Abstract
The world of regulatory RNAs is fast expanding into mainstream molecular biology as both a subject of intense mechanistic study and as a tool for functional characterization. The RNA world is one of complex structures that carry out catalysis, sense metabolites and synthesize proteins. The dynamic and structural nature of RNAs presents a whole new set of informatics challenges to the computational community. The ability to relate structure and dynamics to function will be key to understanding this complex world. I review several important classes of structured RNAs that present our community with a series of biologically novel informatics challenges. I also review available informatics tools that have been recently developed in the field.
Keywords: RNA, Folding, Informatics, Riboswitch, Ribosome, RNAi
Central to many of the regulatory networks in the cell, RNA's role in regulating both transcription and translation is garnering much attention from both wet and dry labs[1-3]. The fact that RNA generally exists as a single-stranded molecule, and the additional conformational flexibility of the sugar-phosphate backbone, allows RNA to adopt stunningly complex three-dimensional structures[4]. These structures confer function to the RNA including catalysis, metabolite sensing, and transcriptional control[5-7]. The unique molecular architecture of RNA, combined with the wide range in size of functional RNAs, presents our community with a series of novel opportunities in RNA informatics[8].
Much like proteins, RNAs have elements of primary, secondary and tertiary structure[9]. The linear nature of the RNA biopolymer (containing Guanine, Cytosine, Uracil, and Adenine bases) and the ability of bases to form Watson-Crick (or canonical) base-pairs (G with C and A with U) makes the prediction of RNA secondary structure from primary sequence a contemporary challenge in RNA informatics. Significant advances in the field of RNA secondary structure have been made in the last two years (for a review see [8] as well as [10]), but are not the main focus of this review. I will rather focus on emerging informatics challenges in the area of structured RNAs, and their dynamic behavior. Specifically this review addresses challenges and opportunities related to RNA molecules for which the three-dimensional structure has been determined experimentally. Furthermore, I will review recent RNA/Protein (RNP) complexes of particular significance in the emerging field of RNA interference (RNAi) [6, 11].
Functionally important structured RNAs
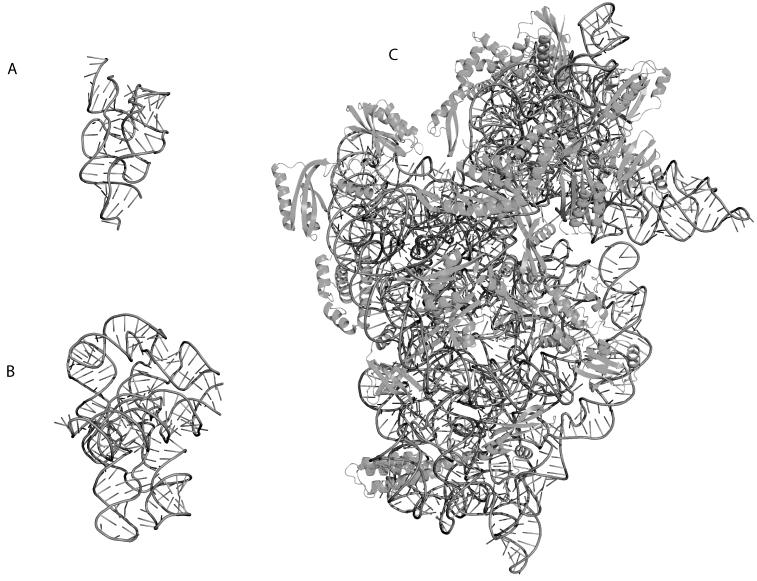
Figure 1 illustrates the difference in size of three functionally important classes of structured RNAs. Riboswitches are small (100 nucleotides, Figure 1A) single stranded RNAs that act as metabolite sensors in Bacteria[5]. They are generally found upstream of the Gene they control (in this case Thiamine synthesis) and undergo a conformational change in the presence of the metabolite (Thiamine)[12]. When the metabolite is bound to the Riboswitch, it generally inhibits translation thereby down regulating the production of the corresponding protein product. Self-splicing introns are transcribed with the mRNA and fold into a catalytically active conformation (Figure 1B) that auto-splices itself out of the mRNA effectively processing the message for translation[2]. The Ribosomal RNAs (rRNAs) are molecular machines (Figure 1C illustrates the 30S rRNA subunit) that perform the translation reaction in the cell (synthesis of protein). RNAs therefore come in many shapes and sizes that each have specialized functions within the cell.
Figure 1.
Three-dimensional crystal structure cartoon representations of functionally important RNAs rendered in PYMOL (http://pymol.sourceforge.net/). The RNA backbone is indicated as a dark grey tube, while the sticks indicate the orientation of the bases relative to the backbone. A) Structure of a Riboswitch (PDB ID 2H0M) bound to Thiamine. This molecule senses Thiamine and undergoes a conformational change when the ligand is present, thereby down-regulating Thiamine synthesis. B) Structure of the Twort group I intron (PDB ID 1Y0Q). This catalytic RNA auto-splices itself thereby processing the mRNA for translation. C) Structure of the T. thermophilus 30S Ribosomal Subunit (PDB ID 1J5E). The proteins are indicated in dark grey and drawn using cartoon representations. This rRNA forms a molecular machine with the 50S subunit that captures the mRNA and translates it into the functional protein.
Common to all three RNAs illustrated in Figure 1 are dynamic conformational changes that enable and regulate the function of the biopolymer. Riboswitches undergo significant conformational changes upon ligand binding, while catalytic RNAs and rRNAs must fold into active structures. The dynamics of these assembly reactions are critical to the regulatory role of the molecule, as correct sequencing of the reaction directly impacts function. In RNA, the kinetics of folding and assembly are particularly important, as most RNAs that have been studied to date exhibit complex kinetic behavior during their folding reaction.
Structural elements of RNA
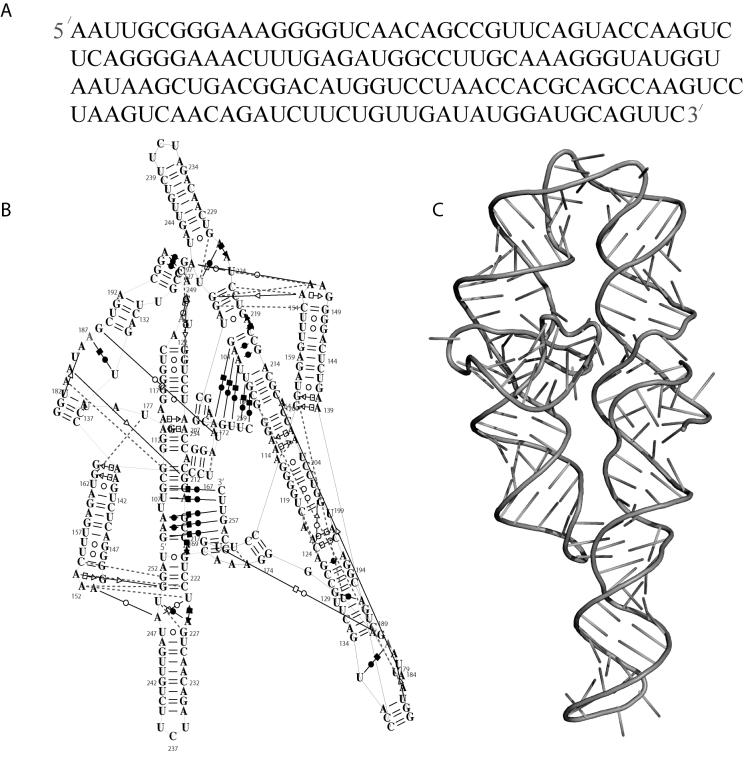
As mentioned above, RNA structures have elements of primary, secondary and tertiary structure. The primary sequence of RNA is comprised of four types of bases (C, G, A and U, Figure 2A). Figure 2B illustrates the secondary structure diagram of the P4P6 subdomain of the L-21 Tetrahymena thermophila group I intron[13]. This diagram differs somewhat from the manually laid out secondary structure diagrams often published with RNAs (see for example Figure 2 in [14] for a particularly aesthetic diagram). The diagram was generated automatically using the RNAMLVIEW software [15], which is now incorporated in S2S [16] (http://bioinformatics.org/S2S/). The layout attempts to best represent the three-dimensional reality (Figure 2C) of the molecule in two dimensions. Helical segments of the molecule are drawn as sequential pairs of stacked bases. Base pairs are indicated with lines, and long-range tertiary contacts are identified by red dotted lines.
Figure 2.
Structural elements of RNA. A) Primary sequence of RNA containing the four nucleotides G, C, A and U for the P4P6 subdomain of the L-21 T. thermophila group I intron[13]. B) Secondary structure of the P4P6 Subdomain as illustrated by RNAMLView[15]. The layout of the structure is automated trying to best represent the three-dimensional reality of the molecule. Long range tertiary contacts are indicated as red dotted lines, while base-pair types are as defined by Leontis and Westhof[17]. C) Three dimensional cartoon representation (similar scheme as Figure 1) of the P4P6 subdomain (PDB ID 1GID).
Canonical base-pairs (known also as Watson-Crick pairs) in RNA will lead to an A-form helix. RNA nucleotides, however, can form non-canonical base-pairs in which different edges of the base form complimentary hydrogen bonds. As a result, stabilizing interactions can form between any two nucleotides in the RNA. A seminal paper by Leontis and Westhof describes all 12 possible base-pairing interactions based on geometrical considerations for each base type[17]. The different symbols connecting the bases in the secondary structure diagram (Figure 2B) represent different types of base-pairing as defined by Leontis and Westhof [17, 18]. S2S and RNAMLVIEW conveniently generate XML annotations of RNA structures (including base-pairing type), greatly reducing the need for specialized code bases to analyze RNA structure[16, 18]. A complete Documentation Type Definition (DTD) for RNA structure has also been developed for the markup language RNAML[19].
The ability of RNA to form 12 different types of base-pairs significantly complicates the prediction of RNA tertiary structure from primary sequence. Nonetheless, remarkably accurate models of large structured RNAs have been generated and later confirmed by x-ray crystallography[20]. These structural models are built in a semi-automated way and rely on the user's intuition and knowledge of RNA structure. Fully automated structure prediction of RNA remains a contemporary challenge in the field with only limited success thus far[21]. The large number of RNA structures that have been solved by crystallography should make knowledge-based structural modeling approaches amenable in the near future. The nucleic acid database curates all new RNA and DNA structures available[22], and SCOR maintains a structural classification[23-26]. Both of these resources offer convenient entry points into the world of structured RNA. Furthermore, global statistical analyses of RNA structure are beginning to identify emergent orientations of RNA helices which can be used to improve automated structure prediction[27].
RNA motifs and potential for homology modeling
Careful analysis of RNA structures has revealed a surprising number of recurring structural motifs across species and function[15, 17, 18, 28-31]. These are three-dimensional orientations of bases that have nearly identical base-pairing and stacking interactions and are believed to constitute building blocks for complex RNA structures[18]. Work in the Levitt lab has shown that it is possible to reconstruct entire RNA structures with a library of clustered di-nucleotides[32]. Alternatively the analysis of the backbone conformation of structured RNA also allows the identification of similar motifs [33-36]. Whether the motifs are automatically or manually identified, there is a general consensus in the field that there most likely exists a set of core RNA building blocks larger than single nucleotides from which all RNA structures can be reconstructed. The identification of these motifs remains a contemporary challenge in the field. It is likely that only consideration of both backbone and base orientations will lead to a complete description of RNA structure[37].
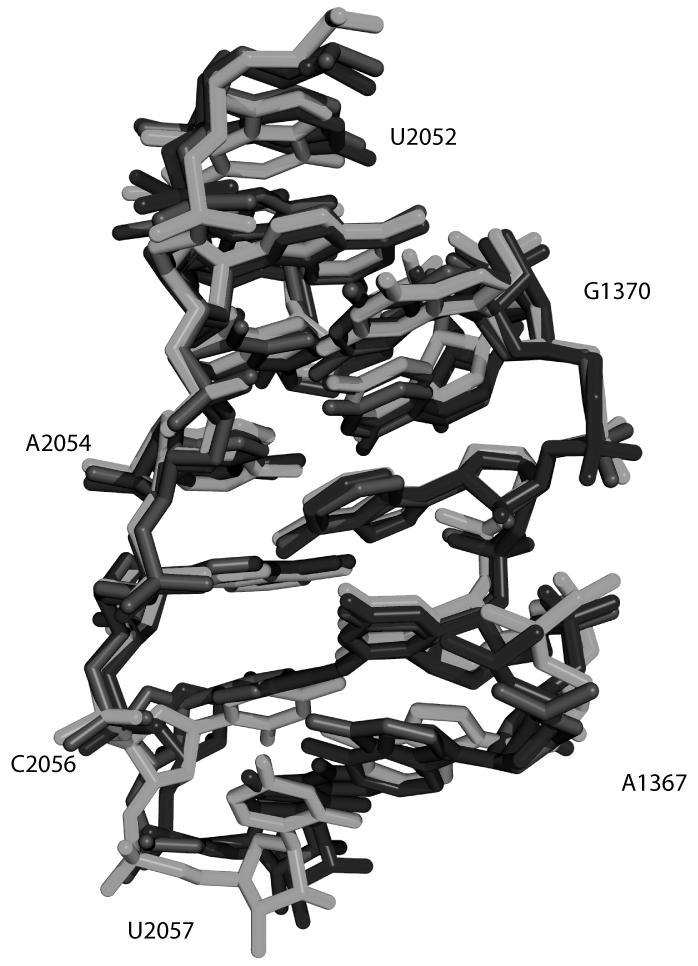
A classic example of an RNA structural motif is illustrated in Figure 3. It is a Sarcin (also known as Sarcin-Ricin) motif, and the three overlaid structures (Red, Green and Blue) occur in different positions within the same structure. The Sarcin motif is clearly a structural motif as the sequences of each strand differ significantly and would not be identified by sequence comparison alone[29]. Approximately 40 such motifs have been identified to date and every year new motifs are discovered. Geometrical definitions for three-dimensional RNA motifs have yet to be finalized as these require consensus building within the community. This has prompted the creation of an RNA Ontology Consortium to develop a structured, community accepted vocabulary for describing RNA structural motifs[38]. The goals of the consortium include providing the informatics community with curated structural annotation databases to facilitate informatics research in RNA (http://roc.bgsu.edu/).
Figure 3.
Three-dimensional representation of a Sarcin motif[29]. The red, green and blue atomic structures correspond to nucleotides 210-229, 1367-2057, and 2689-2705 of the 50S rRNA from H. Marismortui (PDB ID 1S72). The Sarcin loop is an example of a structural motif that has a highly conserved three-dimensional reality not necessarily evident in the sequence of the RNA. Numbering is indicated for black structure.
RNA Folding dynamics
The “RNA Folding Problem” is not limited to predicting the tertiary structure of an RNA from its primary sequence (Figure 2). A vast majority of biologically active RNAs have a propensity to naturally misfold, and require changes in conditions (e.g. heating) or exogenous molecules (e.g. RNA chaperones) to resolve into the active structure[7, 39, 40]. The propensity of RNAs to misfold suggests important regulatory roles related to the dynamics of the molecule and its interactions with other cellular constituents. Understanding RNA misfolding biophysically and phenomenologically is at the heart of the RNA folding problem and presents a series of novel informatics and modeling challenges.
The ability to easily synthesize RNA from DNA (using RNA polymerase) and the use of quantitative RT (Reverse Transcriptase) reactions for the analysis of RNA containing solutions has enabled the development of many RNA specific experimental modalities[41, 42]. The relative simplicity of such “footprinting” experiments (compared to analogous modalities in Proteins) and the development of specialized software for the analysis of the results has created a new paradigm in terms of the throughput and quality of dynamic information available for large structured RNAs[43, 44]. These new experimental modalities have in turn required the development of novel software for the analysis and modeling of RNA folding reactions[45, 46].
Hydroxyl radical footprinting experiments determine the accessible surface area of a molecule by measuring the reactivity of individual nucleotides when exposed to hydroxyl radicals. Nucleotides that are buried in the structure are less likely to react with a hydroxyl radical [41] than those on the outside [47-49]. Given the fast reaction kinetics of the Hydroxyl radical with the RNA, and recent improvements in mixer technology, this technique can now resolve folding transitions with millisecond accuracy[44].
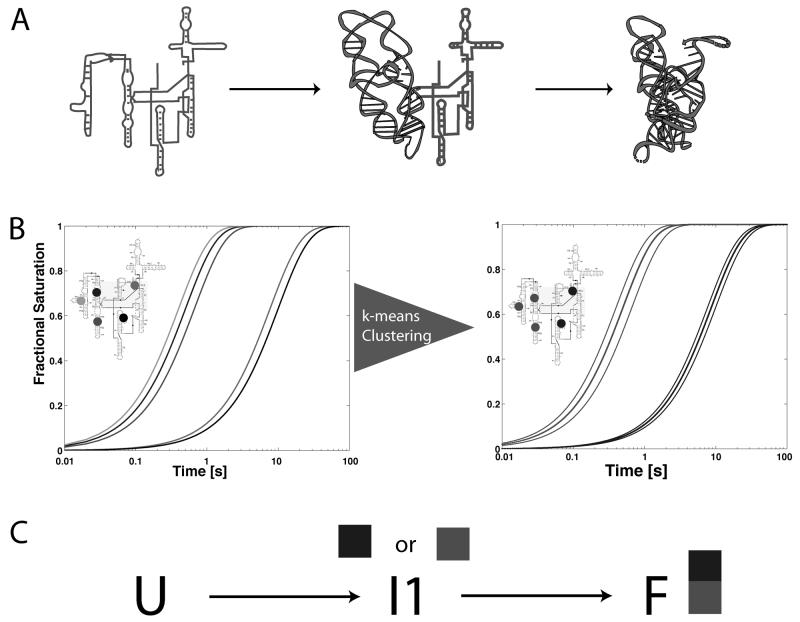
Hydroxyl radical footprinting experiments are unique in that they provide local probes of structure, as opposed to global probes (for example, the change in Radius of Gyration determined by analytical ultracentrifugation). Time-resolved hydroxyl radical footprinting experiments monitor the change in protection of each nucleotide in the RNA as a function of time. The information content of such data is sufficient to allow the determination of the underlying kinetic parameters of the system. Figure 4 illustrates the basic premise of the KinFold algorithm, which was developed to automatically model the folding reactions of large RNAs based on time- resolved hydroxyl radical footprinting experiments[45, 46]. If footprinting data were collected on the folding reaction illustrated in Figure 4A, the time-progress curves would look like those illustrated in Figure 4B. Given that the three nucleotides indicated in red, green and blue, respectively are all on the subdomain of the molecule that folds first, protection would initially be seen for these three nucleotides. Once the intermediate has accumulated, the black and magenta nucleotides would report protection as the concentration of folded molecule increases.
Figure 4.
Basic premise of the KinFold algorithm for automated modeling of RNA folding dynamics. A) Folding pathway of the RNA with one intermediate in which a subdomain of the molecule forms initially followed by formation of the folded active conformation. B) Illustration of the time-progress curves that would be observed for five nucleotides on the RNA were the folding reaction shown in A measured experimentally using time-resolved hydroxyl radical footprinting. Curve colors indicate corresponding nucleotide positions in the RNA as shown in the inset. KinFold [46]initially analyzes the data by k-means clustering of the progress curves. C) Reconstruction of the kinetic model from an analysis of the clustered time-progress curves shown in B.
The KinFold algorithm analyzes this type of data in a two-step process. Initially, the time-progress curves are clustered using k-means clustering to establish the subdomains of the molecule that form in a concerted manner. In this case the clustering would identify the fast folding subdomain of the molecule by clustering the red, green and blue nucleotides together as illustrated in the right pane of Figure 4B. Once the time-progress curves are clustered, the data is kinetically modeled as illustrated in Figure 4C. In this case two alternative models need to be tested, as the intermediate can either be associated with the red or blue clusters. It is important to note that a-priori the kinetic model shown in Figure 4A is not known, but that it can be automatically deduced from the analysis of the kinetic folding data illustrated in Figure 4B. This type of approach is particularly useful when studying the effects of mutation and solution conditions on the folding kinetics of RNA[45].
At the heart of the RNA folding problem is the relationship between the three-dimensional structure of the molecule and the folding dynamics. The timescales of RNA folding reactions (often on the order of seconds to minutes) makes the problem of simulating these large molecules computationally challenging at the atomic scale [50, 51]. Nonetheless, several groups have studied the structure/dynamic relationships using simplified representations of RNA (such as contact order) and come up with good correlations to experimentally observed folding rates[40, 52].
Nucleic Acid recognition in RNA interference
RNA interference (or RNAi) is fundamentally changing the world of molecular biology as it is both a technology that enables specific regulation of gene expression, and a mechanism by which cells regulate their genes[6, 11]. RNAi refers to the general mechanism by which double stranded RNAs are processed into single stranded RNAs with complimentary sequences to mRNAs. These single-stranded RNAs bind to the mRNA and down regulate the associated gene. MicroRNAs (miRNAs) are naturally occurring hairpin structures that the cell uses for genetic regulation. Small interfering RNAs (siRNA) are double-stranded synthetic molecules that are used in molecular biology to target specific genes for down-regulation. Both miRNAs and siRNAs are processed by a series of proteins for which the three-dimensional structures have very recently been solved (a comprehensive review of RISC (RNA induced silencing complex) structures can be found in [53].
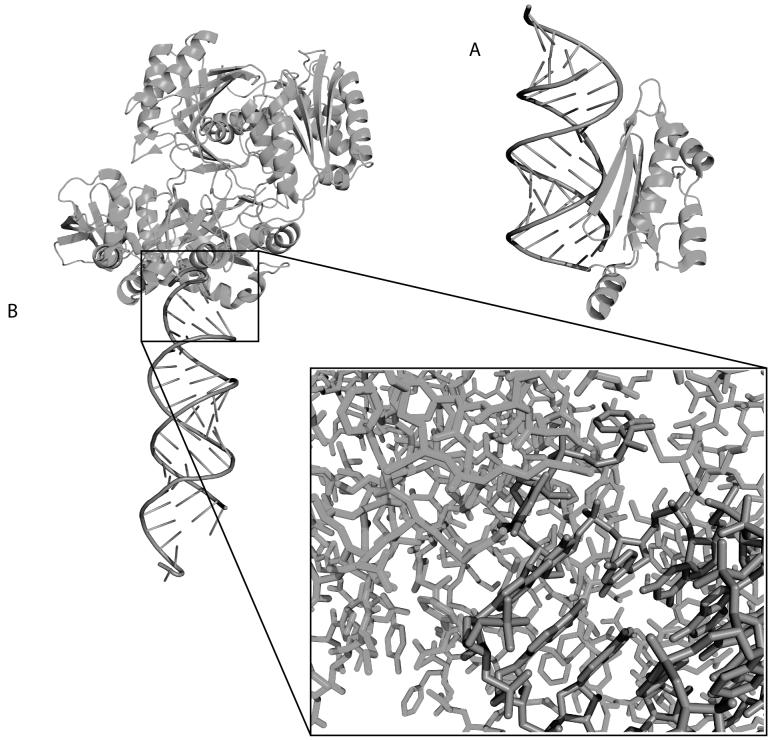
RNA-protein interactions are critical to the regulation of RNAi. The importance of these regulatory networks is such that even viruses have evolved proteins to interact with siRNAS. The viral proteins inhibit the silencing activity of siRNAs thereby disabling the defense mechanisms in the cell. One such example is the p19 protein virus from tombusvirus (Figure 5A) which is able to recognize double stranded RNAs of length 22 by projecting two alpha-helical “reading heads” that contain terminal tryptophans that bind to the two ends of the siRNA[54].
Figure 5.
Cartoon representations of the structures of RNA/Protein complexes important in RNA interefence. A) Crystal structure of the p19 protein viral suppressor bound to double-stranded RNA (PDB ID 1R9F). The alpha-helical arm binds to the bottom of the siRNA and is capped with Tryptophan residues that detect the exposed nucleotide bases at the ends of double-stranded RNAs. B) Crystal structure of the Argonaute protein bound to an siRNA (PDB ID 2F8S). The inset illustrates the complexity of the RNA/protein interface.
The Argonaute class of proteins (Figure 5B) are part of RISC and provide catalytic and structural function to the complex[55-57]. The atomic reality of the recognition of the double-stranded RNA (Figure 5B, inset) by the protein illustrates the challenges associated with predicting and understanding RNAi function. The interface between the protein and siRNA involves at least 7 nucleotides and approximately 30 amino acids. The fact that the protein recognizes both strands makes it unlikely that sequence analysis alone will be able to predict the specificity of the interaction. Furthermore, several domains in the protein are involved in binding to the RNA and this suggests that sequence motif type analyses alone will not be successful in predicting these types of interactions.
The most significant informatics contribution to the world of RNAi to date has been the development of algorithms to detect miRNAs in genomic sequences (see for a complete review [58]). miRNAs are generally expressed as 2×22 nucleotide long hairpins, and this sequence pattern can readily be identified in non-coding genomic sequences[55]. A more interesting informatics challenge, however, is the identification and characterization of miRNA promoters, which allows a systematic analysis of the interaction networks within RNAi controlled genes[59]. Nonetheless, only a physics-based understanding of the molecular details of the RISC complex in the context of the cellular milieu will enable a comprehensive prediction of RNAi function.
The Future of RNA informatics
The linear nature of DNA has made it very amenable to sequence based informatics analyses. Furthermore, genomic technologies have produced an overwhelming amount of genetic information ideal for informatics analyses. The Rfam database (RNA families) provides a comprehensive repository of structured RNAs [60, 61]. These RNAs are identified from genomic sequences based on their sequence similarity to known structured RNAs. The REF 500,000 structured RNAs in Rfam demonstrate the ubiquitous nature of these fascinating molecules and their biological importance.
In 1994, Stanley and co-workers identified patterns similar to language in what was then termed “junk DNA” [62] using statistical and linguistics approaches[63-65]. Incredibly, in little over a decade, we now know that much of the “junk DNA” is in fact transcribed into RNA, and that these molecules play critical regulatory roles. The physical reality of these RNAs is necessarily three-dimensional, and only a geometric understanding of their shape and dynamics will yield insight into their function. The future of RNA informatics will require integration of this four-dimensional reality (time and structure). The systematic and integrative approach to biology will require interdisciplinary collaborations across multiple fields of computation including the engineering and modeling communities.
Acknowledgements
I wish to thank Jesse Stombaugh for providing the atomic coordinates for Figure 3. A.L. is a Damon Runyon Cancer Foundation Research Fellow. This work was funded by the NIH through NIGMS K99-GM079953, P01-GM66275 and an NIH Roadmap grant U54 GM072970 for the National Centers for Biomedical Computation and the NSF 0443508 for the RNA Ontology Consortium.
Biography
Alain Laederach is a post-doctoral fellow in the Genetics Department at Stanford University. His main interests lie in applying informatics and modeling to the RNA folding problem.
Footnotes
- 1.) RNA is both a messenger of genetic information and a molecule capable of carrying out reactions in the cell.
- 2.) The function of RNA is intimately related to its structure and dynamic behavior.
- 3.) RNA molecules interact with proteins in the cell to regulate gene expression.
- 4.) Structured RNAs are comprised of motifs that are found in many structures.
Bibliography
- 1.Grundy FJ, Henkin TM. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit Rev Biochem Mol Biol. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- 2.Doherty EA, Doudna JA. Ribozyme structures and mechanisms. Annu Rev Biophys Biomol Struct. 2001;30:457–475. doi: 10.1146/annurev.biophys.30.1.457. [DOI] [PubMed] [Google Scholar]
- 3.Konarska MM, Query CC. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- 4.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 5.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Ying SY, Chang DC, Miller JD, et al. The microRNA: overview of the RNA gene that modulates gene functions. Methods Mol Biol. 2006;342:1–18. doi: 10.1385/1-59745-123-1:1. [DOI] [PubMed] [Google Scholar]
- 7.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 8.Reeder J, Hochsmann M, Rehmsmeier M, et al. Beyond Mfold: recent advances in RNA bioinformatics. J Biotechnol. 2006;124:41–55. doi: 10.1016/j.jbiotec.2006.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 10.Do CB, Woods DA, Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics. 2006;22:e90–98. doi: 10.1093/bioinformatics/btl246. [DOI] [PubMed] [Google Scholar]
- 11.Ying SY, Lin SL. Current perspectives in intronic micro RNAs (miRNAs) J Biomed Sci. 2006;13:5–15. doi: 10.1007/s11373-005-9036-8. [DOI] [PubMed] [Google Scholar]
- 12.Edwards TE, Ferre-D'Amare AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Cate JH, Gooding AR, Podell E, et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 14.Shcherbakova I, Gupta S, Chance MR, et al. Monovalent ion-mediated folding of the Tetrahymena thermophila ribozyme. J Mol Biol. 2004;342:1431–1442. doi: 10.1016/j.jmb.2004.07.092. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Jossinet F, Leontis N, et al. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jossinet F, Westhof E. Sequence to Structure (S2S): display. manipulate and interconnect RNA data from sequence to structure, Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- 17.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waugh A, Gendron P, Altman R, et al. RNAML: a standard syntax for exchanging RNA information. Rna. 2002;8:707–717. doi: 10.1017/s1355838202028017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lescoute A, Westhof E. The interaction networks of structured RNAs. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popenda M, Bielecki L, Adamiak RW. High-throughput method for the prediction of low-resolution, three-dimensional RNA structures. Nucleic Acids Symp Ser (Oxf) 2006:67–68. doi: 10.1093/nass/nrl033. [DOI] [PubMed] [Google Scholar]
- 22.Berman HM, Westbrook J, Feng Z, et al. The nucleic acid database. Methods Biochem Anal. 2003;44:199–216. [PubMed] [Google Scholar]
- 23.Huang HC, Nagaswamy U, Fox GE. The application of cluster analysis in the intercomparison of loop structures in RNA. Rna. 2005;11:412–423. doi: 10.1261/rna.7104605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klosterman PS, Hendrix DK, Tamura M, et al. Three-dimensional motifs from the SCOR, structural classification of RNA database: extruded strands, base triples, tetraloops and U-turns. Nucleic Acids Res. 2004;32:2342–2352. doi: 10.1093/nar/gkh537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura M, Hendrix DK, Klosterman PS, et al. SCOR: Structural Classification of RNA, version 2.0. Nucleic Acids Res. 2004;32:D182–184. doi: 10.1093/nar/gkh080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klosterman PS, Tamura M, Holbrook SR, et al. SCOR: a Structural Classification of RNA database. Nucleic Acids Res. 2002;30:392–394. doi: 10.1093/nar/30.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laederach A, Chan JM, Schwartzman A, et al. Coplanar and coaxial orientations of RNA bases and helices. Rna. 2007 doi: 10.1261/rna.381407. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leontis NB, Hills MT, Piotto M, et al. Helical stacking in DNA three-way junctions containing two unpaired pyrimidines: proton NMR studies. Biophys J. 1995;68:251–265. doi: 10.1016/S0006-3495(95)80182-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leontis NB, Stombaugh J, Westhof E. Motif prediction in ribosomal RNAs Lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie. 2002;84:961–973. doi: 10.1016/s0300-9084(02)01463-3. [DOI] [PubMed] [Google Scholar]
- 30.Leontis NB, Westhof E. Analysis of RNA motifs. Curr Opin Struct Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 31.Lescoute A, Leontis NB, Massire C, et al. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sykes MT, Levitt M. Describing RNA structure by libraries of clustered nucleotide doublets. J Mol Biol. 2005;351:26–38. doi: 10.1016/j.jmb.2005.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wadley LM, Pyle AM. The identification of novel RNA structural motifs using COMPADRES: an automated approach to structural discovery. Nucleic Acids Res. 2004;32:6650–6659. doi: 10.1093/nar/gkh1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pang PS, Jankowsky E, Wadley LM, et al. Prediction of functional tertiary interactions and intermolecular interfaces from primary sequence data. J Exp Zoolog B Mol Dev Evol. 2005;304:50–63. doi: 10.1002/jez.b.21024. [DOI] [PubMed] [Google Scholar]
- 35.Duarte CM, Wadley LM, Pyle AM. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. Nucleic Acids Res. 2003;31:4755–4761. doi: 10.1093/nar/gkg682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray LJ, Arendall WB, 3rd, Richardson DC, et al. RNA backbone is rotameric. Proc Natl Acad Sci U S A. 2003;100:13904–13909. doi: 10.1073/pnas.1835769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore PB. Structural motifs in RNA. Annu Rev Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- 38.Leontis NB, Altman RB, Berman HM, et al. The RNA Ontology Consortium: an open invitation to the RNA community. Rna. 2006;12:533–541. doi: 10.1261/rna.2343206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell R, Zhuang X, Babcock HP, et al. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci U S A. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorin EJ, Nakatani BJ, Rhee YM, et al. Does native state topology determine the RNA folding mechanism? J Mol Biol. 2004;337:789–797. doi: 10.1016/j.jmb.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 41.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 42.Gross P, Arrowsmith CH, Macgregor RB., Jr. Hydroxyl radical footprinting of DNA complexes of the ets domain of PU.1 and its comparison to the crystal structure. Biochemistry. 1998;37:5129–5135. doi: 10.1021/bi972591k. [DOI] [PubMed] [Google Scholar]
- 43.Das R, Laederach A, Pearlman SM, et al. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. Rna. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shcherbakova I, Mitra S, Beer RH, et al. Fast Fenton footprinting: a laboratory-based method for the time-resolved analysis of DNA, RNA and proteins. Nucleic Acids Res. 2006;34:e48. doi: 10.1093/nar/gkl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laederach A, Shcherbakova I, Jonikas M, et al. Distinct contribution of electrostatics, initial conformational ensemble and macromolecular stability in RNA folding. PNAS. 2007 doi: 10.1073/pnas.0608765104. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laederach A, Shcherbakova I, Liang M, et al. Local kinetic measures of macromolecular structure reveal partitioning among multiple parallel pathways from the earliest steps in the folding of a large RNA molecule. J Mol Biol. 2006;358:1179–1190. doi: 10.1016/j.jmb.2006.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brenowitz M, Chance MR, Dhavan G, et al. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical “footprinting”. Curr Opin Struct Biol. 2002;12:648–653. doi: 10.1016/s0959-440x(02)00366-4. [DOI] [PubMed] [Google Scholar]
- 48.Brenowitz M, Senear DF, Shea MA, et al. “Footprint” titrations yield valid thermodynamic isotherms. Proc Natl Acad Sci U S A. 1986;83:8462–8466. doi: 10.1073/pnas.83.22.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brenowitz M, Senear DF, Shea MA, et al. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 50.Su LJ, Waldsich C, Pyle AM. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res. 2005;33:6674–6687. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Sosnick TR, Pan T. Reduced contact order and RNA folding rates. J Mol Biol. 2004;342:1359–1365. doi: 10.1016/j.jmb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 53.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 54.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Preall JB, Sontheimer EJ. RNAi: RISC gets loaded. Cell. 2005;123:543–545. doi: 10.1016/j.cell.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 56.Miyoshi K, Tsukumo H, Nagami T, et al. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ikeda K, Satoh M, Pauley KM, et al. Detection of the argonaute protein Ago2 and microRNAs in the RNA induced silencing complex (RISC) using a monoclonal antibody. J Immunol Methods. 2006;317:38–44. doi: 10.1016/j.jim.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang B, Pan X, Wang Q, et al. Computational identification of microRNAs and their targets. Comput Biol Chem. 2006;30:395–407. doi: 10.1016/j.compbiolchem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Zhou X, Ruan J, Wang G, et al. Characterization and Identification of MicroRNA Core Promoters in Four Model Species. PLoS Comput Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Griffiths-Jones S, Moxon S, Marshall M, et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffiths-Jones S, Bateman A, Marshall M, et al. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flam F. Hints of a language in junk DNA. Science. 1994;266:1320. doi: 10.1126/science.7973718. [DOI] [PubMed] [Google Scholar]
- 63.Mantegna RN, Buldyrev SV, Goldberger AL, et al. Linguistic features of noncoding DNA sequences. Phys Rev Lett. 1994;73:3169–3172. doi: 10.1103/PhysRevLett.73.3169. [DOI] [PubMed] [Google Scholar]
- 64.Peng CK, Buldyrev SV, Goldberger AL, et al. Statistical properties of DNA sequences. Physica A. 1995;221:180–192. doi: 10.1016/0378-4371(95)00247-5. [DOI] [PubMed] [Google Scholar]
- 65.Mantegna RN, Buldyrev SV, Goldberger AL, et al. Systematic analysis of coding and noncoding DNA sequences using methods of statistical linguistics. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1995;52:2939–2950. doi: 10.1103/physreve.52.2939. [DOI] [PubMed] [Google Scholar]