Abstract
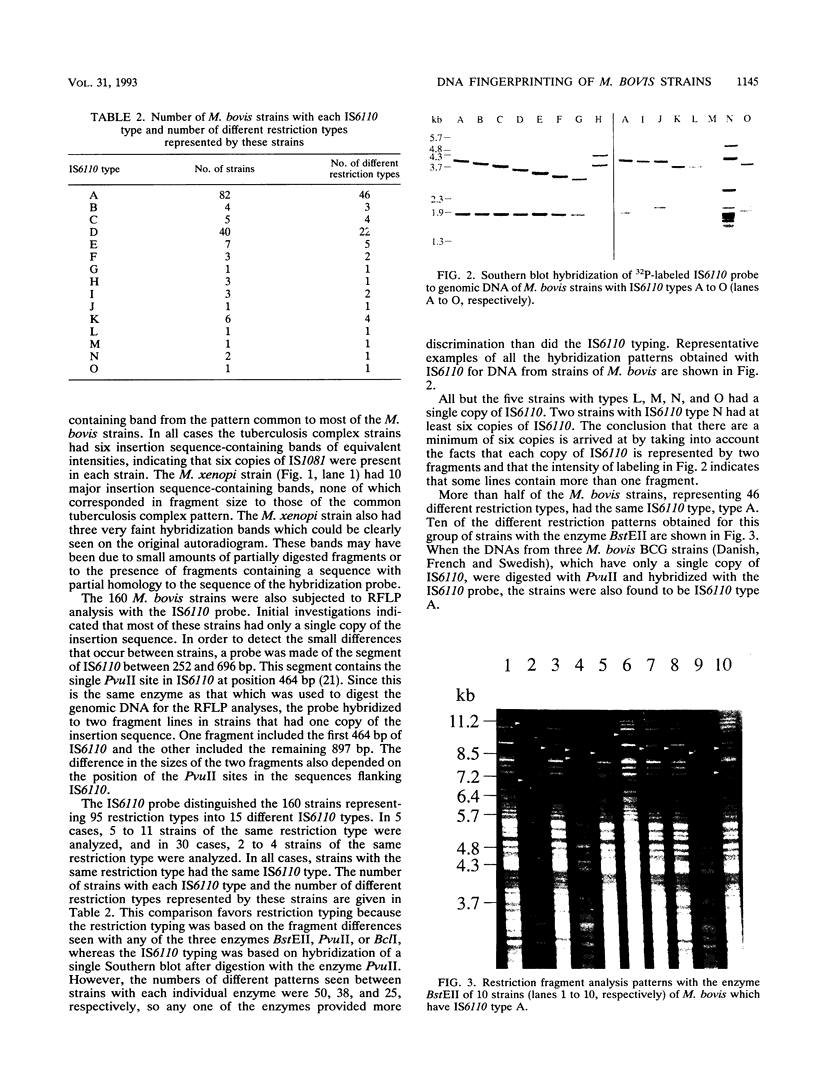
Strains of Mycobacterium bovis, the causative organism of bovine tuberculosis, can be clearly distinguished from each other by restriction fragment analysis. This method of DNA fingerprinting has been used for many epidemiological studies in New Zealand, but the technique presents practical difficulties that hinder its widespread use. The insertion element IS6110 is being widely used as a DNA probe for distinguishing restriction fragment polymorphisms among strains of Mycobacterium tuberculosis. Both this element and another recently sequenced element, IS1081, are also present in M. bovis. We assessed the usefulness of these two elements for distinguishing between 160 strains of M. bovis. These strains, most of which were isolated in New Zealand, were selected to be representative of the 95 different types that were identified among 530 strains that were previously typed by restriction fragment analysis. Fifteen IS6110 types were identified, but more than half of the strains representing 46 restriction types had the same IS6110 type. Virtually all M. bovis strains as well as strains of M. tuberculosis and Mycobacterium africanum had the same IS1081 type. The results indicate that for M. bovis, IS1081 cannot be used to type strains, IS6110 can be used to distinguish strains into broad groups, but only restriction fragment analysis is sufficiently sensitive for detailed epidemiological studies. An investigation of the host range of IS1081 revealed that, apart from its presence in species of the tuberculosis complex, it is also present in a strain of Mycobacterium xenopi.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baess I. Deoxyribonucleic acid relatedness among species of slowly-growing mycobacteria. Acta Pathol Microbiol Scand B. 1979 Aug;87(4):221–226. doi: 10.1111/j.1699-0463.1979.tb02430.x. [DOI] [PubMed] [Google Scholar]
- Collins D. M., De Lisle G. W. BCG identification by DNA restriction fragment patterns. J Gen Microbiol. 1987 Jun;133(6):1431–1434. doi: 10.1099/00221287-133-6-1431. [DOI] [PubMed] [Google Scholar]
- Collins D. M., De Lisle G. W. DNA restriction endonuclease analysis of Mycobacterium bovis and other members of the tuberculosis complex. J Clin Microbiol. 1985 Apr;21(4):562–564. doi: 10.1128/jcm.21.4.562-564.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins D. M., De Lisle G. W. DNA restriction endonuclease analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG. J Gen Microbiol. 1984 Apr;130(4):1019–1021. doi: 10.1099/00221287-130-4-1019. [DOI] [PubMed] [Google Scholar]
- Collins D. M., De Lisle G. W., Gabric D. M. Geographic distribution of restriction types of Mycobacterium bovis isolates from brush-tailed possums (Trichosurus vulpecula) in New Zealand. J Hyg (Lond) 1986 Jun;96(3):431–438. doi: 10.1017/s0022172400066201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins D. M., Gabric D. M., de Lisle G. W. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J Clin Microbiol. 1990 Jul;28(7):1591–1596. doi: 10.1128/jcm.28.7.1591-1596.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins D. M., Gabric D. M., de Lisle G. W. Typing of Mycobacterium bovis isolates from cattle and other animals in the same locality. N Z Vet J. 1988 Mar;36(1):45–46. doi: 10.1080/00480169.1988.35476. [DOI] [PubMed] [Google Scholar]
- Collins D. M., Stephens D. M. Identification of an insertion sequence, IS1081, in Mycobacterium bovis. FEMS Microbiol Lett. 1991 Sep 15;67(1):11–15. doi: 10.1016/0378-1097(91)90435-d. [DOI] [PubMed] [Google Scholar]
- Eisenach K. D., Crawford J. T., Bates J. H. Genetic relatedness among strains of the Mycobacterium tuberculosis complex. Analysis of restriction fragment heterogeneity using cloned DNA probes. Am Rev Respir Dis. 1986 Jun;133(6):1065–1068. doi: 10.1164/arrd.1986.133.6.1065. [DOI] [PubMed] [Google Scholar]
- Fomukong N. G., Dale J. W., Osborn T. W., Grange J. M. Use of gene probes based on the insertion sequence IS986 to differentiate between BCG vaccine strains. J Appl Bacteriol. 1992 Feb;72(2):126–133. doi: 10.1111/j.1365-2672.1992.tb01813.x. [DOI] [PubMed] [Google Scholar]
- Hermans P. W., van Soolingen D., Bik E. M., de Haas P. E., Dale J. W., van Embden J. D. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991 Aug;59(8):2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans P. W., van Soolingen D., Dale J. W., Schuitema A. R., McAdam R. A., Catty D., van Embden J. D. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990 Sep;28(9):2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek G. H., Cave M. D., Eisenach K. D., Wallace R. J., Jr, Bates J. H., Crawford J. T. Chromosomal DNA fingerprint patterns produced with IS6110 as strain-specific markers for epidemiologic study of tuberculosis. J Clin Microbiol. 1991 Sep;29(9):2030–2033. doi: 10.1128/jcm.29.9.2030-2033.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otal I., Martín C., Vincent-Lévy-Frebault V., Thierry D., Gicquel B. Restriction fragment length polymorphism analysis using IS6110 as an epidemiological marker in tuberculosis. J Clin Microbiol. 1991 Jun;29(6):1252–1254. doi: 10.1128/jcm.29.6.1252-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R., Kvach J. T., Mounts P. Isolation and restriction endonuclease analysis of mycobacterial DNA. J Gen Microbiol. 1986 Feb;132(2):541–551. doi: 10.1099/00221287-132-2-541. [DOI] [PubMed] [Google Scholar]
- Ross B. C., Raios K., Jackson K., Sievers A., Dwyer B. Differentiation of Mycobacterium tuberculosis strains by use of a nonradioactive Southern blot hybridization method. J Infect Dis. 1991 Apr;163(4):904–907. doi: 10.1093/infdis/163.4.904. [DOI] [PubMed] [Google Scholar]
- Schowalter D. B., Sommer S. S. The generation of radiolabeled DNA and RNA probes with polymerase chain reaction. Anal Biochem. 1989 Feb 15;177(1):90–94. doi: 10.1016/0003-2697(89)90019-5. [DOI] [PubMed] [Google Scholar]
- Thierry D., Cave M. D., Eisenach K. D., Crawford J. T., Bates J. H., Gicquel B., Guesdon J. L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990 Jan 11;18(1):188–188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoli E., Simonetti M. T., Labardi C., Lopes Pegna A., Meli E., Stanflin N., Susini S. Mycobacterium xenopi isolation from clinical specimens in the Florence area: review of 46 cases. Eur J Epidemiol. 1991 Nov;7(6):677–681. doi: 10.1007/BF00218681. [DOI] [PubMed] [Google Scholar]
- Wards B. J., Collins D. M., de Lisle G. W. Restriction endonuclease analysis of members of the Mycobacterium avium-M. intracellulare-M. scrofulaceum serocomplex. J Clin Microbiol. 1987 Dec;25(12):2309–2313. doi: 10.1128/jcm.25.12.2309-2313.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne L. G. Microbiology of tubercle bacilli. Am Rev Respir Dis. 1982 Mar;125(3 Pt 2):31–41. doi: 10.1164/arrd.1982.125.3P2.31. [DOI] [PubMed] [Google Scholar]
- Wayne L. G., Sramek H. A. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin Microbiol Rev. 1992 Jan;5(1):1–25. doi: 10.1128/cmr.5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mazurek G. H., Cave M. D., Eisenach K. D., Pang Y., Murphy D. T., Wallace R. J., Jr DNA polymorphisms in strains of Mycobacterium tuberculosis analyzed by pulsed-field gel electrophoresis: a tool for epidemiology. J Clin Microbiol. 1992 Jun;30(6):1551–1556. doi: 10.1128/jcm.30.6.1551-1556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lisle G. W., Collins D. M., Loveday A. S., Young W. A., Julian A. F. A report of tuberculosis in cats in New Zealand, and the examination of strains of Mycobacterium bovis by DNA restriction endonuclease analysis. N Z Vet J. 1990 Apr;38(1):10–13. doi: 10.1080/00480169.1990.35606. [DOI] [PubMed] [Google Scholar]
- van Soolingen D., Hermans P. W., de Haas P. E., Soll D. R., van Embden J. D. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991 Nov;29(11):2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D., Hermans P. W., de Haas P. E., van Embden J. D. Insertion element IS1081-associated restriction fragment length polymorphisms in Mycobacterium tuberculosis complex species: a reliable tool for recognizing Mycobacterium bovis BCG. J Clin Microbiol. 1992 Jul;30(7):1772–1777. doi: 10.1128/jcm.30.7.1772-1777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]