Abstract
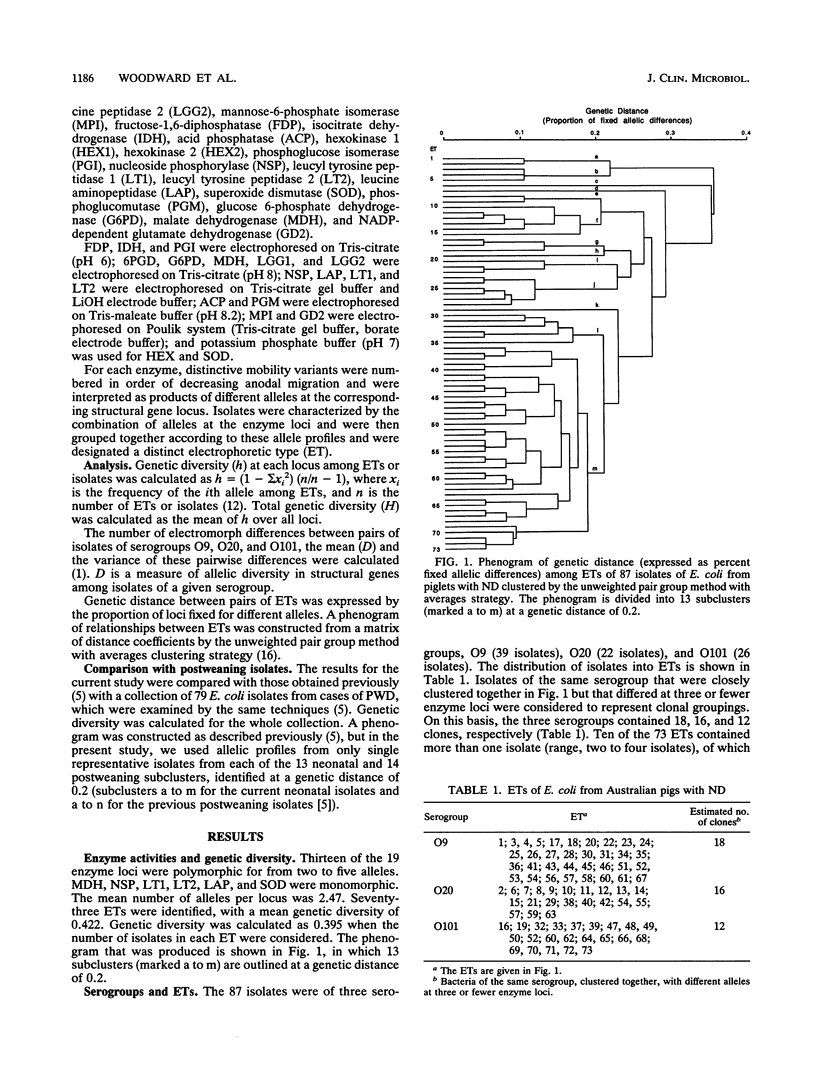
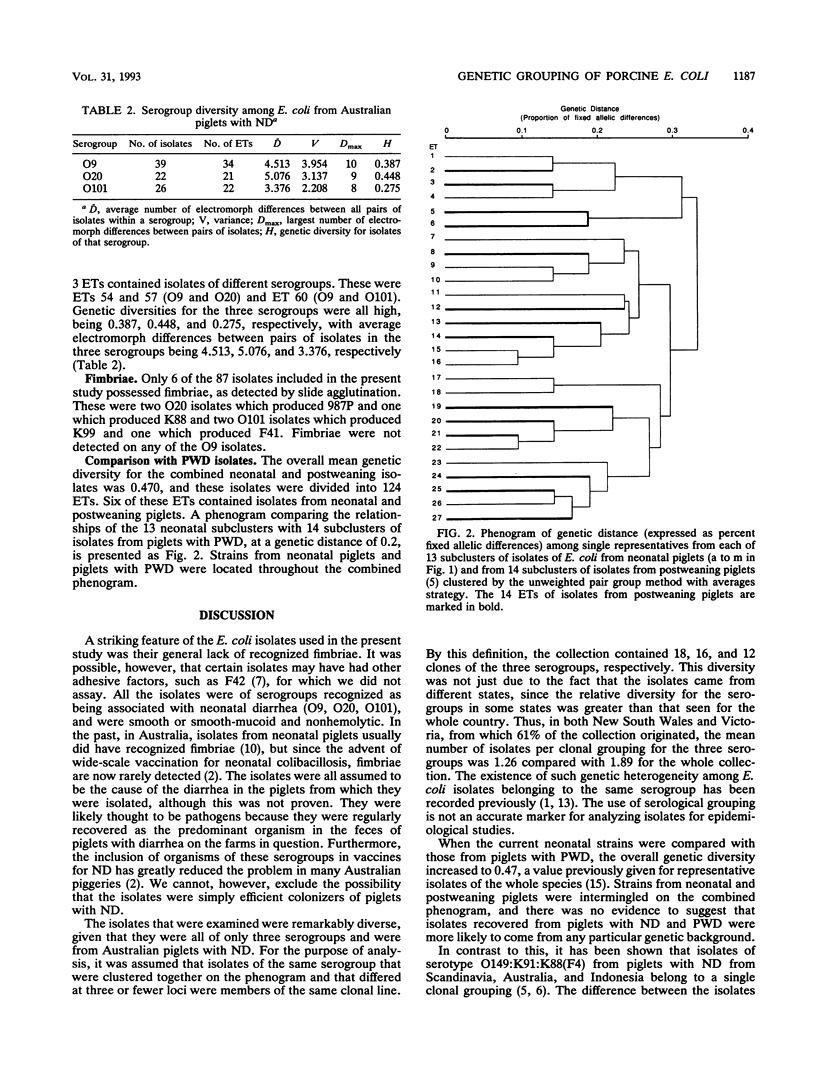
The genetic diversity of 87 isolates of Escherichia coli recovered from Australian pigs with neonatal diarrhea was examined by multilocus enzyme electrophoresis. The isolates were of serogroups O9, O20, and O101, and although most isolates lacked K88(F4), K99(F5), 987P(F6), and F41 fimbriae, they were considered to be involved in the etiology of the diarrhea. The isolates were extremely diverse, considering their origin from a single pathological condition in one country. There were estimated to be 18, 16, and 12 clones of the three respective serogroups in the collection, with serogroup diversities of 0.387, 0.448, and 0.275, respectively. Comparison with the results previously obtained for isolates from piglets with postweaning diarrhea suggested that bacteria from piglets with these two conditions did not come from any particular common genetic background. The overall genetic diversity for the combined collection was the same as that reported by others for representative isolates selected from throughout the species (0.47). The current results indicate that if isolates of these O groups are involved in porcine diarrhea, their pathogenicity is directly linked to their O somatic antigen type and is not simply due to the wide distribution of a small number of virulent clones.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Caugant D. A., Levin B. R., Orskov I., Orskov F., Svanborg Eden C., Selander R. K. Genetic diversity in relation to serotype in Escherichia coli. Infect Immun. 1985 Aug;49(2):407–413. doi: 10.1128/iai.49.2.407-413.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn I., Franklin A., Söderlind O., Möllby R. Phenotypic variations among enterotoxinogenic Escherichia coli from Swedish piglets with diarrhoea. Med Microbiol Immunol. 1985;174(3):119–130. doi: 10.1007/BF02298122. [DOI] [PubMed] [Google Scholar]
- Leite D. S., Yano T., Pestana de Castro A. F. Production, purification and partial characterization of a new adhesive factor (F42) produced by enterotoxigenic Escherichia coli isolated from pigs. Ann Inst Pasteur Microbiol. 1988 May-Jun;139(3):295–306. doi: 10.1016/0769-2609(88)90021-x. [DOI] [PubMed] [Google Scholar]
- Monckton R. P., Hasse D. Detection of enterotoxigenic Escherichia coli in piggeries in Victoria by DNA hybridisation using K88, K99, LT, ST1 and ST2 probes. Vet Microbiol. 1988 Mar;16(3):273–281. doi: 10.1016/0378-1135(88)90031-4. [DOI] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978 Jul;89(3):583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Levin B. R. Genetic diversity and structure in Escherichia coli populations. Science. 1980 Oct 31;210(4469):545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- Söderlind O., Thafvelin B., Möllby R. Virulence factors in Escherichia coli strains isolated from Swedish piglets with diarrhea. J Clin Microbiol. 1988 May;26(5):879–884. doi: 10.1128/jcm.26.5.879-884.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]


