Abstract
The formation of disulfides within proteins entering the secretory pathway is catalyzed by the protein disulfide isomerase family of endoplasmic reticulum localized oxidoreductases. One such enzyme, ERp57, is thought to catalyze the isomerization of non-native disulfide bonds formed in glycoproteins with unstructured disulfide-rich domains. Here we investigated the mechanism underlying ERp57 specificity toward glycoprotein substrates and the interdependence of ERp57 and the calnexin cycle for their correct folding. Our results clearly show that ERp57 must be physically associated with the calnexin cycle to catalyze isomerization reactions with most of its substrates. In addition, some glycoproteins only require ERp57 for correct disulfide formation if they enter the calnexin cycle. Hence, the specificity of ER oxidoreductases is not only determined by the physical association of enzyme and substrate but also by accessory factors, such as calnexin and calreticulin in the case of ERp57. These conclusions suggest that the calnexin cycle has evolved with a specialized oxidoreductase to facilitate native disulfide formation in complex glycoproteins.
The ability to form disulfide bonds within proteins entering the secretory pathway is essential for cell survival and occurs within the endoplasmic reticulum (ER).3 For proteins with few disulfides, the process can be catalyzed by oxidation of cysteine residues to form the correct, native disulfide; however, for proteins with several disulfides, an isomerization reaction is also required to correct non-native disulfides formed following oxidation (1). Both these reactions are catalyzed by a group of ER-resident proteins that belong to the protein disulfide isomerase (PDI) family, which comprises over 17 members (2). It is well established that PDI and several other family members are able to catalyze the formation and isomerization of disulfides in vitro, although the exact function of each of the family members in vivo is unknown. It is still an open question as to whether they all catalyze similar reactions and have distinct substrate specificities or whether they have distinct enzymatic functions related to the breaking and formation of disulfides.
For one member of the PDI family, the function and substrate specificity is a little clearer. ERp57 has been shown previously to interact specifically with glycoproteins during their folding (3). The enzyme is physically associated with either calnexin or calreticulin (4) and is therefore ideally placed to catalyze correct disulfide formation within proteins entering the calnexin/calreticulin cycle (referred to subsequently just as the calnexin cycle). In addition, the ability of ERp57 to catalyze the refolding of substrates in vitro is greatly enhanced if the substrate is bound to calnexin (5). Recently, substrates for the reduction or isomerization reaction catalyzed by ERp57 have been identified by trapping mixed disulfides between enzyme and substrate (6). Strikingly, there was an overrepresentation of substrate proteins with cysteine-rich domains containing little secondary structure, suggesting that the main function of ERp57 is in the isomerization of non-native disulfides. ERp57 has also been shown to function independently from the calnexin cycle. It is a component of the MHC class I loading complex where it forms a disulfide-linked complex with tapasin and is thought to either stabilize the complex or facilitate correct assembly of class I molecules (7, 8). Recently, ERp57 has been demonstrated to isomerize interchain disulfides in the major capsid protein, VP1, of simian virus 40 (9). The ability to dissociate VP1 pentamers by ERp57 does not require the substrate to interact with the calnexin cycle. Hence, it is still unclear how ERp57 recognizes its substrates, and in particular, whether this recognition is solely determined by an interaction with the calnexin cycle.
The recognition of substrates by PDI is somewhat clearer in that one particular domain within the protein (the b′ domain) has been shown to be primarily responsible for substrate recognition and peptide binding (10). The corresponding domain within ERp57 has been shown to be responsible for interaction with the calnexin cycle (11), suggesting that for ERp57, substrate recognition must occur outside this domain or is determined solely by substrate interaction with calnexin via its oligosaccharide side chain. Hence, the aim of our study was to evaluate the necessity of the calnexin cycle both for ERp57 to recognize its substrates and for correct folding of glycoproteins. ERp57 was found to be required for the efficient folding of one substrate, influenza virus hemagglutinin (HA), but only when it entered the calnexin cycle. HA did not require ERp57 to fold if it was blocked from entering the calnexin cycle. In contrast, β1-integrin does not fold efficiently either if ERp57 was depleted or if ERp57 is blocked from entering the calnexin cycle (6). Although ERp57 may be dispensable for the folding of some glycoproteins, the interaction with calnexin commits them to an ERp57-dependent fate. We also found that the majority of ERp57 substrates need to enter the calnexin cycle to be acted upon by the enzyme, demonstrating that substrate specificity is primarily dependent upon substrate entry into the calnexin cycle.
EXPERIMENTAL PROCEDURES
Antibodies and Cell Lines—Mouse monoclonal antibody (8E3) to β1-integrin was a gift from Martin Humphries (University of Manchester, Manchester, UK). A rabbit polyclonal antibody to calnexin was as described previously (12). Rabbit polyclonal anti-calreticulin was purchased from Stressgen (Ann Arbor, MI), mouse monoclonal anti-V5 antibody was from Invitrogen, PaSta.1 (mouse monoclonal to tapasin) and R.RING.4C (purified rabbit serum to TAP-1) were from Peter Cresswell (Yale University School of Medicine, New Haven, CT), and V5-agarose beads were from Sigma. ERp57-/- cells and mouse fibroblasts were a gift from Natalio Garbi and Günter Hämmerling, (German Cancer Research Center, Heidelberg, Germany).
Transcription and Translation in Vitro—A cDNA clone encoding influenza virus HA strain A/Japan/305/57 (H2N2) was a gift from Mary-Jane Gething (University of Melbourne, Melbourne, Australia). A cDNA coding for tapasin was a gift from Peter Cresswell (Yale University School of Medicine). Transcription and translation in semipermeabilized (SP) cells were performed essentially as described previously (13). HA and tapasin cDNA were linearized with BamHI and transcribed using T7 polymerase. β1-integrin, Ero1α, and HLA-B35 mRNA were prepared exactly as described previously (6, 14, 15). Transcripts were translated using rabbit reticulocyte lysate (Flexilysate, Promega) with SP cells added as required. Where appropriate, SP cells were preincubated with 1 mm castanospermine for 5 min or with γ-interferon (200 units/ml) for 48 h. Initiation of protein synthesis was allowed to proceed for 5 min at 30 °C before inhibition with 1 mm ATCA (Sigma) followed by incubation at 30 °C to allow elongation and post-translational modification. At specified times, 25 mm NEM was added at 4 °C to prevent disulfide exchange. β1-integrin translation products were immunoisolated prior to electrophoresis; otherwise, SP cells were isolated and resuspended in SDS-PAGE sample buffer (31.25 mm Tris-HCl, pH 6.8, 2% w/v SDS, 5% v/v glycerol, 0.01% w/v bromphenol blue).
Electrophoresis and Western Blotting—Samples for SDS-PAGE were resuspended in SDS-PAGE sample buffer, and dithiothreitol (50 mm) was added to reduce samples where indicated. All samples were boiled for 5 min before electrophoresis through 7.5% acrylamide gels, and proteins were transferred to nitrocellulose for Western blotting or fixed in 10% (v/v) acetic acid and 10% (v/v) methanol and then dried. Radiolabeled products were visualized by autoradiography using Kodak Biomax MR film (GRI, Essex, UK). For Western blotting, nitrocellulose was blocked with 3% milk in Tris/Tween-buffered saline (10 mm Tris, 150 mm NaCl, pH 7.5, 0.1% Tween 20). Primary antibody incubations were performed for 1 h at 22 °C with 3% milk, whereas secondary antibodies (polyclonal goat anti-rabbit or rabbit anti-mouse immunoglobulins conjugated to horseradish peroxidase (Dako, Ely, UK)) were diluted 1:2000 in Tris/Tween-buffered saline and again incubated at 22 °C for 1 h. Products were visualized using enhanced chemiluminescent substrate (Perbio, Northumberland, UK) and Fuji Super RX film (Fujifilm UK, Bedford, UK).
Creation of ERp57 R282A Stable Cell Lines—Human ERp57 was mutated to change arginine 282 to alanine (primer sequences available on request). Plasmids were linearized with SspI before transfecting into subconfluent HT1080 human fibroblasts with FuGENE 8 (Roche Applied Science) according to the manufacturer's instructions. Stable cell lines were selected with G418 (Sigma) for 14 days before colonies were isolated and screened for expression of ERp57-V5.
AMS Treatment—HA mRNA was translated in the presence of wild-type or ERp57-/- SP cells. ATCA (1 mm) was added after 5 min, and NEM (25 mm) was added after 30 min. Cells were pelleted, lysed cellular debris was removed, and cleared lysates were boiled with 1% SDS for 3 min. Tris[2-carboxyethyl]phosphine (10 mm) was added and incubated at room temp for 5 min, and then AMS (30 mm) was added for 1 h at room temperature. For reduced control, dithiothreitol was added to 20 mm for 3 min at 30 °C prior to NEM addition. For oxidized control, dipyridyl disulphide was added to 1 mm for the same period.
BMH Cross-linking—HT1080 cells in adherent culture were treated for 10 min with 1 mm BMH (Pierce) at 4 °C, and cross-linking was quenched by the addition of 10 mm dithiothreitol. Cells were subsequently lysed, and V5-tagged ERp57 was immunoisolated prior to SDS-PAGE and Western blotting.
Two-dimensional Gel Electrophoresis—Non-reducing/reducing two-dimensional electrophoresis and substrate identification were performed exactly as described previously (6).
RESULTS AND DISCUSSION
Disulfide Bond Formation in Substrates Entering the Calnexin Cycle—The requirement for ERp57 to facilitate efficient disulfide formation varies depending upon the substrate protein (16). For some proteins like HA and β1-integrin, there is a clear defect of folding in an ERp57 knockout cell line; however, for others such as Semliki forest virus E1 and p62 proteins, there is no such defect (6, 16). Such a deficiency in the folding of HA in the absence of ERp57 could be due to ERp57 being the only oxidoreductase that can catalyze correct disulfide formation in HA, or it could be a consequence of HA entering the calnexin cycle, thereby preventing access by other PDI family members.
To address this question, we followed disulfide formation of HA in a SP cell in vitro translation system. In such a system, SP cells prepared from cells grown in culture are added to a reticulocyte lysate in the presence of an RNA transcript and radiolabeled amino acid (13). The reticulocyte lysate does not contain added reducing agent; therefore, disulfide bond formation can occur co- and post-translationally within proteins translocated into the ER of the SP cells (17). The electrophoretic mobility of the reduced, native, and disulfide-bonded intermediates of HA have been characterized extensively elsewhere (18), allowing us to follow folding by assessing the mobility of translation products on non-reducing gels. When the mobility of translation products was analyzed following reduction, two distinct bands were observed (Fig. 1A, lane 6). We have previously demonstrated that these bands correspond to glycosylated (upper) and unglycosylated (lower) HA (13). The unglycosylated material is not translocated as we have shown previously that it can be digested with proteinase K (13). When the same translation products were separated under non-reducing conditions (Fig. 1A, lane 5), the glycosylated HA migrated as a distinct band migrating faster through the gel, demonstrating that disulfide bonds had formed. The unglycosylated protein also migrated faster through the gel but appeared as a smear, indicating that a heterogeneous population of disulfide-bonded species had formed. Hence, in our SP cell system, HA was translated and translocated, and the glycosylated protein was folded into a form with a mobility corresponding to that of the fully oxidized, correctly folded protein. The unglycosylated material formed disulfide bonds, but these were heterogeneous in nature, indicating that, as described previously (19), in the absence of glycosylation, HA does not fold correctly.
FIGURE 1.
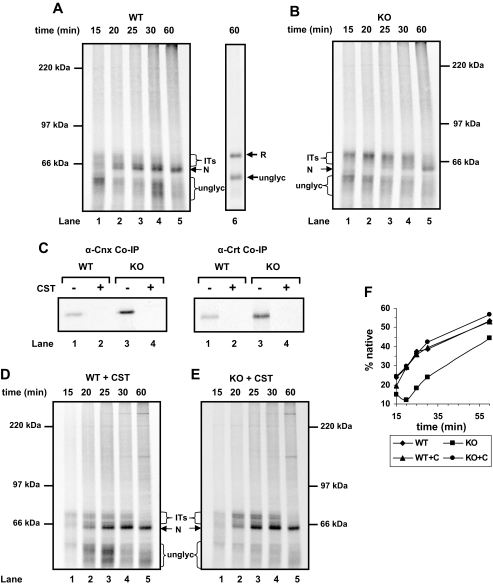
Requirement for ERp57 in HA maturation is dependent upon interaction with the calnexin cycle. A, B, D, and E, translation was performed in the presence of SP cells from wild-type (WT) or ERp57-/- knockout (KO) mouse fibroblasts (A and B) and the same cells preincubated with castanospermine (CST) (D and E). Translation initiation was blocked with ATCA after 5 min, and then NEM was added and cells were harvested at the indicated times before non-reducing SDS-PAGE. All samples were run under non-reducing conditions apart from the gel inset (A, lane 6). ITs, intermediates in native disulfide formation; R, reduced protein; unglyc, unglycosylated protein; N, correctly disulfide-bonded protein. C, HA was translated as in A, B, D, and E, lane 4. Cells were harvested and lysed, and endogenous calnexin was immunoisolated. Co-precipitated (Co-IP) HA was analyzed under reducing conditions. Cnx, calnexin; Crt, calreticulin. F, for each time point in each folding time course, the total amount of glycosylated HA was calculated by densitometry. The fully folded material is given as a percentage of the total. Each time point is an average from three separate time courses.
To determine the consequence of an absence of ERp57 on disulfide bond formation within HA, we carried out a time course in the presence of either wild-type or ERp57-/- cells. At early time points, disulfide-bonded intermediates in the folding pathway were evident (Fig. 1, A and B, lanes 1 and 2). When the time courses are compared between wild-type and ERp57-/- cells, it was clear that the absence of ERp57 slowed down the formation of the fully oxidized faster migrating product, indicating that the native long range disulfides had not formed efficiently (Fig. 1, A, B, and F). Also, there was an accumulation of disulfide-bonded intermediates at early time points in the ERp57-/- cells (Fig. 1B, lanes 1–5). These results confirm those published previously (16) and demonstrate that our SP cell system can reproduce the folding defect caused by a lack of ERp57.
In the absence of ERp57, HA would still interact with the calnexin cycle, but the absence of the oxidoreductase could have led to the slowdown in correct disulfide formation. To investigate this possibility, we carried out a time course in the presence of the glucosidase inhibitor castanospermine. Glycoproteins can be prevented from entering the calnexin cycle by inhibiting glucose trimming of the oligosaccharide side chain. To demonstrate the effectiveness of this treatment, we showed that HA was co-immunoprecipitated with antibodies to calnexin or calreticulin following translation in the presence of wild-type or ERp57-/- cells. However, when castanospermine was included in the translation, no HA was co-immunoprecipitated, demonstrating that castanospermine blocked the interaction between HA and calnexin or calreticulin (Fig. 1C). In addition, in the presence of castanospermine, HA formed native disulfide bonds with similar kinetics in both wild-type and ERp57-/- cells (Fig. 1, D and E, lanes 1–5, and F). Similar results were obtained previously when HA was folded post-translationally in isolated microsomal membranes in the presence of castanospermine (20). Hence, the lack of correct disulfide formation in the ERp57-/- cells was due to the targeting of the protein to the calnexin cycle where the protein becomes sequestered away from other oxidoreductases.
Under normal physiological conditions, HA would enter the calnexin cycle where it requires ERp57 for efficient folding. It has been previously suggested that the main advantage for HA to engage with the calnexin cycle is to slow down folding, thereby ensuring high yield of correctly folded protein and to prevent premature targeting of the protein for degradation (20, 21). Our results strongly suggest that HA can fold efficiently even in the absence of ERp57 if it is prevented from engaging with the calnexin cycle, suggesting that once HA has been dissociated from calnexin, other oxidoreductases can catalyze correct disulfide formation.
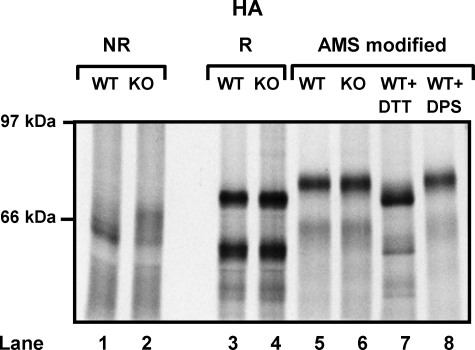
Is the Defect in Disulfide Formation Due to a Lack of Oxidation?—The defect in disulfide formation seen with HA in ERp57-/- cells could be a result of a lack of oxidation or isomerization of non-native disulfides. To determine whether there is a defect in oxidation, we measured the extent of disulfide oxidation by carrying out modification with the alkylating agent AMS. In this experiment, SP cells were treated with NEM to block free thiols and then washed to remove NEM, reduced, and treated with AMS. AMS reacts with free cysteine residues formed, and in doing so, increases the molecular mass by 0.5 kDa per cysteine modified. Hence, if disulfides had formed during translation, the cysteine residues involved would be modified with AMS, resulting in a slower migrating product. When HA was translated in the presence of wild-type or ERp57-/- cells and the translation products were modified with AMS, the resulting product migrated with the fully oxidized protein (Fig. 2, lanes 5, 6, and 8). These results clearly indicate that the defect observed with the ERp57-/- cells is not due to diminished oxidation and is most likely to be due to a lack of isomerization of disulfides. The implication is that an alternate oxidoreductase, possibly PDI, catalyzes the formation of both native and non-native disulfides and that ERp57 then reshuffles the non-native disulfides. The formation of non-native disulfides is likely to be more prevalent in substrates with complicated patterns of disulfides. These results provide further evidence that the essential function of ERp57 is as an isomerase rather than oxidase. Such a clear demarcation of roles for ERp57 and other oxidoreductases suggests that they are able to catalyze distinct reactions within the ER.
FIGURE 2.
ERp57 is not required for full oxidation of glycosylated substrates. HA mRNA was translated in the presence of wild-type (WT) (lanes 1, 3, 5, 7, and 8) or ERp57-/- knockout (KO) (lanes 2, 4, and 6) SP cells. ATCA was added after 5 min, and free thiols were alkylated with NEM after 30 min. Cells were harvested, and translated products were analyzed under non-reducing (NR) (lanes 1 and 2) and reducing conditions (R) (lanes 3–8). In addition, samples of each were treated with Tris[2-carboxyethyl]phosphine and AMS to modify disulfide linked thiols (lanes 5 and 6). As controls for AMS modification, the process was repeated with each protein translated with wild-type SP cells and cysteine residues fully reduced (dithiothreitol (DTT)) (lane 7) or fully oxidized (dipyridyl disulfide (DPS)) (lane 8) prior to NEM addition.
Can ERp57 Form Mixed Disulfides with Substrates When Not in Association with Calnexin?—It is known that the b′ domain of PDI contains a polypeptide binding site (10) and that the binding of ERp57 to calnexin is mediated by its b′ domain (11, 22). To prevent ERp57 associating with calnexin or calreticulin, we aimed to abolish the interaction between these proteins by introducing a single point mutation (R282A) into the b′ domain of ERp57, which has previously been shown to completely abrogate binding to calnexin in vitro (22). To assay for ERp57 substrate interactions, we took advantage of the fact that during the initial reduction of a non-native disulfide, a mixed disulfide between the enzyme and substrate can be trapped if the second cysteine in the oxidoreductase active site is mutated to alanine. We generated stable cell lines that were expressing V5-tagged versions of ERp57, namely wild-type, R282A, cys2,7 (where both CXXC active sites have been mutated to CXXA), and cys2,7 R282A. Using such an approach, we were able to determine whether preventing an association of ERp57 with calnexin and calreticulin prevented the trapping of mixed disulfides between enzyme and glycoprotein substrate.
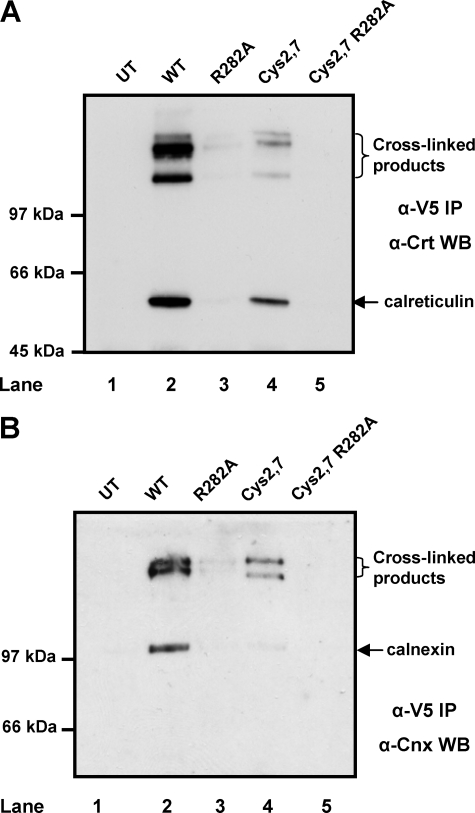
To verify that the R282A mutation prevented complex formation between ERp57 and calnexin or calreticulin in vivo, we added a cross-linking agent to intact cells, lysed the cells, and then analyzed V5-immunoisolated material for the presence of calreticulin (Fig. 3A) or calnexin (Fig. 3B). It has been demonstrated previously that calnexin and calreticulin form complexes with ERp57 that can be stabilized by cross-linking with BMH (4). Complexes between ERp57 and calreticulin (Fig. 3A) or calnexin (Fig. 3B) were isolated from stable cell lines expressing V5-tagged wild-type and cys2,7 ERp57 (Fig. 3, A and B, lanes 2 and 4). Some non-cross-linked calreticulin, and to a lesser extent, calnexin was co-immunoprecipitated with the V5 antibody from the wild-type and cys2,7 ERp57 cell line. Calnexin and calreticulin were absent from control immunoisolated material from untransfected cells demonstrating the specificity of the interaction with ERp57 (Fig. 3, A and B, lanes 1, 2, and 4). The R282A and the cys2,7 R282A ERp57 did not associate with calreticulin or calnexin (Fig. 3, lanes 3 and 5). Hence, we were able to create stable cell lines expressing versions of ERp57 that are not associated with calreticulin or calnexin and have the potential to form mixed disulfides with substrates proteins.
FIGURE 3.
The R282A mutation in ERp57 abolishes its interaction with calnexin and calreticulin. HT1080 cells expressing various V5-tagged ERp57 constructs (lanes 2–5) or untransfected (UT) cells (lane 1) were cross-linked with BMH and lysed, and the ectopically expressed V5-tagged ERp57 was immunoisolated with a V5-specific antibody. The resulting immunoisolated material was separated on a reducing SDS-PAGE gel, and calreticulin (Crt) (A) or calnexin (Cnx) (B) was identified following Western blotting (WB). WT, wild type; IP, immunoprecipitation.
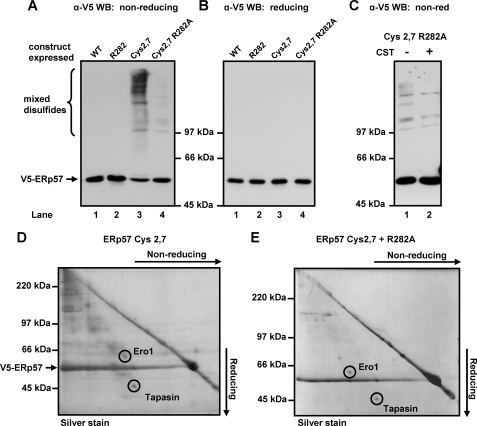
When cell lysates from the cys2,7 cell line were separated under non-reducing conditions and probed with the V5 antibody, several mixed disulfides between ERp57 and substrate proteins were observed (Fig. 4A, lane 3). When the cys2,7 R282A cell line was analyzed under identical conditions, a dramatic reduction in the number and amount of mixed disulfides was observed (Fig. 4A, lane 4). The addition of castanospermine to the cys2,7 R282A cell line prior to cell lysis did not alter these residual mixed disulfides between ERp57 and substrate proteins, confirming that these interactions did not require association of ERp57 with calnexin (Fig. 4C). The fact that the population of mixed disulfides did not increase upon castanospermine treatment also indicates that the decrease in mixed disulfide formation is not due to the substrates being unable to interact with cys2,7 R282A ERp57 due to their sequestration in the calnexin cycle. The identity of the V5-reactive slower migrating bands as mixed disulfides was confirmed by their disappearance when separated under reducing conditions (Fig. 4B).
FIGURE 4.
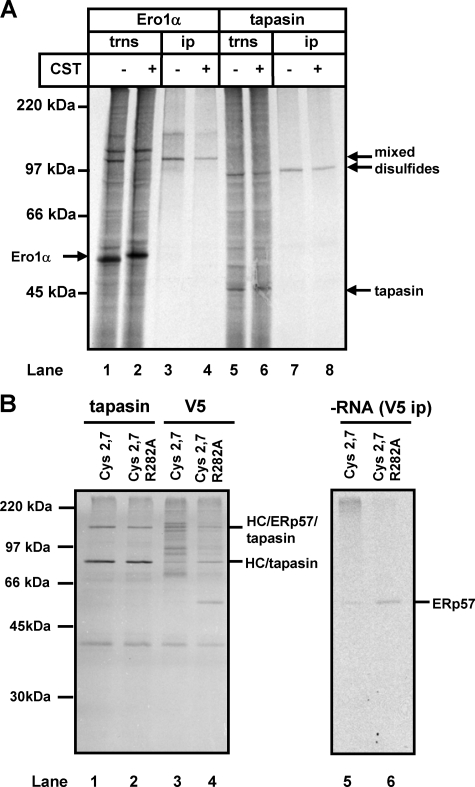
The R282A mutation in ERp57 dramatically reduces mixed disulfide formation. Stable HT1080 cell lines expressing various V5-tagged ERp57 constructs were treated with 25 mm NEM (A and B) or with NEM following treatment with castanospermine (CST) for 16 h (C) and lysed prior to SDS-PAGE and Western blotting (WB) with V5-specific antibody. Lysates were separated under non-reducing (non-red) (A and C) or reducing conditions (B). WT, wild type. V5-tagged ERp57 and mixed disulfides were immunoisolated from either the cys2,7 cell line (D) or the cys2,7 R282A cell line (E) with V5-specific antibody immobilized on agarose beads. Immunoisolated protein was eluted with SDS-PAGE buffer and separated by two-dimensional gel electrophoresis, first dimension non-reducing, second dimension reducing before silver staining. Spots corresponding to Ero1α and tapasin were identified by mass spectrometry.
We have previously been able to identify substrates of ERp57 by immunoisolation of the mixed disulfides followed by two-dimensional gel analysis, silver staining, and mass spectrometry (6). We carried out a similar analysis with the mixed disulfides isolated from the cys2,7 R282A cell line (Fig. 4E). A two-dimensional non-reducing/reducing gel of mixed disulfides from the cys2,7 cell line is included for comparison (Fig. 4D). Only a few spots were visible following silver staining from the cys2,7 R282A cell line, and unfortunately, the amount of product isolated was insufficient for mass spectrometry analysis for most of the substrates; however, two proteins, Ero1α and tapasin, were identified. It would appear that the ability of ERp57 to form mixed disulfides with these proteins is independent of its association with calnexin, a conclusion that is consistent with what is already known concerning the interaction of ERp57 with tapasin (8).
As an alternative approach, we also translated either Ero1α or tapasin mRNA into SP cells isolated from the ERp57 cys 2,7 cell line. Both proteins were able to form mixed disulfides with ERp57, as judged by immunoisolation with the V5 antibody of complexes between the radiolabeled translation product and ERp57 (Fig. 5A, lanes 3 and 7). The complexes were still seen when the translations were carried out in the presence of castanospermine (Fig. 5A, lanes 4 and 8). The slight decrease in mobility of the ero1α and tapasin translation products indicates the inhibition of glucose trimming in the presence of castanospermine (Fig. 5A, lanes 2 and 6). These results clearly demonstrate that these particular client proteins do not need to enter the calnexin cycle to bind to ERp57.
FIGURE 5.
ERp57 forms mixed disulfides with tapasin and Ero1α in the absence of an interaction with calnexin. A, Ero1α or tapasin mRNA was translated in the presence of SP cells from the ERp57 cys 2,7 cell line. Translations (trns) were carried out in the presence or absence of castanospermine (CST) as indicated. Products of translation were separated directly under non-reducing conditions (lanes 1, 2, 5, and 6) or were first immunoisolated with the V5 antibody (lanes 3, 4, 7, and 8). The mobilities of tapasin, Ero1α, and their mixed disulfides with ERp57 are indicated. ip, immunoprecipitation. B, MHC class I heavy chain mRNA was translated in the presence of SP cells from either the cys 2,7 or the cys 2,7 R282A cell line as indicated. Translation products were immunoisolated with either tapasin (lane 1 and 2) or the V5 antibody (lanes 4 and 5). A translation was also carried out in the absence of added mRNA and immunoisolated with the V5 antibody (lanes 5 and 6). HC, heavy chain.
To investigate further the requirement for ERp57 to enter the calnexin cycle to recognize substrate proteins, we translated MHC class I heavy chain in the presence of cys 2,7 or cys 2,7 R282A SP cells and immunoisolated mixed disulfides with either tapasin or V5 antibodies (Fig. 5B). We have previously demonstrated that MHC class I heavy chain forms a number of complexes when translated in the presence of SP cells expressing V5-tagged ERp57 cys2,7 (14). These have been characterized and correspond to a heavy chain-tapasin and a heavy chain-tapasin-ERp57 complex (Fig. 5B) and are part of the peptide-loading complex. In addition, a number of mixed disulfides are formed between heavy chain and ERp57, which can be distinguished by their immunoisolation with the V5 antibody (Fig. 5B, lane 3) and by the fact that they occur outside the peptide-loading complex (14, 23). When heavy chain was translated in the presence of the ERp57 R282A cys2,7 cell line, the heavy chain-tapasin and the heavy chain-tapasin ERp57 complexes were still seen (Fig. 5B, lane 2). However, under identical conditions, the mixed disulfides between heavy chain and ERp57 were abolished (Fig. 5B, lane 4). The radiolabeled product seen at 57 kDa in lane 4 corresponds to endogenous ERp57 translated in this cell line as it is also present when no RNA is added to the translation reaction (Fig. 5B, lane 6). These results demonstrate that the tapasin-ERp57 interaction occurs irrespective of the ERp57 association with calnexin. In contrast, the interaction between ERp57 and calnexin is necessary for ERp57 to form mixed disulfides with heavy chain. These results further demonstrate that the ability of ERp57 to bind and act upon most substrates requires an initial interaction between the substrate glycoprotein and calnexin.
The main consequence of introducing a point mutation in ERp57, which reduces its affinity for calnexin, is to practically eliminate the formation of mixed disulfides with substrates. This observation suggests that for ERp57 to function as an isomerase or reductase, it must be associated with the calnexin cycle. Conversely, the ability of ERp57 to interact with a small defined set of proteins does not seem to be abrogated by the R282A mutation or by blocking glucose trimming, suggesting that the interaction with these proteins is not mediated by the same site as that required for calnexin binding. This conclusion would explain how ERp57 can form a stable interaction with both calreticulin and tapasin within the MHC class I peptide-loading complex (24).
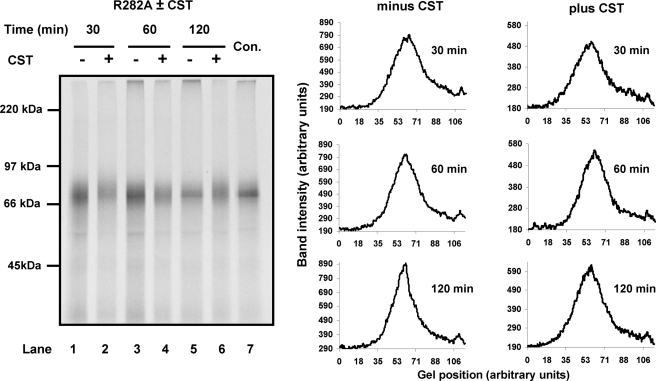
Can ERp57 Assist the Folding of Proteins When It Is Not in Association with Calnexin?—The results obtained with HA would suggest that the function of ERp57 is redundant if proteins are prevented from entering the calnexin cycle. However, we have shown previously that when β1-integrin is prevented from entering the calnexin cycle, it no longer folds efficiently (6). These experiments were carried out by including castanospermine in the translation reaction, which prevented β1-integrin entering the calnexin cycle and caused a defect in folding (6). To determine whether this defect was due to ERp57 being sequestered into the calnexin cycle, we investigated whether β1-integrin could fold correctly in the ERp57 R282A cell line. In this cell line, β1-integrin would be expected to fold correctly as endogenous ERp57 is present and interacts with the calnexin cycle. However, when the translations are carried out in the presence of castanospermine, β1-integrin does not interact with the calnexin cycle, so we can test whether any defect in folding can be reversed by the presence of the ERp57 R282A mutant that is not in association with calnexin. β1-integrin is a complex glycoprotein with over 20 disulfides and 12 glycosylation sites. β1-integrin migrates as a distinct band when the correctly disulfide-bonded protein is separated under non-reducing conditions (6) (Fig. 6, lane 7). When castanospermine was included during translation of β1-integrin in the presence of ERp57 R282A cells, a clear defect in disulfide formation was observed (Fig. 6, lanes 2, 4, and 6). A diffuse pattern indicative of heterogeneous, non-native disulfides was seen throughout the time course in the presence of castanospermine, whereas the protein folded into a more compact structure in the absence of castanospermine. Densitometry scans of the each lane confirmed that a more compact band was observed in the absence of castanospermine (Fig. 6). Hence, for this substrate, there is a requirement to engage with the calnexin cycle to ensure correct disulfide formation; blocking such an interaction does not allow correct folding by providing access to other oxidoreductases including non-calnexin-associated ERp57. Presumably, the combination of unstructured cysteine-rich domains and the extensive N-linked glycosylation preclude correct folding in the absence of calnexin. The crucial role of ERp57 in the folding of β1-integrin illustrates the requirement for an oxidoreductase to be associated with calnexin to facilitate disulfide isomerization within complex glycoproteins.
FIGURE 6.
β1-integrin folding requires both ERp57 and the calnexin cycle. β1-integrin was translated in the presence of HT1080 SP cells stably expressing V5-tagged ERp57 R282A and preincubated in the presence (lanes 2, 4, and 6) or absence (lanes 1, 3, and 5) of castanospermine (CST). The control lane (Con) shows β1-integrin translated for 120 min in the presence of untransfected HT1080 SP cells. Lanes 1–6 were also analyzed by densitometry, and the band intensity was plotted versus the gel position for each time point.
Summary—The presence of multiple oxidoreductases in the ER lumen suggests that each enzyme has the ability to act on a specific subset of substrates or that each enzyme catalyzes a specific type of reaction (2). It is clear from our results with ERp57 that a combination of these two possibilities can occur. Substrate specificity is directly related to the sequestration of glycoproteins into the calnexin cycle, but this specificity is not a consequence of a direct interaction of ERp57 with substrates; rather, it is mediated by the presence of a monoglucosylated oligosaccharide side chain. In addition, the essential function of ERp57 is as an isomerase as oxidation of substrates can occur in the absence of the enzyme. Hence, one of the reasons for the proliferation of ER oxidoreductases is the fact that the substrates for disulfide formation can be sequestered into subcomplexes, which require both spatial and temporal coordination with oxidoreductases to catalyze oxidation, isomerization, or reduction. The calnexin cycle and ERp57 is one example of such a partitioning of substrates, but it may also be the case that substrates associating with BiP require specific oxidoreductases to associate and facilitate disulfide reactions. Indeed, P5 (25) and ERdj5 (26, 27) are known to associate with BiP and may well facilitate disulfide exchange reactions within such a complex.
The absence of a specific oxidoreductase for glycoprotein folding within lower eukaryotes (28) suggests that substrates entering the calnexin cycle in these organisms already contain the correct disulfides. The calnexin cycle may have initially evolved to fulfil a function distinct from protein folding such as retention of unfolded or incompletely assembled proteins (29). An inevitable consequence of the sequestration of glycoproteins would appear to be a slowing of the rate of folding, particularly if the glycoprotein has a complex set of disulfides. Hence, an evolutionary advantage would be afforded to the association of an oxidoreductase with the calnexin cycle to allow the isomerization of non-native disulfides, thereby accelerating folding. Unlike an oxidoreductase that is not associated with the calnexin cycle, ERp57 does not need to bind directly to the glycoprotein as it would be brought into close proximity to its substrate by its binding to calnexin. Hence, the substrate-binding function of PDI, which is mediated via the b′ domain, is lost in ERp57 so that when the binding to calnexin is abrogated, it can no longer facilitate disulfide isomerization even with its authentic substrates. Thus, the specificity of ER oxidoreductases may in part be explained by their interaction with other proteins rather than a direct interaction between enzyme and substrate out with the active site.
Acknowledgments
We acknowledge Natalio Garbi, Günter Hämmerling, Martin Humphries, Peter Cresswell, and Mary Jane Gething for reagents used during this work. We also thank David Knight and Emma Keevil for the mass spectrometry.
The work was supported by grants from the Wellcome Trust (Reference 074081) and the Biotechnology and Biological Sciences Research Council (Reference D00764). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; PDI, protein disulfide isomerase; ATCA, aurin tricarboxylic acid; AMS, 4-acetamido-4′maleimidylstilbene-2,2′-disufhonic acid; BMH, bismaleimidohexane; HA, influenza virus hemagglutinin; NEM, N-ethylmaleimide; SP cells, semipermeabilized cells; MHC, major histocompatibility complex.
References
- 1.Jansens, A., van Duijn, E., and Braakman, I. (2002) Science 298 2401-2403 [DOI] [PubMed] [Google Scholar]
- 2.Ellgaard, L., and Ruddock, L. W. (2005) EMBO Rep. 6 28-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliver, J. D., van der Wal, F. J., Bulleid, N. J., and High, S. (1997) Science 275 86-88 [DOI] [PubMed] [Google Scholar]
- 4.Oliver, J. D., Roderick, H. L., Llewellyn, D. H., and High, S. (1999) Mol. Biol. Cell 10 2573-2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zapun, A., Darby, N. J., Tessier, D. C., Michalak, M., Bergeron, J. J., and Thomas, D. Y. (1998) J. Biol. Chem. 273 6009-6012 [DOI] [PubMed] [Google Scholar]
- 6.Jessop, C. E., Chakravarthi, S., Garbi, N., Hammerling, G. J., Lovell, S., and Bulleid, N. J. (2007) EMBO J. 26 28-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick, T. P., Bangia, N., Peaper, D. R., and Cresswell, P. (2002) Immunity 16 87-98 [DOI] [PubMed] [Google Scholar]
- 8.Peaper, D. R., Wearsch, P. A., and Cresswell, P. (2005) EMBO J. 24 3613-3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schelhaas, M., Malmstrom, J., Pelkmans, L., Haugstetter, J., Ellgaard, L., Grunewald, K., and Helenius, A. (2007) Cell 131 516-529 [DOI] [PubMed] [Google Scholar]
- 10.Pirneskoski, A., Klappa, P., Lobell, M., Williamson, R. A., Byrne, L., Alanen, H. I., Salo, K. E., Kivirikko, K. I., Freedman, R. B., and Ruddock, L. W. (2004) J. Biol. Chem. 279 10374-10381 [DOI] [PubMed] [Google Scholar]
- 11.Russell, S. J., Ruddock, L. W., Salo, K. E., Oliver, J. D., Roebuck, Q. P., Llewellyn, D. H., Roderick, H. L., Koivunen, P., Myllyharju, J., and High, S. (2004) J. Biol. Chem. 279 18861-18869 [DOI] [PubMed] [Google Scholar]
- 12.Allen, S., and Bulleid, N. J. (1997) Biochem. J. 328 113-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson, R., Allen, A. J., Oliver, J., Brookman, J. L., High, S., and Bulleid, N. J. (1995) Biochem. J. 307 679-687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chambers, J. E., Jessop, C. E., and Bulleid, N. J. (2008) J. Biol. Chem. 283 1862-1869 [DOI] [PubMed] [Google Scholar]
- 15.Cabibbo, A., Pagani, M., Fabbri, M., Rocchi, M., Farmery, M. R., Bulleid, N. J., and Sitia, R. (2000) J. Biol. Chem. 275 4827-4833 [DOI] [PubMed] [Google Scholar]
- 16.Solda, T., Garbi, N., Hammerling, G. J., and Molinari, M. (2006) J. Biol. Chem. 281 6219-6226 [DOI] [PubMed] [Google Scholar]
- 17.Bulleid, N. J., Wilson, R., and Lees, J. F. (1996) Biochem. J. 317 195-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maggioni, M. C., Liscaljet, I. M., and Braakman, I. (2005) Nat. Struct. Mol. Biol. 12 258-263 [DOI] [PubMed] [Google Scholar]
- 19.Roberts, P. C., Garten, W., and Klenk, H. D. (1993) J. Virol. 67 3048-3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hebert, D. N., Foellmer, B., and Helenius, A. (1996) EMBO J. 15 2961-2968 [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, W., Helenius, J., Braakman, I., and Helenius, A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 6229-6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozlov, G., Maattanen, P., Schrag, J. D., Pollock, S., Cygler, M., Nagar, B., Thomas, D. Y., and Gehring, K. (2006) Structure (Camb.) 14 1331-1339 [DOI] [PubMed] [Google Scholar]
- 23.Kienast, A., Preuss, M., Winkler, M., and Dick, T. P. (2007) Nat. Immunol. 8 864-872 [DOI] [PubMed] [Google Scholar]
- 24.Hughes, E. A., and Cresswell, P. (1998) Curr. Biol. 8 709-712 [DOI] [PubMed] [Google Scholar]
- 25.Meunier, L., Usherwood, Y. K., Chung, K. T., and Hendershot, L. M. (2002) Mol. Biol. Cell 13 4456-4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anelli, T., and Sitia, R. (2008) EMBO J. 27 315-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ushioda, R., Hoseki, J., Araki, K., Jansen, G., Thomas, D. Y., and Nagata, K. (2008) Science 321 569-572 [DOI] [PubMed] [Google Scholar]
- 28.Simons, J. F., Ebersold, M., and Helenius, A. (1998) EMBO J. 17 396-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellgaard, L., Molinari, M., and Helenius, A. (1999) Science 286 1882-1888 [DOI] [PubMed] [Google Scholar]