methionine-biosynthesis pathway and are therefore called met2.
cg0961 was an obvious candidate for the met2 gene of C. glutamicum. However, while the metX gene of C. glutamicum (cg0754 in KEGG) has been unambiguously demonstrated both to play a role in methionine biosynthesis (Rückert et al., 2003 ▶) and to encode a bona fide HAT enzyme (Park et al., 1998 ▶), neither of these has been demonstrated for cg0961. On the contrary, the possibility that cg0961 encodes an enzyme that catalyzes a step in the main or in an alternative route of the methionine pathway has been ruled out by deletion and complementation experiments (Rückert et al., 2003 ▶). Thus, the annotation of the cg0961 gene and the corresponding protein as stored in the aforementioned databases is incorrect.
We detected a probably more reliable hint concerning the function of the cg0961-derived protein in the Conserved Domain Database (Marchler-Bauer et al., 2007 ▶), which is also part of the NCBI database system. An inquiry in this database identifies the protein as a member of the esterase/lipase superfamily. This information derives from the fact that the cg0961-encoded protein has a sequence identity of about 60% to the esterase MekB from Pseudomonas veronii, which is involved in the degradation of alkyl methyl ketones by this strain (Onaca et al., 2007 ▶). In fact, like MekB, the cg0961-derived protein catalyzes the in vitro hydrolysis of p-nitrophenyl acetates. Hence, it cannot be excluded that the protein operates as an esterase rather than as a transferase in vivo.
Taken together, information retrieval from the literature and from standard life-science databases about the function of the cg0961/met2 gene leads to incomplete, partly contradictory and thus confusing results. To summarize current knowledge about the gene product, we refer to it as HAT-like esterase from C. glutamicum (cgHle) from here on. The cgHle crystallization we describe here is part of a comprehensive effort to clarify the function of the enzyme using enzymological and structural methods. The latter approach will involve a detailed structural comparison of cgHle with genuine HAT enzymes, two of which have been structurally characterized to date (Mirza et al., 2005 ▶; Wang et al., 2007 ▶).
2. Materials and methods
2.1. Expression of the cg0961 gene
The cg0961 gene was amplified by PCR with genomic DNA from C. glutamicum ATCC13032 serving as template. The gene-specific primers S4709, 5′-AAACATATGCTCGACAATAGTTTTTAC-3′, and S4711, 5′-GCAAGCTTAGCTCTCGAACAGC-3′, were used in this reaction, which introduced an additional NdeI restriction site at the putative N-terminal cg0961 sequence and a HindIII restriction site just behind the putative stop codon. The 1052 bp fragment was inserted into the l-rhamnose-inducible expression vector JOE2702 (Volff et al., 1996 ▶), which was cut with the appropriate restriction enzymes to create the plasmid pHWG771. Escherichia coli JM109 transformed with pHWG771 was used for the expression of cgHle. Cells were grown at 310 K to an OD600 of 0.3 in 200 ml 2×YT medium containing 100 µg ml−1 ampicillin. CgHle production was induced by the addition of 0.1%(w/v) rhamnose and cultivation continued for 4 h at 303 K. Cells were harvested by centrifugation, washed and resuspended in 20 ml 10 mM potassium phosphate buffer pH 6.5 and stored at 253 K.
2.2. Purification of cgHle
Recombinant E. coli cells were lysed by passing them twice through a French press cell at 7 MPa. Crude cell-free extract was obtained from the supernatant following centrifugation at 11 950g for 30 min at 277 K. The proteins in the supernatant were fractionated by anion-exchange chromatography. 40 mg crude cell-free extract was applied onto a Fractogel EMD-DMAE 150-10 column (Merck) pre-equilibrated with 10 mM potassium phosphate buffer pH 6.5. Bound proteins were eluted with an NaCl gradient (0–1 M) in the same buffer. 1 ml fractions were collected and tested for esterase activity. Active fractions were combined and further purified on a MonoQ HR 5/5 column (GE Healthcare) applying the same buffer gradient and tested further for purity by SDS–PAGE. The fractions containing the purified protein were combined, dialyzed against 10 mM HEPES buffer pH 7.0 and concentrated to a final protein concentration of 12.9 mg ml−1 by ultrafiltration on Centricon YM-10 devices (Millipore). The protein concentration was determined by the method of Bradford (1976 ▶) with bovine serum albumin as a standard.
2.3. Gel-filtration chromatography
We determined the native molecular weight of cgHle by gel-filtration chromatography with a Superdex 200 10/300 GL column (GE Healthcare). The column was equilibrated with a solution containing 150 mM NaCl, 5%(v/v) glycerol, 10 mM imidazole pH 6.5, which also served for elution. A calibration run was performed with ferritin (440 kDa), catalase (240 kDa), bovine serum albumin (66 kDa) and cytochrome c (12.4 kDa) as molecular-weight marker proteins.
2.4. Enzyme-activity assay
The esterase activity of cgHle was determined by measuring the rate of p-nitrophenyl acetate (pNp-Ac) hydrolysis. The assay was performed in 1 ml 0.1 M sodium phosphate buffer pH 6.5 at 298 K with 1 mM pNp-Ac. The reaction was started by adding the enzyme solution. The progress of pNp-Ac hydrolysis was followed by measuring the absorption at 410 nm in 10 s intervals for a total period of 2 min. One unit is defined as the amount of enzyme required to hydrolyse 1 µmol of pNp-Ac per minute.
2.5. Crystallization
For crystallization, the concentrated cgHle stock solution was diluted with 1 mM tris-(2-carboxyethyl)phosphine hydrochloride (TCEP) solution to a final protein concentration of 5 mg ml−1. All crystallization experiments were performed at 293 K using the sitting-drop variant of the vapour-diffusion method. For initial screening, we used various sparse-matrix and grid screens from Hampton Research.
2.6. Collection and processing of X-ray diffraction data
Crystals were prepared for diffraction experiments by flash-freezing in liquid nitrogen. Depending on whether cryoprotectants were present in sufficient concentrations in the mother liquor, the crystals were transferred to liquid nitrogen either directly from the crystallization drops (crystals 3 and 4 in Table 1 ▶) or after a quick soak in a cryoprotectant solution (crystals 1 and 2 in Table 1 ▶). X-ray diffraction data were collected either on beamline X12 of the EMBL Outstation in Hamburg (crystals 1 and 2 in Table 1 ▶) or on beamline BL14.2 of BESSY, Berlin (crystals 3 and 4 in Table 1 ▶). A MAR 224 CCD detector was mounted on both beamlines. The data-collection temperature was 100 K. The raw diffraction data were indexed and integrated with DENZO and scaled with SCALEPACK (Otwinowski et al., 1997 ▶). Wilson scaling and transformation to structure-factor amplitudes were performed using TRUNCATE from the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶).
Table 1. Crystallization of cgHle and X-ray diffraction of cgHle crystals.
Values in parentheses are for the highest resolution shell.
| Crystal No. | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Reservoir composition | 24%(w/v) PEG 8000, 0.2 M magnesium acetate, 0.1 M sodium cacodylate pH 6.6 | 24%(w/v) PEG 8000, 0.2 M magnesium acetate, 0.1 M sodium cacodylate pH 6.6 | 24%(w/v) PEG 4000, 20%(v/v) glycerol, 0.16 M MgCl2, 80 mM Tris–HCl pH 8.5 | 25.5%(w/v) PEG 4000, 15%(v/v) glycerol, 0.17 M lithium sulfate, 85 mM Tris–HCl pH 8.5 |
| Drop composition before equilibration | 0.8 µl cgHle solution, 0.4 µl reservoir solution, 0.4 µl 20 mM acetyl-CoA, 0.48 µl 30%(v/v) ethylene glycol | 0.8 µl cgHle solution,0.4 µl reservoir solution, 0.1 µl 30%(v/v) ethylene glycol | 0.8 µl cgHle solution, 0.4 µl reservoir solution | 0.8 µl cgHle solution, 0.4 µl reservoir solution |
| Cryoprotectant solution | 24%(w/v) PEG 8000, 0.2 M magnesium acetate, 0.1 M sodium cacodylate, 20%(v/v) ethylene glycol pH 6.6 | 24%(w/v) PEG 8000, 0.2 M magnesium acetate, 0.1 M sodium cacodylate, 20%(v/v) ethylene glycol pH 6.6 | — | — |
| Beamline | X12, EMBL, Hamburg | X12, EMBL, Hamburg | 14.1, BESSY, Berlin | 14.1, BESSY, Berlin |
| Wavelength (Å) | 0.90000 | 0.90000 | 0.91841 | 0.91841 |
| Space group | P21 | P212121 | P212121 | P3121 or P3221 |
| Unit-cell parameters (Å, °) | a = 56.79, b = 71.79, c = 91.14, β = 92.82 | a = 72.47, b = 98.96, c = 105.41 | a = 44.15, b = 89.58, c = 322.62 | a = 143.57, b = 143.5, c = 197.86 |
| VM (Å3 Da−1) | 2.39 | 2.43 | 2.05 | 7.52 |
| Solvent content (%) | 48.6 | 49.5 | 40.1 | 83.7 |
| Resolution range (Å) | 20.0–2.0 (2.07–2.00) | 51.2–1.2 (1.24–1.20) | 45.0–1.9 (1.97–1.90) | 48.5–3.3 (3.42–3.30) |
| Rmerge (%) | 17.0 (44.6) | 11.5 (62.8) | 17.7 (76.7) | 12.9 (79.2) |
| Completeness (%) | 97.2 (93.9) | 98.3 (96.9) | 92.6 (93.0) | 100 (100) |
| Redundancy | 3.1 (2.9) | 6.9 (5.2) | 9.4 (9.5) | 9.4 (8.7) |
| Signal-to-noise ratio [I/σ(I)] | 6.3 (2.2) | 16.0 (2.3) | 11.8 (2.3) | 18.8 (2.4) |
| Wilson B factor (Å2) | 10.4 | 7.8 | 16.8 | 100.1 |
2.7. Solvent-content analysis and Patterson searches
V M values (Matthews, 1968 ▶) were calculated for the cgHle crystals in order to estimate the numbers of protomers per asymmetric unit and the solvent contents of the crystals. Furthermore, in order to detect local twofold rotation axes we computed self-rotation functions with GLRF (Tong & Rossmann, 1997 ▶) in polar-angle mode and restricted to a κ angle of 180°. The resolution range was 15–4 Å in all cases. For some crystal forms additional native Patterson functions were calculated and analysed with corresponding programs from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The high-resolution limit of these maps was 6 Å.
3. Results and discussion
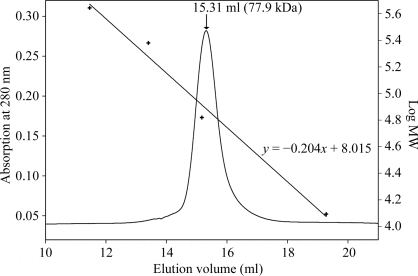
The bacterial expression system for the cg0961 gene was highly effective: according to an SDS–PAGE analysis (not shown), approximately 20% of the cellular protein was cgHle. On the basis of this overexpression, two successive anion-chromatography steps with different column materials were sufficient to purify cgHle to at least 95% homogeneity. The chromatogram of a final analytical gel-filtration run shown in Fig. 1 ▶ documents the high quality of the preparation.
Figure 1.
Molecular-weight determination of cgHle by gel-filtration chromatography. The figure shows an overlay of the chromatogram with a calibration curve obtained using four proteins (ferritin, 440 kDa; catalase, 240 kDa; bovine serum albumin, 66 kDa; cytochrome c, 12.4 kDa). The cgHle peak corresponds to a molecular weight of 77.9 kDa.
The catalytic activity remained stable over a period of a few months and was also not significantly impaired by freezing. According to the gel-filtration run (Fig. 1 ▶), the native molecular weight of cgHle is 77.9 kDa, which corresponds almost perfectly to a dimeric quaternary structure similar to those of the homologous homoserine acetyltransferases (Mirza et al., 2005 ▶; Wang et al., 2007 ▶).
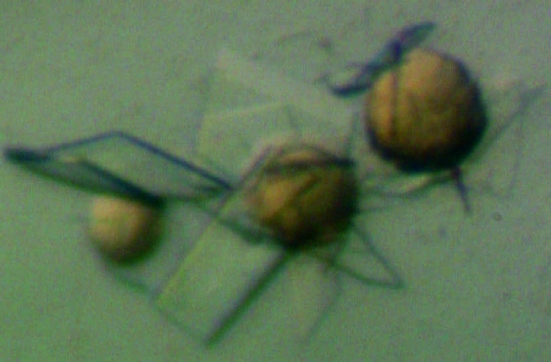
Four different crystal forms of cgHle could be obtained. The optimized crystallization conditions are listed in Table 1 ▶. The growth of crystal forms 1 and 2 was strictly dependent on the presence of magnesium acetate in the crystallization mixture. In many cases, we observed a kind of Ostwald ripening phenomenon (Ostwald, 1897 ▶): after 1 d of equilibration brownish spherulites appeared in the crystallization drops from which larger crystals grew after a while (Fig. 2 ▶). In many cases the spherulites finally converted completely to single crystals which could be used for diffraction experiments.
Figure 2.
Crystals of cgHle during growth from initially formed spherulites. The crystals belong to the orthorhombic form 2 (Table 1 ▶).
All crystals were frozen in liquid nitrogen either after transfer to cryoprotectant solutions (crystal forms 1 and 2; Table 1 ▶) or after direct harvesting from the crystallization drop (crystal forms 3 and 4). In the case of crystal forms 1 and 2 an annealing procedure comprising several rounds of thawing and refreezing (Harp et al., 1999 ▶) was helpful in improving the quality of the X-ray diffraction images.
X-ray diffraction data sets were collected from all four crystal forms; their characteristics are documented in Table 1 ▶. The best resolution (1.2 Å) was achieved with crystal form 2, which promises a cgHle structure of high quality. Scaling of the diffraction data from crystals 1 and 3 led to R merge values of more than 17% (Table 1 ▶). Plausible explanations of these relatively large values are the high redundancy of the data set in the case of crystal form 3 (Table 1 ▶) and impurities in the diffraction images of crystal form 1, i.e. strange spots that did not fit into the main diffraction pattern and originated from small twin crystals.
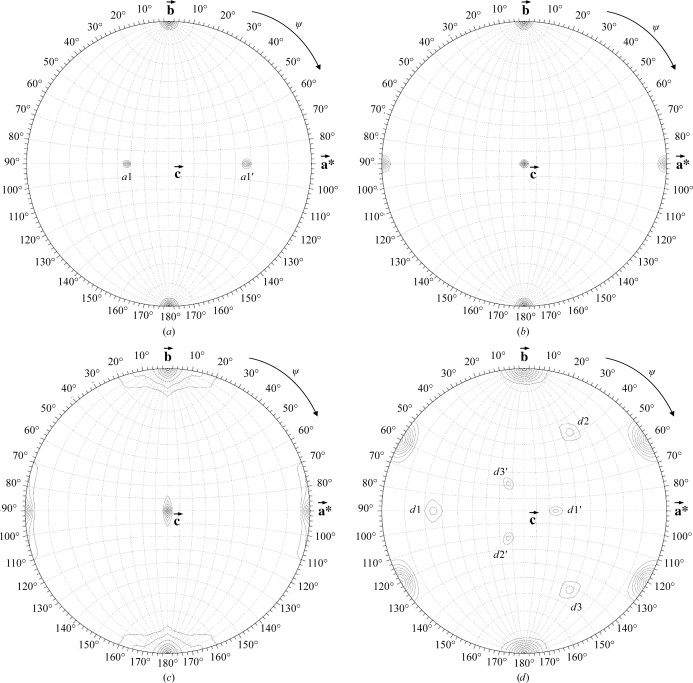
The dimeric nature of cgHle led us to calculate self-rotation functions and to search for local twofold-rotation axes (Fig. 3 ▶). We detected such a dyad in the monoclinic crystal form 1; it corresponds either to peak a1 or to peak a1′ in Fig. 3 ▶(a) and is perpendicular to the twofold axis of the monoclinic crystal symmetry. This particular arrangement of a crystallographic and a proper noncrystallographic axis results in a pseudo-rotation axis which is perpendicular to both [peak a1′ in Fig. 3 ▶(a) in the case that the arbitrary assignment of peak a1 to the proper noncrystallographic axis was correct].
Figure 3.
Search for 180° rotation axes in the four cgHle crystal forms listed in Table 1 ▶. (a) Crystal form 1, (b) crystal form 2, (c) crystal form 3, (d) crystal form 4.
In the trigonal crystal form (Fig. 3 ▶ d), we observed a similar 222 system consisting of a proper noncrystallographic twofold axis (peak d1), a crystallographic twofold axis and a pseudo-rotation axis (peak d1′). In this case, however, the whole arrangement occurs three times owing to the presence of threefold symmetry in the c direction (Fig. 3 ▶ d).
Taken together, the self-rotation functions of crystal forms 1 and 4 can be completely explained on the basis of the presence of one cgHle dimer per asymmetric unit, although in the case of the trigonal crystal form 4 this conclusion led to a V M value and a solvent content that were rather high (7.52 Å3 Da−1 and 83.7%; Table 1 ▶). Since we did not determine crystal densities experimentally, this is a preliminary result that needs to be confirmed by the complete structure solution; however, the low protein content of this crystal form is consistent with the fact that its diffraction power (the high-resolution limit is 3.3 Å) was much poorer than those of the three other crystal forms (Table 1 ▶). Moreover, a search of the Protein Data Bank (Berman et al., 2000 ▶) showed that the solvent content of 83.7% we determined for crystal form 4 is plausible as we detected more than 40 entries with solvent contents higher than 83%.
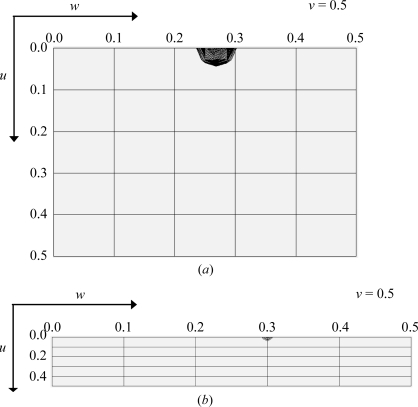
In the orthorhombic crystal form 2, no noncrystallographic twofold rotation axis could be found (Fig. 3 ▶ b). This could mean that the local dyad relating the protomers of a cgHle dimer is parallel to one of the three crystallographic 21 axes. In this case, the unit cell should contain pairs of cgHle dimers with identical orientations related by pure translation vectors. To check this possibility, we calculated a native Patterson function in the resolution range 50–6 Å in which we detected a pseudo-origin peak with fractional coordinates 0, 0.5, 0.27, i.e. located on the Harker section v = 0.5 (Fig. 4 ▶ a). This Patterson peak has a height of 24.1σ above the mean, which is about 50% of the height of the origin peak (46.2σ above the mean). Thus, the local twofold axis of the cgHle dimer must be parallel to the crystallographic 21 axis in the b direction.
Figure 4.
Native Patterson functions of the two orthorhombic cgHle crystal forms: (a) crystal form 2 and (b) crystal form 3. The Harker sections at v = 0.5 are drawn. In both cases the Patterson peak visible on the Harker section is the second highest of the whole Patterson function after the origin peak.
In the second orthorhombic crystal form (crystal form 3 in Table 1 ▶), again no noncrystallographic twofold rotation axis was obvious in the self-rotation map (Fig. 3 ▶ c). However, in this case an analysis of the native Patterson functions was less clear: we calculated such maps in various resolution ranges, but two weak pseudo-origin peaks were only visible under low-resolution conditions. One of these peaks has a height of 8.1σ above the mean, compared with 66.9σ above the mean for the origin peak, and lies on the v = 0.5 Harker section (Fig. 4 ▶ b). It is probable that the point-symmetry axes of the two corresponding cgHle dimers are only approximately parallel to the crystallographic b axis. A further Patterson peak with a height of 3.28σ above the mean, significantly higher than the noise peaks, appeared at 0.28, 0, 0, i.e. not on a Harker section. It may be that this peak relates two cgHle dimers of similar orientation within the asymmetric unit. Taken together, this would mean that the asymmetric unit of crystal form 3 contains four cgHle protomers. The corresponding packing density is relatively high (40.1% solvent content), which correlates with the relatively good diffraction power of these crystals (Table 1 ▶).
In summary, the outlined crystal-packing analyses are plausible; whether they are really correct will be become apparent from the corresponding crystal structures, which we are currently determining.
Acknowledgments
We are grateful to the staff of the EMBL Outstation in Hamburg and of BESSY in Berlin for support during X-ray diffraction experiments and to Sabine Lohmar for excellent technical assistance.
References
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res.28, 235–242. [DOI] [PMC free article] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Harp, J. M., Hanson, B. L., Timm, D. E. & Bunick, G. J. (1999). Acta Cryst. D55, 1329–1334. [DOI] [PubMed]
- Hermann, T. (2003). J. Biotechnol.104, 155–172. [DOI] [PubMed]
- Ikeda, M. & Nakagawa, S. (2003). Appl. Microbiol. Biotechnol.62, 99–109. [DOI] [PubMed]
- Kalinowski, J. et al. (2003). J. Biotechnol.104, 5–25. [DOI] [PubMed]
- Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., Katayama, T., Kawashima, S., Okuda, S., Tokimatsu, T. & Yamanishi, Y. (2008). Nucleic Acids Res.36, D480–D484. [DOI] [PMC free article] [PubMed]
- Marchler-Bauer, A. et al. (2007). Nucleic Acids Res.35, D237–D240.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mirza, I. A., Nazi, I., Korczynska, M., Wright, G. D. & Berghuis, A. M. (2005). Biochemistry, 44, 15768–15773. [DOI] [PubMed]
- Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. J., Silman, I., Schrag, J., Sussman, J. L., Verschueren, K. H. G. & Goldman, A. (1992). Protein Eng.5, 197–211. [DOI] [PubMed]
- Onaca, C., Kieninger, M., Engesser, K.-H. & Altenbuchner, J. (2007). J. Bacteriol.189, 3759–3767. [DOI] [PMC free article] [PubMed]
- Ostwald, W. (1897). Z. Phys. Chem.22, 289–330.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Park, S. D., Lee, J. Y., Kim, Y., Kim, J. H. & Lee, H. S. (1998). Mol. Cells, 8, 286–294. [PubMed]
- Rückert, C., Pühler, A. & Kalinowski, J. (2003). J. Biotechnol.104, 213–228. [DOI] [PubMed]
- The UniProt Consortium (2008). Nucleic Acids Res.36, D190–D195. [DOI] [PMC free article] [PubMed]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol.276, 594–611. [PubMed]
- Volff, J. N., Eichenseer, C., Viell, P., Piendl, W. & Altenbuchner, J. (1996). Mol. Microbiol.21, 1037–1047. [DOI] [PubMed]
- Wang, M., Liu, L., Wang, Y., Wei, Z., Zhang, P., Li, Y., Jiang, X., Xu, H. & Gong, W. (2007). Biochem. Biophys. Res. Commun.363, 1050–1056. [DOI] [PubMed]