Abstract
Cells use a variety of intercellular structures, including gap junctions and synapses, for cell–cell communication. Here, we present recent advances in the understanding of thin membrane bridges that function in cell–cell signaling and intercellular transport. Cytonemes or filopodial bridges connect neighboring cells via mechanisms of adhesion, which enable ligand-receptor-mediated transfer of surface-associated cargoes from cell to cell. By contrast, tunneling nanotubes establish tubular conduits between cells that provide for the exchange of both cell-surface molecules and cytoplasmic content. We propose models for the biogenesis of both types of membrane bridges and describe how viruses use these structures for the purpose of cell-to-cell spread.
Introduction
Life depends on the ability of cells to communicate with each other. Much of this crosstalk occurs at cell–cell contacts and is regulated by complex structural interfaces. Neurological or immunological synapses transmit cell–cell signals through the extracellular space, relying on mechanisms of ligand-receptor signaling across tight cell–cell junctions. By contrast, gap junctions in animal cells and plasmodesmata in plants provide portals for the direct exchange of cytoplasmic contents and can efficiently propagate an intracellular signal from cell to cell (Figure 1). In addition to these well-established examples of cell–cell communication, advances in fluorescence-based imaging have recently illuminated thin, fragile and elongated intercellular membrane bridges [1–3]. These membrane bridges probably represent different biological structures that are formed, maintained and disassembled by different mechanisms. Given the diversity of these mechanisms, we review this emerging field in the context of well-characterized cell–cell contacts such as synapses and gap junctions (Figure 1).
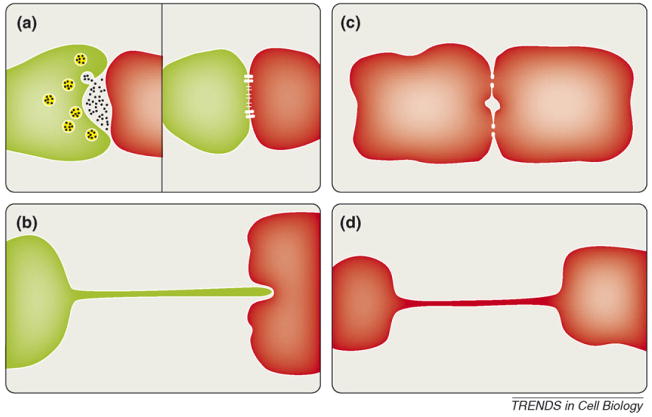
Figure 1.
Cytonemes and nanotubes in comparison to classical forms of cell–cell communication. (a) Neurological and immunological synapses transmit cell–cell signals through the extracellular space, relying on neurotransmitter release and receptor signaling through a tight cell–cell junction. (b) Cytonemes as thin filopodial bridges topologically identical to stretched out synaptic contacts. (c) By contrast, cells communicating using gap junctions establish direct connectivity between two cytoplasms. (d) Tunneling nanotubes represent thin cell–cell contacts with communicating cytoplasms.
Cytonemes and tunneling nanotubules
In a broad sense, two distinct types of thin membrane bridges have been described, which are distinguished by the ability of cytosolic content to traffic within the interior of the filament from cell to cell (tubular) versus contacts in which no cytoplasmic connection is made (non-tubular) (Figure 1). For the non-tubular bridges, two membranes are tightly juxtaposed at the site of cell–cell contact. Signals must be transduced using molecules resident at the outer surface of the membrane. By contrast, tubular membrane bridges provide an open conduit for the transfer of cytoplasmic material (Figure 1). Signals or cargo can be delivered through the central channel and along the continuous outer membrane. For the purpose of this review, bridges that do not connect cytoplasm but that still enable the transport of ligands across the outer surface are referred to as cytonemes or filopodial bridges. By contrast, structures that physically connect two cytoplasms are referred to as tunneling nanotubes (Figure 1). The term ‘bridge’ reflects the movement of cargo along the outside of the structure [4], whereas ‘tube’ is more consistent with the observed exchange of material via the interior of the tunnel [5,6].
The first description of a non-tubular bridge was by Ramirez-Weber and Kornberg, who observed actin-rich filopodia-like structures of up to 100 μm extending between the anterior and posterior compartments of the imaginal disc in fruit flies [7]. They named these structures ‘cytonemes’ after the Latin for ‘cell thread’. Cytonemes have been proposed to operate as long-range structures that enable a morphogen-patterning gradient for signaling molecules that are not readily soluble but that are membrane associated [1,7,8]. In support of this idea, morphogen receptors such as Decapentaplegic have been visually detected migrating along the length of the cytoneme, perhaps indicating actin-based trafficking of surface-bound signaling ligands [9].
Cytoneme structure and function require two critical factors: an intact actin cytoskeleton and cognate transmembrane receptor-ligand interactions that strengthen, lengthen and anchor the fiber between cells. In our laboratory, we have recently demonstrated the formation of structures between cells infected with retroviruses and uninfected target cells similarly dependent on actin and receptor-ligand interactions [4]. These ‘viral cytonemes’ have been shown to form only during reciprocal expression of a retroviral envelope (Env) glycoprotein on the surface of the infected cell and the appropriate viral receptor on the uninfected cell. Time-lapse microscopy and correlative fluorescent and scanning electron microscopy have demonstrated that viruses move directionally along the outer surface of these filopodial bridges to infect neighboring cells – vividly demonstrating that the establishment of membrane bridges is rapidly coupled to actin-mediated transport of cargo from one cell to another (Figure 2).
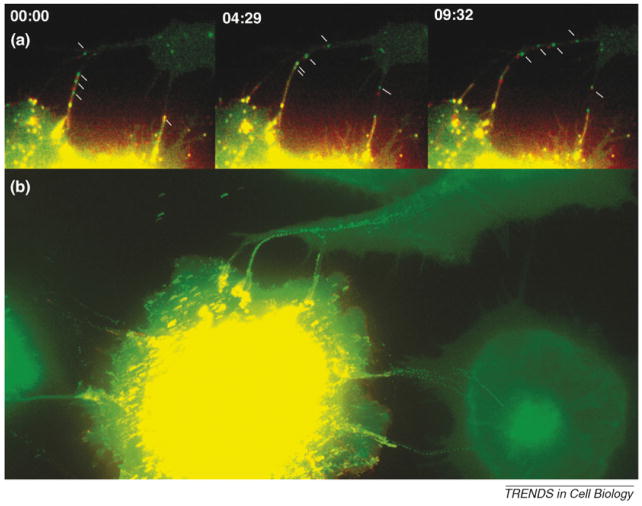
Figure 2.
Retroviruses establish filopodial bridges for the purpose of cell-to-cell transmission. (a) Filopodial bridges between virally infected cells (yellow) and target cells (green) support the transport of fluorescently labeled retroviruses from cell to cell. Time intervals are given in min:sec. (b) All frames from the time-lapse video depicted in (a) are superimposed into a single image to illustrate the individual tracks of viruses as they ‘walked’ along a filopodial bridge to infect neighboring cells.
Of a tubular nature, the structures termed ‘tunneling nanotubes’ were first described during live imaging of cultures of the neuron-like pheochromocytoma cell line PC12 [5]. Like cytonemes, tunneling nanotubes contain filamentous actin, can extend over several cell lengths and are capable of mediating the exchange of surface-membrane material. As an important new feature, tunneling nanotubes could also provide for the actin-based transit of endolysosomal cargo via the tubular interior. Similar nanotubes have since been identified in cultures of dendritic cells with monocytes that rapidly transmit a calcium signal from a single stimulated cell to multiple surrounding cells [6]. Additional ‘nanotubes’ have been described in cultures of human embryonic kidney 293T and normal rat kidney cells and for primary macrophages, T cells, activated B cells and human glioblastoma cells [5,10–12]. Thicker membrane bridges formed between macrophages can, in addition to actin filaments, contain microtubules that transport vesicles through their interior from cell to cell [10]. Finally, thin membrane structures that are also called ‘nanotubes’ can connect primary T cells and function as highways for the spread of HIV-1 virions from cell to cell [13]. Interestingly, these T-cell nanotubes do not enable the spread of a calcium signal, and electron micrographs have confirmed that their cytoplasms are not connected [13]. Consequently, these structures fall into a ‘non-tubular’ category, but are distinguished from viral cytonemes described in fibroblasts because linkage formation is not dependent on the viral Env glycoprotein [2,13].
Biogenesis of membrane bridges
The biogenesis of membrane bridges can probably proceed by either of two general mechanisms [1–3,14]. In the first, linkages form after a cell extends a de novo filopodial process that is then bound and tightly anchored to a neighboring cell (Figure 3a,b). Filopodia are dynamic exploratory and sensory organelles that can redirect cell migration towards specific sources, including nearby cells. These structures can be maintained to form a long-lived stable filopodial bridge or, alternatively, function as structural intermediates to more complex cell–cell interfaces (Figure 3c). In a second model of bridge formation, topologically similar structures result when two associated cells that are tightly adhered pull apart but retain a residual filamentous linkage [Figure 3, (c⃗a) or (d⃗f)]. As such, the membrane bridge or nanotube results from the ‘stretching out’ of a tight contact zone.
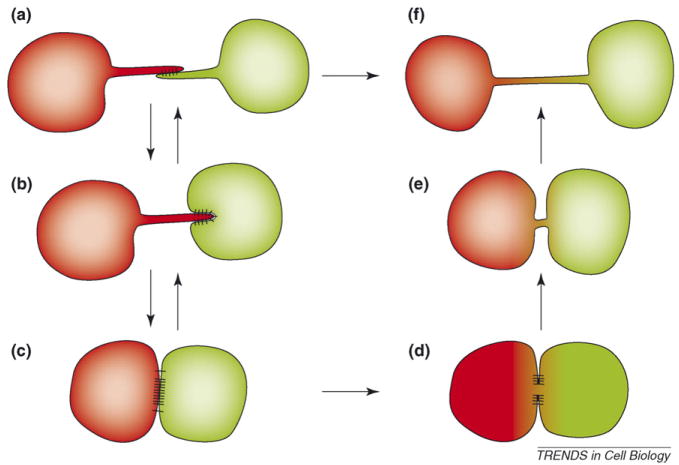
Figure 3.
Biogenesis of cytonemes and nanotubes in relationship to tight cell–cell contacts. Transient filopodial contacts (a) can stabilize to form elongated stable cytonemes (b). In the presence of additional adhesion proteins that favor strong cell–cell interfaces, cytonemes could mature into broad synaptic cell–cell interfaces (c). Downregulation of these cell–cell contacts could lead to cytoneme regeneration (c⃗a). At some synapses, cytoplasmic connectivity is established [61,62] (d). Downregulation of these cell–cell interfaces generates tunneling nanotubes with connected cytoplasms (e,f). Under specific circumstances, filopodial contacts might directly mature into tunneling nanotubes (a or b⃗f transition).
Filopodial intermediates in cell adhesion and synaptogenesis
Increasing evidence in various cell–cell adhesion systems points to an important role for filopodia in the establishment of cell–cell contacts. Our system, which uses virally infected cells expressing the viral Env glycoprotein as an adhesion protein, has demonstrated that ‘fingertip’ filopodial contacts between cells can lead to anchorage of filopodia from the uninfected target cell at the infected cell surface [4]. Maintenance of anchored bridges is dependent on the continued pulling of target-cell filopodia into the infected cell, probably representing a form of ‘frustrated phagocytosis’. Indeed, endocytic factors including dynamin and caveolin specifically accumulate at these sites of anchorage. Furthermore, tips of anchored filopodia are frequently torn off and internalized into endocytic compartments. As such, the actin–endocytic machinery has a key role in the formation and maintenance of these structures and can forcefully mediate the exchange of cell membranes.
Interestingly, there is substantial evidence accumulating that the binding and anchoring of filopodia is actually a prerequisite to the formation of broad cell–cell interfaces and synapses. During synaptogenesis between neurons, filopodial contacts precede the formation of a mature synapse [15–18]. Similarly, when primary keratinocytes reform a continuous cell layer after calcium signaling, they initially form a meshwork of intertwined filopodia called an ‘adhesion zipper’ [19]. The adhesion zipper then proceeds to form a tight epithelial sheet of cells that are tethered to each other by adherence junctions [19]. Finally, during the formation of the immunological synapse between T cells and antigen-presenting cells, filopodia precede the formation of a full synaptic cleft and might even persist as functional ‘probes’ operating in the cell–cell interface [20]. We predict that the crucial transition from a filopodial intermediate to a tighter cell–cell contact depends on the recruitment of additional adhesion proteins and intracellular adaptors that initiate the assembly of macromolecular cell–cell interfaces. For example, clusters of E-cadherin are recruited to nascent cell–cell contacts to establish adherence junctions [19]. In the case of neuronal synapses, synaptic adhesion proteins such as neuroligin and Syn-CAM are sufficient to induce presynaptic specialization upon contact in vitro [21,22]. Likewise, adhesion factors such as intercellular adhesion molecule 1 (ICAM-1) and lymphocyte function-associated antigen 1 (LFA-1) are crucial progenitors to the generation of immunological synapses [23]. As a corollary, we have predicted that in the absence of specialized epithelial or synaptic adhesion proteins, for example in our fibroblast cultures, a filopodial intermediate is stabilized to form a prolonged and elongated filopodial bridge.
Downregulation of tight cell–cell contacts
In contrast to the formation of filopodial bridges by de novo outgrowth, membrane bridges or nanotubes can also form when tight cell–cell contacts are downregulated (Figure 3). Time-lapse microscopy has monitored the formation of nanotubes and filopodial bridges during the separation of both macrophages and T cells [10,13]. Importantly, the formation of T-cell nanotubes requires a prolonged period of T-cell interaction, indicating a kinetic requirement for the accumulation of adhesion factors [13]. It should be noted that many of the residual membrane bridges described could, in fact, bear more similarity to retraction fibers, as observed for migrating or mitotic cells, than to de novo formed filopodia [24]. Retraction fibers differ from filopodia in subtle ways that could have molecular consequences for intercellular crosstalk. For instance, myosin II is located at the base of bona fide filopodia, but is found throughout the length of retraction fibers ([25] and references therein).
To summarize, based on these mechanisms we propose a general model for bridge and nanotube or synapse and junction formation regulated by cell-adhesion factors (Figure 3). Filopodial contacts form between cells, providing a thin interface for the exchange of membrane signals. Over time, these contacts proceed to a tight synapse provided that the cells express an appropriate repertoire of cellular-adhesion factors needed for synapse formation. The establishment of tight cell–cell contact is accompanied by a remodeling of the underlying actin cytoskeleton. Downregulation of adhesion factors, coupled with cell motility or retraction, then returns cells to a ‘tethered’ conformation. We find this model particularly appealing for the establishment and recycling of immunological synapses, in which the interaction between antigen-presenting cells and T cells must be transient [26]. Importantly, it has been observed that in the absence of additional adherence factors, filopodial bridges persist for extended periods of time and do not proceed to a tighter cell–cell interface [4,13].
It must be noted that there is striking visual evidence indicating that cellular processes can, in some cases, fuse at or near the fingertip to form an open-ended nanotubular conduit [5]. As such, filopodial linkages might proceed directly to an open conduit [Figure 3, (a⃗f) instead of (a⃗b)]. This observation is of some interest because it implies that, like in the case of virus–cell fusion, filopodial membranes could exhibit the capacity to promote intercellular lipid mixing and resolution of the apposed membrane bilayers. Furthermore, it poses an interesting problem to cells, in that they must either maintain sufficient physical distance or use an alternative molecular mechanism to prevent cell–cell fusion and the generation of a multinucleated syncytium.
Nanotubes and cytonemes in vivo
Because cell–cell adhesion profiles are vital to intercellular signaling, it follows that in vitro investigations, in which cells are cultured in the absence of their natural tissue context, can be misleading. The limitations of ‘suspension immunology’ and importance of intravital imaging have been widely recognized [27]. For instance, in vivo, dendritic cells are maintained in a wide interconnected web [28]. When cultured in vitro, they clump together, a feature that depends on the expression of E-cadherin [29]. Despite this clumping, in vitro cultured dendritic cells do form a nanotubular web, which probably reflects the in vivo tendency of these cells to form a continuous web. Despite this degree of uncertainty, evidence that nanotubes and cytonemes exist in vivo has been mounting. As discussed, cytonemes were first observed in the imaginal disc of flies [7]. Similar structures have been observed in mouse blastocysts and in mouse cornea [30,31]. Increases in the resolution of intravital imaging techniques should facilitate the study of nanotubes in vivo. Of course, in vitro model systems will continue to have a crucial role in probing the molecular mechanisms by which these structures are formed, maintained and downregulated.
Bridging the void: viral exploitation of membrane bridges
In culture, viral infection is thought to be several orders of magnitude more efficient under conditions of physical cell–cell contact between infected and uninfected cells [32–34]. In vivo, it is likely that direct cell–cell spread predominates in the dense and dynamic environment of an animal tissue. Viral spread is probably enhanced at cell–cell contacts because the mechanisms of viral egress and entry can be directly coupled in space and time [35,36]. However, only recently has live imaging enabled visual monitoring of these events, revealing a variety of elegant strategies by which viruses subvert the peripheral actin cytoskeleton to establish and/or exploit a filopodial or synaptic interface.
Mechanisms of cell–cell spread using membrane bridges are universally actin-dependent, but vary in terms of virus–cell topology and relative contribution from either the host or the target cell. As discussed, retroviruses including murine leukemia virus and HIV-1 can drive the formation of stable intracellular bridges whereby the viral envelope glycoprotein functions as an adhesion protein to anchor target-cell filopodia in the infected cell [4] (Figure 4a,b). For HIV-1-infected primary T cells, this activity could be ancillary because T-cell-membrane tethers also form in the absence of Env, presumably owing to alternative adherence factors [13]. After budding, viruses ‘surf’ directionally along the outer surface of the linkage toward the uninfected target cell before fusion and entry at the cell body (Figure 4b). Surfing is driven by retrograde F-actin flow, a myosin II-driven process occurring constitutively in filopodia that delivers bound substrates inward toward the cell center [25]. Because surfing is both Env and receptor dependent, engagement of the underlying actin filament from across the membrane probably involves signaling through cytoplasmic tails, as has been observed for the movement of epidermal growth factor (EGF)–EGF receptor complexes along filopodia [37,38]. Importantly, these studies demonstrated that interactions between the viral Env glycoprotein and its receptor go beyond target specificity and virus–cell fusion, but are also crucial for the establishment and manipulation of cell–cell contacts and the directional movement of viral particles from cell to cell.
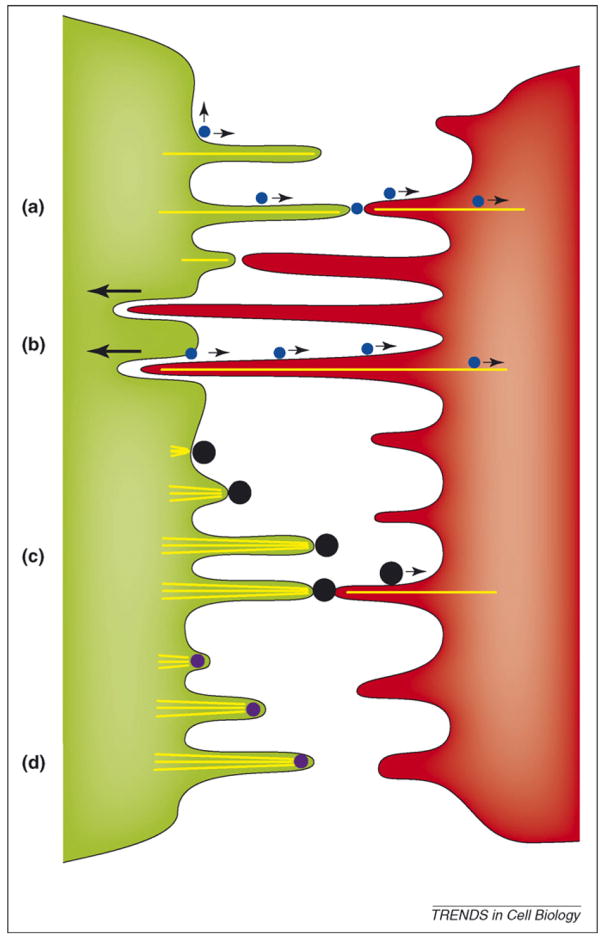
Figure 4.
Viral use and manipulation of filopodia for the purpose of viral spread. (a) Viruses (blue circles) accumulating at the surface of infected cells (green) can be passed on by transient contact to filopodia of target cells (red) where they use the underlying retrograde flow of filamentous actin to infect cells at the cell body [25]. (b) Continued filopodial contact leads to the internalization of target cell filopodia into the infected cell [4]. Viruses then bud at the infected–target cell interface and move along the filopodial bridge powered by the underlying actin flow (actin is depicted by yellow lines). (c) Vaccinia virus (black circles) induces the formation of actin comet tails beneath the released virus particle, from ‘outside in’ to propel itself towards the target cell [39]. Once in contact with the target cell, particles can still engage the underlying actin flow to further advance towards the cell body [45]. (d) By contrast, African swine fewer virus induces the formation of propulsive filopodia while still residing inside the cytoplasm of the infected cell [40]. The mechanism by which the virus is transferred to target cells is not known.
Given the importance of filopodia to the establishment of cell–cell contacts, it is not surprising that large DNA viruses such as African swine fever and the poxvirus vaccinia encode viral factors that actively induce the generation of filopodia to propel virus particles toward neighboring cells [39,40] (Figure 4d). African swine fever virus capsids traffic to the cytosolic face of the plasma membrane before signaling actin polymerization that projects the capsid outward [40]. Vaccinia uses a more elaborate strategy whereby, upon release at the cell surface, the extracellular cell-associated enveloped vaccinia intermediate (CEV) triggers assembly of an actin comet tail from the ‘outside in’ [41,42]. Comet-tail formation requires phosphorylation of viral factor A36R, an event that initiates signaling required for the recruitment of the actin-related 2/3 complex responsible for the nucleation of actin polymerization [43,44]. Interestingly, released vaccinia particles can still undergo surfing on filopodia during their entry into target cells [45]. This observation demonstrates that filopodial-trafficking mechanisms can be crucial to opposing stages of the viral life cycle. In an alternative strategy, Ebola and Marburg filoviruses form dramatic ‘filopodia-like’ filamentous viral particles that could facilitate transmission to neighboring cells [46–48]. Pseudorabiesvirus and herpes simplex virus are also capable of inducing extended projections on infected cells and these structures are implicated in viral spread [49–51].
As discussed, the biogenesis of membrane bridges is closely related to the formation of cell–cell synapses (Figure 3). Consequently, viral manipulation of synaptic structures follows similar principles. Pseudorabies and herpes viruses spread throughout the nervous system by means of neuronal synapses [52]. Others, such as HIV-1 and HTLV-1 use ‘virological synapses’ in infected lymphocytes, so named in analogy to immunological synapses [53–55]. Like viral cytonemes, the formation of virological synapse requires Env–receptor interactions and an intact cytoskeleton [34,54]. Virological synapses represent an elegant example of how viruses can manipulate an otherwise antiviral interface to favor their dissemination. HIV-1 expresses a viral effector protein, Nef, which interferes with the formation of immunological synapses and, consequently, prevents full T-cell activation [56–59]. Although HIV interferes with the formation of efficient immunological synapses, it uses part of the synaptic-core machinery, the tyrosine family kinase Zap70, to promote viral spread [60]. Thus, synapse modulation can function to both favor viral spread and provide for viral evasion of the host immune response.
Concluding remarks and future perspectives
Cells communicate with each other using a variety of distinct structures. As is the case for classical, well-characterized interfaces such as synapses and gap junctions, it is clear that mechanisms of contact and communication by membrane bridges are diverse and multi-faceted. In this review, we have emphasized cytoplasmic connectivity as a defining criterion and summarized testable models for the biogenesis and downregulation of membrane bridges. Thorough studies assessing the ability of contacts to transmit calcium or exchange endolysosomes through the interior or, alternatively, transport cargo along the outside of a membrane bridge would facilitate the comparison of datasets obtained in different systems [4,6,13]. Real-time kinetic analyses will be vital in characterizing the formation of these dynamic structural linkages and in determining the mechanistic details of their cell–cell signaling. In this sense, viruses and other obligate intracellular organisms will continue to serve as invaluable tools for further probing aspects of membrane-bridge structure and function.
Additionally, the field must ultimately define the molecular determinants that define various membrane bridges as distinct. Given the importance of adhesion molecules, their identification is of great importance to the understanding of bridge biogenesis. The establishment of criteria to distinguish filopodia from retraction fibers and tunneling nanotubes is crucial. Common criteria and a unifying terminology would be beneficial for the field. Finally, pushing imaging techniques toward higher resolution to visualize thin membrane bridges in vivo would clarify to what extend in vitro data will translate into in vivo relevance. Further understanding of membrane bridges will lead to insights into the basic understanding of cellular crosstalk and to the identification of new targets with relevance to therapeutic and, especially, antimicrobial strategies.
Acknowledgments
We thank Jolynne Roorda for art work and Thomas Biederer and Jing Jin for critical reading of the manuscript. The work was supported by EMBO long-term fellowship (ALTF 176–2007) to N.S. and a NIH grant (CA098727) to W.M.
References
- 1.Ramirez-Weber FA, Kornberg TB. Signaling reaches to new dimensions in Drosophila imaginal discs. Cell. 2000;103:189–192. doi: 10.1016/s0092-8674(00)00111-2. [DOI] [PubMed] [Google Scholar]
- 2.Davis DM, Sowinski S. Membrane nanotubes: dynamic long-distance connections between animal cells. Nat Rev Mol Cell Biol. 2008;9:431–436. doi: 10.1038/nrm2399. [DOI] [PubMed] [Google Scholar]
- 3.Gerdes HH, Carvalho R. Intercellular transfer mediated by tunneling nanotubes. Curr Opin Cell Biol. 2008;20:470–475. doi: 10.1016/j.ceb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Sherer NM, et al. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rustom A, et al. Nanotubular highways for intercellular organelle transport. Science. 2004;303:1007–1010. doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- 6.Watkins SC, Salter RD. Functional connectivity between immune cells mediated by tunneling nanotubules. Immunity. 2005;23:309–318. doi: 10.1016/j.immuni.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Ramirez-Weber FA, Kornberg TB. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 1999;97:599–607. doi: 10.1016/s0092-8674(00)80771-0. [DOI] [PubMed] [Google Scholar]
- 8.De Joussineau C, et al. δ-promoted filopodia mediate long-range lateral inhibition in Drosophila. Nature. 2003;426:555–559. doi: 10.1038/nature02157. [DOI] [PubMed] [Google Scholar]
- 9.Hsiung F, et al. Dependence of Drosophila wing imaginal disc cytonemes on Decapentaplegic. Nature. 2005;437:560–563. doi: 10.1038/nature03951. [DOI] [PubMed] [Google Scholar]
- 10.Onfelt B, et al. Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J Immunol. 2006;177:8476–8483. doi: 10.4049/jimmunol.177.12.8476. [DOI] [PubMed] [Google Scholar]
- 11.Onfelt B, et al. Cutting edge: membrane nanotubes connect immune cells. J Immunol. 2004;173:1511–1513. doi: 10.4049/jimmunol.173.3.1511. [DOI] [PubMed] [Google Scholar]
- 12.Gupta N, DeFranco AL. Visualizing lipid raft dynamics and early signaling events during antigen receptor-mediated B-lymphocyte activation. Mol Biol Cell. 2003;14:432–444. doi: 10.1091/mbc.02-05-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sowinski S, et al. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat Cell Biol. 2008;10:211–219. doi: 10.1038/ncb1682. [DOI] [PubMed] [Google Scholar]
- 14.Gurke S, et al. The art of cellular communication: tunneling nanotubes bridge the divide. Histochem Cell Biol. 2008;129:539–550. doi: 10.1007/s00418-008-0412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knott GW, et al. Spine growth precedes synapse formation in the adult neocortexin vivo. Nat Neurosci. 2006;9:1117–1124. doi: 10.1038/nn1747. [DOI] [PubMed] [Google Scholar]
- 16.Fiala JC, et al. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci. 1998;18:8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niell CM, et al. In vivo imaging of synapse formation on a growing dendritic arbor. Nat Neurosci. 2004;7:254–260. doi: 10.1038/nn1191. [DOI] [PubMed] [Google Scholar]
- 18.Ziv NE, Smith SJ. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron. 1996;17:91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]
- 19.Vasioukhin V, et al. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 20.Williams GS, et al. Membranous structures transfer cell surface proteins across NK cell immune synapses. Traffic. 2007;8:1190–1204. doi: 10.1111/j.1600-0854.2007.00603.x. [DOI] [PubMed] [Google Scholar]
- 21.Scheiffele P, et al. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 22.Biederer T, et al. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 23.Dustin ML, et al. Membranes as messengers in T cell adhesion signaling. Nat Immunol. 2004;5:363–372. doi: 10.1038/ni1057. [DOI] [PubMed] [Google Scholar]
- 24.Mitchison TJ. Actin based motility on retraction fibers in mitotic PtK2 cells. Cell Motil Cytoskeleton. 1992;22:135–151. doi: 10.1002/cm.970220207. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann MJ, et al. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mempel TR, et al. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427:154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 27.Sumen C, et al. Intravital microscopy: visualizing immunity in context. Immunity. 2004;21:315–329. doi: 10.1016/j.immuni.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Lindquist RL, et al. Visualizing dendritic cell networks in vivo. Nat Immunol. 2004;5:1243–1250. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- 29.Jiang A, et al. Disruption of E-cadherin-mediated adhesion induces a functionally distinct pathway of dendritic cell maturation. Immunity. 2007;27:610–624. doi: 10.1016/j.immuni.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chinnery HR, et al. Cutting edge: membrane nanotubes in vivo: a feature of MHC class II+ cells in the mouse cornea. J Immunol. 2008;180:5779–5783. doi: 10.4049/jimmunol.180.9.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salas-Vidal E, Lomeli H. Imaging filopodia dynamics in the mouse blastocyst. Dev Biol. 2004;265:75–89. doi: 10.1016/j.ydbio.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Dimitrov DS, et al. Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol. 1993;67:2182–2190. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carr JM, et al. Rapid and efficient cell-to-cell transmission of human immunodeficiency virus infection from monocyte-derived macrophages to peripheral blood lymphocytes. Virology. 1999;265:319–329. doi: 10.1006/viro.1999.0047. [DOI] [PubMed] [Google Scholar]
- 34.Chen P, et al. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007;81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips DM. The role of cell-to-cell transmission in HIV infection. AIDS. 1994;8:719–731. doi: 10.1097/00002030-199406000-00001. [DOI] [PubMed] [Google Scholar]
- 36.Johnson DC, Huber MT. Directed egress of animal viruses promotes cell-to-cell spread. J Virol. 2002;76:1–8. doi: 10.1128/JVI.76.1.1-8.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lidke DS, et al. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J Cell Biol. 2005;170:619–626. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lidke DS, et al. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 39.Cudmore S, et al. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 40.Jouvenet N, et al. African swine fever virus induces filopodia-like projections at the plasma membrane. Cell Microbiol. 2006;8:1803–1811. doi: 10.1111/j.1462-5822.2006.00750.x. [DOI] [PubMed] [Google Scholar]
- 41.Smith GL, et al. Vaccinia virus motility. Annu Rev Microbiol. 2003;57:323–342. doi: 10.1146/annurev.micro.57.030502.091037. [DOI] [PubMed] [Google Scholar]
- 42.Newsome TP, et al. SRC mediates a switch from microtubule-to actin-based motility of vaccinia virus. Science. 2004;306:124–129. doi: 10.1126/science.1101509. [DOI] [PubMed] [Google Scholar]
- 43.Frischknecht F, et al. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature. 1999;401:926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- 44.Frischknecht F, Way M. Surfing pathogens and the lessons learned for actin polymerization. Trends Cell Biol. 2001;11:30–38. doi: 10.1016/s0962-8924(00)01871-7. [DOI] [PubMed] [Google Scholar]
- 45.Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 46.Kolesnikova L, et al. Budding of Marburgvirus is associated with filopodia. Cell Microbiol. 2007;9:939–951. doi: 10.1111/j.1462-5822.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- 47.Hartlieb B, Weissenhorn W. Filovirus assembly and budding. Virology. 2006;344:64–70. doi: 10.1016/j.virol.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 48.Noda T, et al. Assembly and budding of Ebolavirus. PLoS Pathog. 2006;2:e99. doi: 10.1371/journal.ppat.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Favoreel HW, et al. Cytoskeletal rearrangements and cell extensions induced by the US3 kinase of an αherpes virus are associated with enhanced spread. Proc Natl Acad Sci U S A. 2005;102:8990–8995. doi: 10.1073/pnas.0409099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.La Boissiere S, et al. Compartmentalization of VP16 in cells infected with recombinant herpes simplex virus expressing VP16-green fluorescent protein fusion proteins. J Virol. 2004;78:8002–8014. doi: 10.1128/JVI.78.15.8002-8014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gill MB, et al. A γ-herpes virus glycoprotein complex manipulates actin to promote viral spread. PLoS ONE. 2008;3:e1808. doi: 10.1371/journal.pone.0001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon H, et al. Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell. 2005;123:669–682. doi: 10.1016/j.cell.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 53.McDonald D, et al. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science. 2003;300:1295–1297. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- 54.Jolly C, et al. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med. 2004;199:283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Igakura T, et al. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science. 2003;299:1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- 56.Thoulouze MI, et al. Human immunodeficiency virus type-1 infection impairs the formation of the immunological synapse. Immunity. 2006;24:547–561. doi: 10.1016/j.immuni.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 57.Wildum S, et al. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J Virol. 2006;80:8047–8059. doi: 10.1128/JVI.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindler M, et al. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell. 2006;125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 59.Fackler OT, et al. Modulation of the immunological synapse: a key to HIV-1 pathogenesis? Nat Rev Immunol. 2007;7:310–317. doi: 10.1038/nri2041. [DOI] [PubMed] [Google Scholar]
- 60.Sol-Foulon N, et al. ZAP-70 kinase regulates HIV cell-to-cell spread and virological synapse formation. EMBO J. 2007;26:516–526. doi: 10.1038/sj.emboj.7601509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stinchcombe JC, et al. The immunological synapse of CTL contains a secretory domain and membrane bridges. Immunity. 2001;15:751–761. doi: 10.1016/s1074-7613(01)00234-5. [DOI] [PubMed] [Google Scholar]
- 62.Neijssen J, et al. Cross-presentation by intercellular peptide transfer through gap junctions. Nature. 2005;434:83–88. doi: 10.1038/nature03290. [DOI] [PubMed] [Google Scholar]