Abstract
Sex-dependent differences in adaptation to famine have long been appreciated, thought to hinge on female versus male preferences for fat versus protein sources, respectively. However, whether these differences can be reduced to neurons, independent of typical nutrient depots, such as adipose tissue, skeletal muscle, and liver, was heretofore unknown. A vital adaptation to starvation is autophagy, a mechanism for recycling amino acids from organelles and proteins. Here we show that segregated neurons from males in culture are more vulnerable to starvation than neurons from females. Nutrient deprivation decreased mitochondrial respiration, increased autophagosome formation, and produced cell death more profoundly in neurons from males versus females. Starvation-induced neuronal death was attenuated by 3-methyladenine, an inhibitor of autophagy; Atg7 knockdown using small interfering RNA; or l-carnitine, essential for transport of fatty acids into mitochondria, all more effective in neurons from males versus females. Relative tolerance to nutrient deprivation in neurons from females was associated with a marked increase in triglyceride and free fatty acid content and a cytosolic phospholipase A2-dependent increase in formation of lipid droplets. Similar sex differences in sensitivity to nutrient deprivation were seen in fibroblasts. However, although inhibition of autophagy using Atg7 small interfering RNA inhibited cell death during starvation in neurons, it increased cell death in fibroblasts, implying that the role of autophagy during starvation is both sex- and tissue-dependent. Thus, during starvation, neurons from males more readily undergo autophagy and die, whereas neurons from females mobilize fatty acids, accumulate triglycerides, form lipid droplets, and survive longer.
Sex-dependent differences in adaptation to famine have long been appreciated (1, 2), thought to hinge on a female preference for fat sources, in contrast to a male preference for protein sources (3). Fatty acid metabolism is different between sexes normally (4) and under conditions of starvation (1, 2). During exercise, in addition to increases in carbohydrate requirement, men increase their need for amino acids, whereas women increase mobilization of fat (5). Furthermore, sex-dependent responses to nutritional stress associated with either self-induced weight loss or illness-related cachexia also exist (6, 7).
An important adaptation to starvation is autophagy (autophagy-associated proteins, abbreviated ATG). Classic, starvation-induced autophagy is initiated by nutrient and amino acid deprivation, glucagon, and cAMP (8, 9). ATG7, a ubiquitin E1-like enzyme, is essential for autophagy, with phosphorylation of preautophagosomal membranes, formation of ATG12-ATG5 complexes, and processing of ATG8/LC3 (microtubule-associated protein light chain-3) as other crucial steps in this process (10). Starvation-induced autophagy is regulated by class III phosphatidylinositol 3-kinase and the Bcl-2-interacting partner, Beclin-1 (11). The autophagosomes then engulf cytoplasmic material and/or organelles, such as mitochondria, the latter sometimes referred to as “mitophagy,” disassembling large proteins and organelles to recycle amino acids and other nutrients, an important response to starvation (12).
It is unknown whether starvation can induce autophagy in the brain; however, there is evidence that critical starvation can result in brain atrophy in humans. It has been reported that ∼30% of people during a prolonged hunger strike (mean of 199 days) will show brain tissue loss (13), and brain shrinkage in patients with anorexia nervosa is well documented (14, 15). Although 48 h of food deprivation does not produce detectable autophagy in brains from mice (16), the aforementioned reports are consistent with long durations of starvation as a bona fide stimulus for autophagy in brain. There are recent studies suggesting that other stimuli can induce autophagy in the brain, such as trauma (17) and ischemia (18), and that autophagy may contribute to neuronal death. There is also evidence for autophagy in the human brain after trauma and critical illness (19), which probably includes both elements of malnutrition and systemic stress. A potential role for brain atrophy as a contributor to neurological morbidity in the critically ill and injured is an emerging topic (20).
EXPERIMENTAL PROCEDURES
Sex-specific Neuron-enriched Cultures—Sex-segregated primary cortical neuron cultures were prepared from embryologic day 17 Sprague-Dawley rats as described (21). Embryologic day 17 male and female rats were distinguished by visualization of the penile vessels and sex cords, segregated, and cultured separately. Dissociated cell suspensions were filtered through a 70-μm Falcon nylon cell strainer and then seeded in 96-well plates at a density of 5 × 104 cells/cm2, in plastic dishes coated with 100 μg/ml poly-d-lysine at a density of 1.3 × 107 cells/cm2, or on glass coverslips coated with poly-d-lysine at a density of 1.3 × 107 cells/cm2. Each contained Neurobasal™ medium supplemented with B27™ (Invitrogen) and GlutaMaxl™ (Sigma) for neuron-enriched cultures (22). Cells are incubated at 37 °C in a humidified chamber containing 5% CO2. On the 2nd and 6th day in vitro the culture medium was replaced with fresh medium. Experiments were done at 8-12 days in vitro, when cultures consist primarily of neurons (>95% MAP2 (microtubule-associated protein-2) immunopositive cells, <5% glial fibrillary acidic protein, GFAP immunopositive cells). Verification of sex segregation of embryos was performed using reverse transcription-PCR for the male rat Sry gene using the primers (left, 5′-GCTCAACAGAATCCCAGCAT-3′; right, 5′-TTTGTTGAGGCAACTTCACG-3′) as previously described (21).
Sex-specific Rat Embryonic Fibroblast Cultures—Sex-segregated fibroblast cultures were prepared from embryologic day 17 Sprague-Dawley rats according to a slight modification of a protocol from CHEMCON International. Nonbrain cells were grown in Dulbecco's modified Eagle's medium (DMEM2; Invitrogen) supplemented with antibiotics (100 units/ml penicillin G, 100 μg/ml streptomycin) and 10% fetal calf serum (FCS; Invitrogen) at 37 °C in humidified air with 5% CO2. After the cultured cells reached semiconfluence, they were harvested using 0.25% trypsin and subcultured with DMEM supplemented with 10% FCS in a 10-cm culture dish. Fibroblasts were cultured in 48-well plates at a density of 6 × 104 cells/well or 6-well plates at a density of 1 × 106 cells/well for various experiments.
In Vitro Starvation Models—For primary cortical neurons, at 8-12 days in vitro, Neurobasal™ medium and supplements were removed and replaced with custom-made medium lacking d-glucose, sodium pyruvate, l-glutamate, l-glutamine, and l-aspartate (Invitrogen). The supplements B27™ and GlutaMaxl™ were not added. Nutrient deprivation times ranged from 2 to 72 h. For embryonic fibroblasts, culture medium was replaced with low glucose (1 g/liter) DMEM in the absence of FCS for 24 h.
3-(4,5-Dimethylthiazol)-2,5-diphenyltetrazolium Bromide (MTT) Assay—MTT (0.5 mg/ml) was added to cells in 96-well plates and incubated for 1 h (21). Duplicate 150-μl samples were aspirated, and formazan crystals were dissolved by DMSO. Optical density was determined using a spectrophotometer (Molecular Devices) at 550 nm test and 690 nm reference wavelengths.
Flow Cytometry—Cells were harvested using trypsin, washed once in ice-cold phosphate-buffered saline (PBS), and resuspended in buffer (10 mm Hepes, pH 7.4, 140 mm NaCl, 2.5 mm CaCl2) (21). Suspensions (100 μl) were transferred into fresh tubes, and 10 μl of propidium iodide (PI) stock solution (50 μg/ml) was added. After 20 min, 400 μl of buffer was added, and samples analyzed on a trilaser FACSCalibur flow cytometer.
Histologic Analysis—Cells grown on glass coverslips were immunostained for the neuronal marker MAP-2, as described (21). Neurons were fixed in 2% paraformaldehyde and then incubated with 5% normal donkey serum and 2% bovine serum albumin in PBS containing 0.2% Triton X-100 for 1 h and then incubated in primary antibodies against MAP-2, followed by appropriate secondary antibodies. Cells were examined using a Leica TCS NT confocal tri-laser-scanning inverted microscope (Wetzlar). A minimum of three separate coverslips were evaluated.
Separation of Subcellular Proteins and Western Blot Analysis—Cells were lysed and resuspended in buffer containing 20 mm Hepes, pH 7.4, 10 mm NaCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 250 mm sucrose in H2O (21). Samples were then centrifuged at 600 × g for 10 min at 4 °C. The supernatants were then centrifuged at 20,000 × g for 50 min at 4 °C. These supernatants contained cytosolic proteins (S1). The pellets (P1) were then lysed in buffer and sonicated until frothy, resuspended in lysis buffer, and centrifuged at 16,000 × g for 25 min at 4 °C. The pellets were then washed in 5 μm CaCl and centrifuged at 1600 × g for 10 min. Pellets (P2) enriched in mitochondria, autophagosomes, and other small organelles were again lysed in buffer and sonicated until frothy. Protein concentrations were determined using a Bradford-based method (Bio-Rad). Western blots were performed using a standard technique using a polyclonal antibody against LC3 (MBL International) at a 1:1000 dilution.
Electron Microscopy—Cells were fixed in 2.5% glutaraldehyde in PBS overnight at 4 °C, pelleted, and resuspended in 3% gelatin in PBS, solidified at 4 °C, and then refixed for 15 min. Pellets were washed three times in PBS and then postfixed in 1% OsO4 and 1% K3Fe(CN)6 for 1 h. After three washes, the pellet will be dehydrated through a series of 30-100% EtOH and then incubated in Polybed 812 epoxy resin (Polysciences) and observed using a JEM 100-B transmission electron microscope (Jeol).
Atg7 RNA Interference—Small interfering RNA (siRNA) targeting rat Atg7 (5′-GCAUCAUCUUUGAAGUGAA-3′; Sigma) and pooled nontargeting control siRNA (5′-AUGAACGUGAAUUGCUCAA-3′, 5′-UAAGGCUAUGAAGAGUAC-3′, 5′-AUGUAUUGGCCUGUAUUAG-3′, and 5′-UAGCGACUAAACACAUCAA-3′; Dharmacon) were purchased commercially. Primary neurons were transfected on day 4, and fibroblasts were transfected at the time of 50% confluence, with 20 nmol of Atg7 or control siRNA using Lipofectamine 2000 (Invitrogen). The efficacy of ATG7 knockdown was assessed by Western blot using an antibody against ATG7 (Sigma). Experiments were performed 72 h after transfection.
Primary Neuron Cultures from LC3-GFP Reporter Mice—Sex-segregrated primary cortical neurons from hemizygous transgenic mice expressing GFP-LC3 (16) were cultured as described above for rat neurons, except only transgenic embryos identified using GFP fluorescent macroscopy were used. GFP-LC3 fluorescence was enhanced using anti-GFP Alexa Fluor® 488-conjugated antibody (Invitrogen) and was visualized using an inverted Olympus Fluoview 1000 microscope. Fluorescent intensity above threshold values measured in arbitrary units was quantified in eight areas (3000 × 3000 pixels) per slide from 4 slides/group by an observer blinded to experimental conditions. Lower threshold values were set as the mean fluorescent intensity of control GFP-LC3 cells.
Time-lapsed Fluorescent Microscopy—Sex-segregrated primary cortical neurons from hemizygous transgenic mice expressing GFP-LC3 (16) were cultured in glass-bottomed 25-mm Petri dishes (Mattek Corp.). Subsequently, they were incubated with or without LysoTracker Red® (Sigma) at a dilution of 1:2000 for 10 min at 37 °C. The cells were then washed, and medium was replaced with nutrient deprivation medium. Dishes were immediately mounted in a Harvard systems PDMI perfusion chamber at 37 °C and mounted in a fully automated Nikon 2000E2 microscope. The cells were focused using a ×60 1.45 numerical aperture Plan Apochromat objective (and maintained in focus using a Nikon Perfect Focus attachment) and 15 stage positions selected in Metamorph (MDC). Images were collected using differential interference contrast, green emission/blue excitation, and red emission/green excitation filter cubes (Chroma) and a Photometrics CoolSnap HQ2 camera every minute from each stage position for 1 h. Movies were generated using Metamorph.
LipidTox™ Staining—The intracellular accumulation of lipids was performed using HCS LipidTox™ Steatosis detection kits (Invitrogen). Cells were incubated for 60 min in LipidTox™ Green to stain for neutral lipid aggregates. The cells were then washed with PBS twice and fixed with 4% paraformaldehyde containing Hoechst 33342 (0.5 μg/ml; Sigma) for 30 min. Lipid aggregates were detected using a Leica microscope equipped for epifluorescence and quantified using Olympus MicroSuite™ software.
Phospholipase A2 (PLA2) Activity Assay—Calcium-dependent, group IV cytosolic PLA2 was measured using a commercially available kit (Cayman). Briefly, 1 × 107 cells per sex and condition were collected using a rubber policeman and centrifuged at 2000 × g for 10 min. The pellets were sonicated in a cold buffer (50 mm Hepes, pH 7.4, containing 1 mm EDTA) and then centrifuged at 10,000 × g for 15 min. Supernatants were pretreated with 1 mm bromoenol lactone for 15 min at 25 °C to prevent residual soluble PLA2 activity. The reaction was initiated by adding 200 μl of substrate solution to 10 μl of sample. The reaction was conducted for 60 min at room temperature and was stopped by adding 10 μl of DTNB/EGTA for 5 min. Absorbance was determined using a spectrophotometer at a reference wavelength of 414 nm.
Treatment with PLA2 Inhibitors—Based on a recent report showing that group IV cytosolic PLA2 is necessary for the synthesis of lipid droplets (23), we treated male and female neurons with the relatively selective PLA2 inhibitors arachidonyl trifluoromethyl ketone (AACOCF3; Calbiochem), methylarachidonyl fluorophosphonate (MAFP; Cayman), and pyrrolidine-2 (Py-2; Calbiochem) and measured cellular respiration using MTT after 24 h of nutrient deprivation. Lipid droplet formation using concentrations of PLA2 inhibitors that exacerbated reductions in MTT was also determined after 24 h of nutrient deprivation.
Preparation of Lipid Extracts, High Performance Thin Layer Chromatography (HPTLC), and Electron Spray Ionization Mass Spectroscopy (ESI-MS) Analysis of Triglycerides and Free Fatty Acids (FFA)—Total lipids were extracted from neuronal cells by the Folch procedure. Neutral lipid classes were separated by one-dimensional HPTLC on silica G plates (5 × 5 cm; Whatman). The plates were developed with a solvent system consisting of hexane/diethyl ether/glacial acetic acid (65:35:2, v/v/v) and then dried with a forced N2 blower to remove the solvent. Neutral lipids were visualized by exposure to iodine vapors and identified by comparison with authentic lipid standards. The spot on the plate corresponding to triglyceride molecular species was recognized by comparison with a triglyceride (T18:1) standard spotted on the side of the same plate. For ESI-MS analysis, triglyceride spots on the silica plates were visualized by spraying the plates with deionized water. After this, the spots were scraped from the silica plates, and triglycerides were extracted by choloroform/methanol/water (20:10:2, v/v/v) and dried under N2. The triglyceride mixtures were resuspended in 250 μl of 1:2 chloroform/methanol, containing 5 mm NH4OAc. ESI-MS was performed by direct infusion into a linear ion trap mass spectrometer (Thermo-Finnigan LXQ), using a syringe pump at a flow rate of 5 μl/min. Triglyceride molecular species were directly ionized in the positive ion mode by ESI and detected as ammonium adduct ions. Typically, a 0.2-min period of signal averaging in the profile mode was employed for each spectrum of a TG sample. Experimental conditions were as follows: 4.97 kV source voltage; 150 °C capillary temperature; 12.0 V capillary voltage; 1.0 m/z ion isolation window. Triglyceride molecular species were quantitated by comparing the peak intensities with that of an internal standard (T12:0). Triglycerides, including trimyristin (T14:0), tripalmitin (T16:0), tristearin (T18:0), and triolein (T18:1), purchased from Supelco (Bellefonte, PA) were used as reference standards. The chemical structure of triglyceride was confirmed by using the Lipid Map Data Base and ChemDraw format (available on the World Wide Web).
Extracted FFAs and phospholipids were divided into aliquots for ESI-MS and phosphorus analysis. ESI-MS analysis of FFA was performed by direct infusion into a linear ion trap mass spectrometer LXQTM, using a syringe pump at a flow rate of 5 ml/min. FFA molecular species were directly ionized in the negative ion mode by ESI and detected as deprotonated ions [M - H]-. FFA molecular species were quantified by comparisons of ion peak intensities with internal standards. FFAs, including C14:0, C16:0, and C18:1, were used as reference standards. The fatty acid composition was calculated on the basis of the ratios of the peak height intensity in the ESI-MS spectra in the analysis of the isotopic effect.
RESULTS
Sex-dependent Sensitivity to Nutrient Deprivation—We first examined the effect of nutrient deprivation on mitochondrial electron transport (respiration) in neurons from males and females using the MTT assay. Sex segregation in neurons was verified using reverse transcription-PCR for the Y chromosome gene Sry as described (21) (Fig. 1A). A time-dependent reduction in mitochondrial respiration was observed during nutrient deprivation (Fig. 1B), with neurons from males being more sensitive than neurons from females at all durations. At 24 h of nutrient deprivation, mitochondrial respiration was 31 ± 10 and 51 ± 17% of control values for neurons from males and females, respectively. Grossly, reduced neuronal numbers were observed during nutrient deprivation using MAP2 staining (Fig. 1C).
FIGURE 1.
Sex-dependent starvation-induced autophagy in neurons. A, detection of the Y chromosome gene Sry by PCR in neurons from males (XY) but not females (XX). M, marker; bl, blank. B, inhibition of mitochondrial respiration during ND, as measured by the conversion of MTT to formazan. XY neurons were more sensitive to ND versus XX neurons at all durations (n > 9/group). C, gross morphology of MAP2-immunostained neurons (green) under control conditions and 24 h after ND. Cells were also stained with Hoechst dye (blue). D, Western blot showing increased LC3-II and reduced cytochrome oxidase subunit IV (COX IV) in XY versus XX neurons in the P2 fraction after 24 h of ND (pooled samples from three independent experiments). Note that LC3-II was detected in the P2 but not the supernatant (S1) fraction, consistent with a shift of cytosolic LC3 into autophagosomes. β-Actin was used as a loading control. E, electron micrographs of XY and XX neurons under control conditions and 24 h after ND. Note the presence of several autophagosomes (a) and swollen mitochondrial fragments (arrows) primarily in XY neurons after 24 h of ND. Also note many lipid droplets (l) after starvation, seen primarily in XX neurons. Results shown are representative of three independent experiments. Bar, 500 nm. F, examples of autophagosomes from XY neurons after 24 h of ND at higher magnification. A double membrane is seen in the right panel (arrowhead). Bar, 500 nm. *, p < 0.05 versus control; †, p < 0.05 versus XY; ANOVA with Dunnett's post hoc test.
Proteolytic cleavage and covalent attachment of phosphatidylethanolamine at the C-terminal glycine of LC3 are essential for cytoplasm-vacuole targeting and are biochemical features of autophagy (24). Lipidation of LC3-I results in a 16-kDa peptide referred to as LC3-II. Western blotting for LC3-I and LC3-II was performed in cell pellets (P2) containing mitochondria and other organelles, including autophagosomes. A relative increase in LC3-II was observed in P2 fractions from neurons from males but not females after 24 h of starvation versus controls (Fig. 1D). At base line, the ratio of LC3-II/LC3-I was 0.17 ± 0.03 versus 0.19 ± 0.06 in neurons from males versus females, respectively. After 24 h of nutrient deprivation, the ratio of LC3-II/LC3-I was 0.31 ± 0.12 versus 0.19 ± 0.04 in neurons from males versus females, respectively (p = 0.1), implying either increased production or reduced turnover of autophagosomes (25). A more prominent reduction in the relative abundance of the mitochondrial protein cytochrome oxidase was also seen in neurons from males versus females after 24 h of starvation (80.3 ± 8.1 versus 98.3 ± 14.2% of control, respectively; p < 0.05; Fig. 1D), consistent with increased mitophagy.
A pathognomonic feature of autophagy is ultrastructural evidence of autophagosomes (26). We therefore examined starved neurons using electron microscopy to qualitatively confirm the presence of autophagy. Ultrastructural analysis after 24 h of nutrient deprivation showed abundant autophagosomes in neurons from males, infrequently observed in neurons from females (Fig. 1, E and F). Swollen, rounded mitochondria were observed in neurons from both sexes after 24 h of nutrient deprivation.
To examine the dynamics of nutrient deprivation-induced autophagy and quantify autophagosome formation, we utilized sex-segregated primary cortical neuron cultures from hemizygous transgenic mice expressing GFP-LC3. GFP-LC3 expression itself does not induce autophagy, but GFP-LC3 localizes to autophagosome membranes similar to endogenous LC3 (26). Rapid formation of autophagosomes was apparent using time-lapsed microscopy, with detection of punctate GFP-enriched vacuoles by 1 h and more so in neurons from males versus females (Fig. 2A and Video S1). For quantification at 2 and 24 h, the fluorescent GFP-LC3 signal was enhanced using anti-GFP antibody. Fluorescence intensity above threshold (defined as mean fluorescence intensity in control GFP-LC3 cells in arbitrary units) was measured in 8 areas (3000 × 3000 pixels)/slide from 4 slides/group. At 2 and 24 h of nutrient deprivation, increased punctate labeling consistent with formation of autophagosomes was seen to a greater degree in neurons from males versus females at 24 h (Fig. 2, B and C). Fusion of GFP-enriched vacuoles with lysosomes was also examined using GFP-LC3-labeled neurons and LysoTracker Red®. Formation of autophagosomes and fusion with lysosomes was observed by 1 h (Fig. 3 and Video S2). To confirm that neurons were undergoing autophagy during nutrient deprivation, knockdown experiments using Atg7 siRNA were performed. Consistent with a sex-dependent increase in autophagy during starvation, transfection of Atg7 siRNA (20 nmol) reduced GFP-LC3 fluorescent intensity in neurons from males after 24 h versus nontargeting scrambled control siRNA (Fig. 2C).
FIGURE 2.
Quantification of starvation-induced autophagy in sex-segregated neurons. A, time-lapsed images of neurons from male (XY) and female (XX) GFP-LC3+/- mice showing formation of GFP-enriched vacuoles consistent with autophagosome formation. Times ranged from 0 to 82 min and were captured from Video S1. B and C, three-dimensional quantification of fluorescent intensity in GFP-LC3+/- mouse cortical neurons transfected with 20 nmol of Atg7 siRNA or a nontargeting scrambled control siRNA 72 h before nutrient deprivation. Atg7 siRNA reduced GFP-LC3 fluorescent intensity in XY neurons at 24 h. AU, arbitrary units; data from 8 regions (3000 pixels2)/slide, 4 slides/group; mean ± S.D.; *, p < 0.05 versus control; †, p < 0.05 versus XY; ‡, p < 0.05 versus control siRNA; ANOVA with Dunnett's post hoc test.
FIGURE 3.
Dynamic fusion of autophagosomes and lysosomes. Time-lapsed images of a GFP-LC3-tagged neuron from male mice showing fusion of GFP-enriched vacuoles with LysoTracker Red-labeled lysosomes. Times ranged from 0 to 54 min and were captured from Video 2. AFU, arbitrary fluorescent units.
Inhibition of Autophagy Reduces Cell Death in Starved Neurons—Nutrient deprivation-induced reduction in mitochondrial respiration was partially inhibited by pretreatment with 3-methyladenine (3-MA; 5 mm; Sigma) in neurons from males but not females at 24 h (Fig. 4A). The amount of cell death occurring after 24 h of nutrient deprivation was quantified using PI labeling and flow cytometry. Nutrient deprivation resulted in 26 ± 2% PI-labeled neurons from males compared with 12 ± 5% neurons from females (Fig. 4, B and C). The effect of 5 mm 3-MA on cell death during nutrient deprivation was also examined using PI, with complete protection from starvation seen in neurons from both sexes. 3-MA is a relatively selective class III phosphatidylinositol 3-kinase inhibitor that inhibits starvation-induced autophagy (9), although 3-MA has other effects, including inhibition of non-class III phosphatidylinositol 3-kinases and promotion of glycogen breakdown in hepatocytes (27).
FIGURE 4.
Inhibition of starvation-induced autophagic reduces neuronal death in vitro. A, ND-induced decreases in mitochondrial respiration in neurons from males (XY) but not females (XX) at 24 h were attenuated by the autophagy inhibitor 3-MA (5 mm; n > 6/group). B, cell death measured using PI labeling showing increased cell death in XY versus XX neurons after 24-h deprivation (n = 3/group). Pretreatment with 5 mm 3-MA prevented ND-induced cell death in both sexes. Representative data from each group are shown in C. D, primary rat neurons transfected with 20 nmol of Atg7 siRNA or a nontargeting scrambled control siRNA 72 h before nutrient deprivation. Western blot showing partially effective knockdown of ATG7 in Atg7 siRNA but not scrambled siRNA-treated neurons (n = 3/group). E, cell death measured using PI labeling showing increased cell death in XY versus XX neurons after 24-h deprivation (n = 3/group). Pretreatment with Atg7 siRNA reduced ND-induced cell death in XY neurons at 2 and 24 h and in XX neurons at 24 h. Mean ± S.D.; *, p < 0.05 versus control, †, p < 0.05 versus vehicle; ‡, p < 0.05 versus control; ANOVA with Dunnett's post hoc test.
To confirm that neurons were dying of autophagy during nutrient deprivation, knockdown experiments using Atg7 siRNA (20 nmol) were performed. Effective knockdown of ATG7 72 h after transfection was verified using Western blot, with a 42.9 and 20.9% reduction of ATG7 in control neurons from males and females, respectively (Fig. 4D). Overall ATG7 knockdown was more effective in neurons from males versus females, and ATG7 appears to be down-regulated by 24 h of nutrient deprivation, particularly in neurons from females. ATG7 knockdown reduced PI-labeled neurons from males at 2 and 24 h and PI-labeled neurons from females at 24 h of starvation (Fig. 3E). Taken together, these data show that inhibition of autophagy can prevent starvation-induced cell death in neurons and that neurons from males are more susceptible to autophagic cell death compared with neurons from females.
Sex-dependent Increases in Lipid Droplets and Triglycerides in Starved Neurons—The ultrastructural appearance of lipid droplets after starvation was also seen (Fig. 1E), predominantly in neurons from females, revealing a potential mechanism underlying their observed tolerance to starvation. To begin to explore this further, starved neurons were stained for neutral lipid deposits. Consistent with the electron microscopy data, an increase in neutral lipid aggregates was observed in neurons from females versus males after starvation (Fig. 5, A and B). Neurons from females but not males were able to maintain lipid stores for up to 72 h of nutrient deprivation, and this appeared to correlate grossly with cell viability.
FIGURE 5.
Sex-specific increases in lipid droplets and triglycerides during starvation in neurons. A, LipidTox™ staining showing increased aggregation of neutral lipids (green) in neurons from females (XX) during ND. B, lipid aggregates were quantified and expressed as aggregates/cell (identified by Hoechst, blue; n = 5 high power fields/group from three independent experiments). C, typical positive ion ESI-mass spectra of TG molecular species isolated from 10 × 106 pooled primary rat cortical neurons before and after ND and MS/MS of TG major ion at m/z 876.8 (52:2) (D). E, TG content in neurons from males (XY) and females before and after ND (mean ± S.D. from five runs from the pooled samples). Mean ± S.D.; *, p < 0.05 versus control; †, p < 0.05 versus XY; ANOVA with Dunnett's post hoc test.
To quantify the amount of neutral lipid in a more robust fashion and to begin to evaluate changes in individual molecular species after nutrient deprivation, we used HPTLC followed by ESI-MS. Since triglycerides (along with cholesterol) are felt to be the primary neutral lipid contained within lipid droplets (28), triglycerides were analyzed. The purified triglyceride fraction obtained after HPTLC from neurons contained a complex mixture of triglyceride molecular species. At least six major ion clusters presented as [M + NH4] were observed between m/z 780 and 1000 (Fig. 5C). The collision induced decomposition of triglyceride ammonium adduct ions [M + NH4]+ resulted in the formation of fragments [M + H]+ and [M + H - RCOOH]+ with characteristic loss of the RCOOH and NH3 from the parent ion as previously reported (29). For example, fragmentation of the major ion at m/z 876 resulted in the formation of fragments with m/z 859, 631.6, 605, 603, 577, 551, and 549, which correspond to the combined loss of NH3 and C14:0, C16:1, C16:0, C18:1, C20:2, or C20:1, respectively. These fragments originated from at least four different triglyceride molecular species as follows: (C14:0)/(C18:1)/(C20:1), (C16:0)/(C16:1)/(C20:1), (C16:0)/(C16:0)/(C20:2), and (C16:0)/(C18:1)/(C18:1) (Fig. 4D). Major triglyceride molecular species are presented in Table 1. Starvation of neurons resulted in increased content not only of triglyceride major molecular species at m/z 822, 848, 850, 876, and 904 but also of several minor molecular species at m/z 926, 932, 950, 954, and 956. The total amount of triglyceride in neuronal cells is shown in Fig. 5E. After starvation, total triglyceride levels increased 12.2-versus 3.5-fold in neurons from females versus males, respectively.
TABLE 1.
Triglyceride molecular species identified from neurons after nutrient deprivation
| Triglyceride molecular speciesa | m/z | Acyl chain composition (acyl/acyl/acyl) |
|---|---|---|
| 48:1 | 822.8 | C14:0/C16:0/C18:1 |
| C16:0/C16:0/C16:1 | ||
| 50:2 | 848.7 | C16:1/C16:0/C18:0 |
| C14:0/C18:0/C18:2 | ||
| 52:2 | 876.8 | C16:0/C16:1/C20:1 |
| C14:0/C18:1/C20:1 | ||
| C16:0/C16:0/C20:2 | ||
| C16:0/C18:1/C18:1 | ||
| C16:0/C18:0/C18:2 | ||
| 54:2 | 904.8 | C16:0/C18:1/C20:1 |
| C18:1/C18:0/C18:1 | ||
| 56:5 | 926.6 | C18:0/C18:1/C20:4 |
| 56:2 | 932.6 | C16:0/C22:1/C18:1 |
| C18:0/C20:1/C18:1 | ||
| 58:7 | 950.6 | C18:0/C18:1/C22:6 |
| 58:5 | 954.6 | C18:0/C18:1/C22:4 |
| C16:0/C20:0/C22:5 | ||
| C18:1/C20:1/C20:3 | ||
| 58:4 | 956.8 | C16:0/C20:0/C22:4 |
| C18:1/C20:0/C20:3 |
Total number of carbons in fatty acyls and the total number of double bonds in fatty acyls.
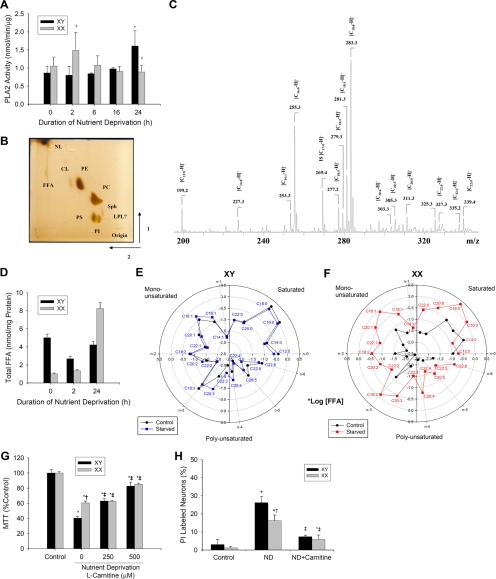
Sex-specific Free Fatty Acid Profile in Starved Neurons—To further detail differences in lipid metabolism of sex-segregated neurons, we performed additional experiments evaluating PLA2 activity as well as the content and molecular species of the two products of phospholipase-catalyzed reactions, FFA and lysophospholipids. PLA2 activity was increased in neurons from females early (2 h) after nutrient deprivation (Fig. 6A). A similar increase in PLA2 activity was observed at a later time point (24 h) after nutrient deprivation in neurons from males. We reasoned that with activation of PLA2, there would be accumulation of FFA and lysophospholipids in neurons after nutrient deprivation. Indeed, ESI-MS analysis revealed a significant accumulation of FFA during nutrient deprivation, a response that was more robust in neurons from females versus males (Fig. 6D). Interestingly, the base-line content of FFA was higher in neurons from males than from females. Overall, however, the level and scale of FFA accumulation was much smaller compared with triglyceride accumulation in neurons from both sexes (compare with Fig. 5). ESI-MS structural analysis of FFA molecular species in the negative ion mode detected a series of deprotonated ions [M - H]- (Fig. 6C). FFA molecular species were quantified by comparisons of ion peak intensities with those of internal standards. The levels of the major types of FFA (saturated, monounsaturated, and polyunsaturated) are presented in Table 2 and Fig. 6, E and F. Obvious differences at base line and during nutrient deprivation between neurons from males and females were observed. A greater accumulation of saturated FFA (C16:0 and C18:0), monounsaturated FFA (C18:1), and polyunsaturated FFA (C18:2, C18:3, C20:4, C22:5, and C22:6) were observed in neurons from females versus males during nutrient deprivation. Treatment with l-carnitine partially restored mitochondrial respiration and reduced cell death in neurons from both sexes, to the point where outcomes between neurons from males and females were similar after 24 h of nutrient deprivation (Fig. 6, G and H).
FIGURE 6.
Sex-specific changes in FFA content and profile during starvation in neurons. A, cytosolic PLA2 activity increased earlier in neurons from females (XX; 2 h) versus males (XY; 24 h) during nutrient deprivation (ND; n = 5/group). B, typical thin layer chromatography separation of lipids (shown are 10 × 106 XX neurons 24 h after ND (CL, cardiolipin; LPL, lysophospholipid; NL, neutral lipid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; Sph, sphingolipid). C, typical negative ion ESI-mass spectra of FFA molecular species isolated from 10 × 106 XX neurons 24 h after ND. D, FFA content from XY and XX neurons before and after ND (mean ± S.D. from five runs from the pooled samples). E and F, polar plots showing individual log[FFA] profiles from XY and XX neurons at base line and after 24 h of ND (refer to Table 2 for more complete data). G, pretreatment with 250 μm l-carnitine partially prevented ND-induced reductions in mitochondrial respiration as measured by the conversion of MTT to formazan in XY neurons versus control at 24 h. Pretreatment with 500 μm l-carnitine partially prevented ND-induced reductions in mitochondrial respiration in both XY and XX neurons versus control at 24 h (n = 10/group). H, cell death measured using PI labeling showing increased cell death in XY versus XX neurons after 24 h (n = 3/group). Pretreatment with 250 μm l-carnitine prevented ND-induced cell death in both sexes. Mean ± S.D.; *, p < 0.05 versus control; †, p < 0.05 versus XY; ‡, p < 0.05 versus vehicle; ANOVA with Dunnett's post hoc test.
TABLE 2.
FFA content (nmol/mg of protein) in neurons from male (XY) and female (XX) rats at base line and at 2 and 24 h of nutrient deprivation
|
Fatty acid
|
XY
|
XX
|
||||
|---|---|---|---|---|---|---|
| 0 h | 2 h | 24 h | 0 h | 2 h | 24 h | |
| C12:0 | 0.399 ± 0.028 | 0.101 ± 0.007 | 0.255 ± 0.018 | 0.114 ± 0.008 | 0.126 ± 0.009 | 0.194 ± 0.014 |
| C14:0 | 0.152 ± 0.011 | 0.058 ± 0.004 | 0.119 ± 0.008 | 0.390 ± 0.003 | 0.047 ± 0.003 | 0.126 ± 0.009 |
| C14:1 | 0.019 ± 0.001 | 0.014 ± 0.001 | 0.017 ± 0.001 | 0.003 ± 0.000 | 0.006 ± 0.001 | 0.044 ± 0.003 |
| C16:0 | 1.144 ± 0.080 | 0.480 ± 0.034 | 0.895 ± 0.063 | 0.225 ± 0.016 | 0.287 ± 0.020 | 1.135 ± 0.079 |
| C16:1 | 0.111 ± 0.008 | 0.096 ± 0.007 | 0.184 ± 0.013 | 0.031 ± 0.002 | 0.044 ± 0.003 | 0.249 ± 0.017 |
| C18:0 | 1.754 ± 0.123 | 0.882 ± 0.062 | 1.250 ± 0.087 | 0.279 ± 0.020 | 0.389 ± 0.027 | 2.305 ± 0.161 |
| C18:1 | 0.302 ± 0.021 | 0.291 ± 0.020 | 0.468 ± 0.033 | 0.075 ± 0.005 | 0.115 ± 0.008 | 0.840 ± 0.059 |
| C18:2 | 0.159 ± 0.011 | 0.096 ± 0.007 | 0.146 ± 0.010 | 0.027 ± 0.002 | 0.036 ± 0.003 | 0.267 ± 0.019 |
| C18:3 | 0.319 ± 0.022 | 0.149 ± 0.010 | 0.186 ± 0.013 | 0.073 ± 0.005 | 0.067 ± 0.005 | 0.757 ± 0.053 |
| C20:0 | 0.111 ± 0.008 | 0.086 ± 0.006 | 0.097 ± 0.007 | 0.028 ± 0.002 | 0.046 ± 0.003 | 0.501 ± 0.035 |
| C20:1 | 0.111 ± 0.008 | 0.044 ± 0.003 | 0.081 ± 0.006 | 0.016 ± 0.001 | 0.035 ± 0.002 | 0.411 ± 0.029 |
| C20:2 | 0.033 ± 0.002 | 0.041 ± 0.003 | 0.051 ± 0.004 | 0.009 ± 0.001 | 0.016 ± 0.001 | 0.137 ± 0.010 |
| C20:3 | 0.038 ± 0.003 | 0.070 ± 0.005 | 0.154 ± 0.011 | 0.013 ± 0.001 | 0.023 ± 0.002 | 0.352 ± 0.025 |
| C20:4 | 0.019 ± 0.001 | 0.031 ± 0.002 | 0.045 ± 0.003 | 0.006 ± 0.001 | 0.010 ± 0.001 | 0.172 ± 0.012 |
| C20:5 | 0.055 ± 0.004 | 0.017 ± 0.001 | 0.027 ± 0.002 | 0.008 ± 0.001 | 0.010 ± 0.001 | 0.046 ± 0.003 |
| C22:0 | 0.097 ± 0.007 | 0.072 ± 0.005 | 0.099 ± 0.007 | 0.027 ± 0.001 | 0.036 ± 0.003 | 0.272 ± 0.019 |
| C22:1 | 0.038 ± 0.003 | 0.045 ± 0.003 | 0.030 ± 0.002 | 0.009 ± 0.001 | 0.016 ± 0.001 | 0.140 ± 0.010 |
| C22:2 | 0.014 ± 0.001 | 0.017 ± 0.001 | 0.013 ± 0.001 | 0.004 ± 0.001 | 0.005 ± 0.001 | 0.062 ± 0.004 |
| C22:3 | 0.010 ± 0.001 | 0.010 ± 0.001 | 0.022 ± 0.002 | 0.002 ± 0.001 | 0.004 ± 0.001 | 0.031 ± 0.002 |
| C22:4 | 0.007 ± 0.001 | 0.007 ± 0.001 | 0.009 ± 0.001 | 0.001 ± 0.001 | 0.002 ± 0.001 | 0.023 ± 0.002 |
| C22:5 | 0.027 ± 0.002 | 0.032 ± 0.002 | 0.013 ± 0.001 | 0.002 ± 0.001 | 0.009 ± 0.001 | 0.029 ± 0.002 |
| C22:6 | 0.091 ± 0.006 | 0.042 ± 0.003 | 0.066 ± 0.005 | 0.017 ± 0.001 | 0.023 ± 0.002 | 0.152 ± 0.011 |
| Saturated | 3.656 | 1.679 | 2.716 | 0.712 | 0.930 | 4.533 |
| Unsaturated | 1.353 | 1.003 | 1.511 | 0.298 | 0.421 | 3.710 |
| Monoenoic | 0.582 | 0.491 | 0.779 | 0.134 | 0.215 | 1.683 |
| Polyenoic | 0.771 | 0.512 | 0.731 | 0.164 | 0.206 | 2.027 |
| Total FFA | 5.009 | 2.682 | 4.227 | 1.010 | 1.351 | 8.243 |
Cytosolic Group IV PLA2-dependent Lipid Droplet Formation in Starved Neurons—The PLA2 inhibitors AACOCF3, MAFP, and Py-2 exacerbated nutrient deprivation-induced reductions in mitochondrial respiration at 24 h as measured by MTT in a dose-dependent manner versus vehicle treatment (Fig. 7, A-C). In contrast to the sex-specific effect of inhibitors of autophagy during nutrient deprivation, the effect of PLA2 inhibitors was more prominent in neurons from females versus males, with reductions in MTT ranging from 19.8 to 24.5% in neurons from females versus 4.0-17.9% in neurons from males for the various inhibitors. Consistent with the report by Gubern et al. (23) showing that cytosolic PLA2 is necessary for the synthesis of lipid droplets, AACOCF3 (100 μm), MAFP (100 μm), and Py-2 (10 μm) inhibited nutrient deprivation (ND)-induced formation of neutral lipid droplets in neurons from both males and females (Fig. 7, D and E). Taken together, these data suggest that formation of lipid droplets during starvation confers a survival benefit.
FIGURE 7.
Cytosolic phospholipase A2 (cPLA2)-dependent lipid droplet formation during starvation in neurons. A-C, treatment with the relatively selective cytosolic phospholipase A2 inhibitors AACOCF3, MAFP, and Py-2 exacerbated ND-induced reductions in mitochondrial respiration as measured by the conversion of MTT to formazan in neurons from females (XX) and to a lesser extent males (XY) versus control at 24 h in a dose-dependent manner (n = 4-8/group). D and E, treatment with AACOCF3 (100 μm), MAFP (100 μm), and Py-2 (10 μm) inhibited ND-induced formation of neutral lipid droplets detected using LipidTox™ staining (green) in both XY and XX neurons (n = 12 measurements/condition). Mean ± S.D.; †, p < 0.05 versus XY; ‡, p < 0.05 versus vehicle; ANOVA with Dunnett's post hoc test.
Inhibition of Autophagy Increases Cell Death in Starved Fibroblasts—In addition to neurons, the sex-dependent susceptibility to nutrient stress was examined in rat embryonic fibroblasts (Fig. 8A). Fibroblasts were cultured in DMEM with 10% FCS or in low glucose (1 g/liter) DMEM without FCS (nutrient deprivation). After 24 h of nutrient deprivation, fibroblasts from males showed a striking increase in secondary lysosomes, multivesicular bodies, and autophagosomes, compared with control fibroblasts (Fig. 8B). Fibroblasts from females also showed an increase in secondary lysosomes and multivesicular bodies but to a much lesser degree than fibroblasts from males, with occasional lipid droplets also seen. Fibroblasts were also transfected with 20 nmol of rat Atg7 or nontargeting scrambled control siRNA. In contrast to neurons, ATG7 knockdown increased PI-labeled fibroblasts from males at 2 and 24 h and PI-labeled fibroblasts from females at 24 h of starvation (Fig. 8C). Thus, although similar sex-dependent sensitivity to nutrient deprivation was observed in fibroblasts and neurons, the role of autophagy in the two distinct cell types appears quite different.
FIGURE 8.
Inhibition of starvation-induced autophagic increases cell death in fibroblasts. A, gross morphology of hemotoxylin and eosin-stained fibroblasts under control conditions and 24 h after ND. B, electron micrographs of fibroblasts from males (XY) and females (XX) under control conditions and 24 h after ND. Note the presence of secondary lysosomes, multivesicular bodies (mv) and autophagosomes (a) more profoundly in XY versus XX neurons after 24 h of ND. Insets, higher magnification (i and ii). Note lipid droplets (l) after starvation seen primarily in XX fibroblasts and an autophagosome with a double membrane (arrowhead). Bar, 500 nm. C, inhibition of autophagy using Atg7 siRNA exacerbated cell death in fibroblasts during ND. Fibroblasts were transfected with 20 nmol of Atg7 siRNA or nontargeting scrambled control siRNA 72 h before ND (n = 3/group). Results are mean ± S.D.; *, p < 0.05 versus control; †, p < 0.05 versus vehicle; ANOVA with Dunnett's post hoc test.
DISCUSSION
The major finding of this study is that during critical nutrient deprivation, neurons from males undergo autophagy, presumably preferring to utilize proteins for fuel, whereas neurons from females have either an increased capacity or preference to store triglycerides, presumably preferring to utilize fats for fuel, allowing for longer survival. This sex-dependent response to starvation appears to be generalizable to nonneuronal cells; however, the role of autophagy and therefore the consequence of its inhibition, appear tissue-dependent. These data are the first to demonstrate that fuel source preferences observed in male and female macro-organisms during nutritional stress can be reduced to individual cells, in isolation from adipocytes, muscle, and the effects of circulating hormones. The relative inability of neurons from males, in contrast to their female counterparts, to utilize lipid stores during starvation may result in increased autophagy and neuronal death. Our data showing that inhibition of autophagy increases tolerance to nutrient deprivation in neurons from males, whereas inhibition of PLA2-dependent lipid droplet formation increases sensitivity to nutrient deprivation in neurons from females, is consistent with this possibility.
To answer the question of whether autophagy may be sex-dependent at the neuronal level, independent of typical nutrient depots, such as glycogen, adipose tissue, and skeletal muscle, we chose to use the classic paradigm of nutrient deprivation in primary neurons. The rationale for examining potential sex differences related to autophagy in neurons was based on our previous work showing gender proclivity in response to cytotoxicity and programmed cell death pathways (21) and the recent development of Atg transgenic mice and other molecular tools that have uncovered a role for autophagy in normal central nervous system development (30) and disease (17, 18, 31, 32). Complete genetic disruption of Atg7 and, therefore, autophagy in mice results in accumulation of ubiquitinated bulk proteins and neurodegenerative disease as the animals age (31). In contrast, these same Atg7-deficient mice are protected from hypoxia-ischemia after unilateral carotid artery ligation and hypoxia (18). Thus, in contrast to chronic neurodegenerative diseases, where autophagy seems to play a preventative role (31), autophagy appears to contribute to neuronal death after acute injury, at least in the developing brain. Whether or not these effects were sex-dependent were not addressed in these reports.
It is likely that the source of fatty acids during nutrient deprivation is endogenous, since the starvation medium is devoid of fatty acids (the B27™ supplement containing fatty acids (22), was withheld), although breakdown of fatty acids from dead cells with intracellular transport cannot be excluded. Coincidentally, intracellular transport of fatty acids is more rapid in females versus males (33). Treatment with l-carnitine, essential for transport of fatty acids into mitochondria, partially restored mitochondrial respiration and reduced cell death in neurons from both sexes, suggesting that neurons from males retain the capacity to utilize FFA for nutrition. These data point to innate sex-dependent differences in lipid transport and/or metabolism at the cellular level in neurons. In addition to serving as fuel sources, formation of lipid droplets during cellular stress may offer other survival advantages. For example, channeling of saturated fatty acids by unsaturated fatty acids into triglycerides reduces their toxicity (34). Mixtures of both unsaturated and saturated FFA residues were identified in triglycerides (Table 1) with a greater capacity to increase triglyceride pools in neurons from females versus males after nutrient deprivation.
Female preferences for fat sources during both normal and nutrient-stressed conditions have long been appreciated (1-5). Although we did not measure lipid metabolism directly, our data suggest that in neurons, the female tolerance to nutrient deprivation is at least related to changes in lipid, triglyceride, and FFA content and/or profile. It should be noted that polyunsaturated and monounsaturated fatty acids have been shown to prevent neuronal death from a number of insults (35-37). Saturated palmitic (C16:0) and stearic (C18:0) fatty acids, on the other hand, have been reported to result in neuronal death in vitro. However, neuronal death observed in these studies occurred at much higher C16:0 and C18:0 concentrations (0.2-0.3 mm) compared with those found in XX neurons during nutrient deprivation (38, 39). The unique, sex-dependent FFA profile before and after starvation certainly appears to merit further investigation. For example, various saturated FFAs have been shown to inhibit platelet phospholipase A2 (40). Thus, it is possible that sex-dependent changes in FFA during starvation may regulate cytosolic PLA2 activity and, therefore, formation of lipid droplets (23).
Using two-dimensional HPTLC and ESI-MS analyses, we were not able to detect any significant accumulation of lysophospholipids during nutrient deprivation (Fig. 6B) in either neurons from either sex. One possible explanation is that nutrient deprivation caused activation of lysophospholipases, resulting in further degradation of lysophospholipids and additional release of FFAs. This is in accordance with a previous report showing high lysophospholipase activity in neurons (41). It is possible that FFAs released from phospholipids via PLA2 and lysophospholipase pathways were further esterified into triacylglycerols, yielding their high content in nutrient-deprived neurons. Alternatively, the lack of detectable lysophospholipids could be attributed to activation of phospholipase C.
As with all in vitro paradigms, direct comparisons of these data with whole organisms warrant caution. It is unknown whether starvation can induce autophagy in the brain; however, there is evidence that critical starvation alone can result in brain atrophy in humans (13-15). It is possible that cellular stress coupled with incomplete nutrient deprivation may be additive stimuli for autophagy in brain. Recent studies show that autophagy is increased in the brain after trauma or ischemia in rodents (17, 18, 42) and trauma or critical illness in humans (19). Results of these studies bring to light the concept of “critical nutrient stress,” where subthreshold degrees of nutrient deprivation in brain tissue, coupled with metabolic disturbances due to injury, may result in brain autophagy. These data may have profound implications for both sexes during times of starvation and may provide clues toward the prevention of neurological morbidities associated with malnourishment, whether due to illness, nutrient deprivation, or famine. These data are also of relevance to diseases where autophagy plays an active role, either adaptive or pathological, and in both males and females.
Supplementary Material
Acknowledgments
We thank Yaming Chen and Sean Alber for expert technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants NS38620, NS30318, HD045968, NS061817, and AG026389. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Videos S1 and S2.
Footnotes
The abbreviations used are: DMEM, Dulbecco's modified Eagle's medium; FCS, fetal calf serum; MTT, 3-(4,5-dimethylthiazol)-2,5-diphenyltetrazolium bromide; PI, propidium iodide; PBS, phosphate-buffered saline; siRNA, small interfering RNA; GFP, green fluorescent protein; PLA2, phospholipase A2; AACOCF3, arachidonyl trifluoromethyl ketone; MAFP, methylarachidonyl fluorophosphonate; Py-2, pyrrolidine-2; HPTLC, high performance thin layer chromatography; ESI, electrospray ionization; MS, mass spectrometry; FFA, free fatty acid; TG, triglyceride; 3-MA, 3-methyladenine; ND, nutrient deprivation; ANOVA, analysis of variance.
References
- 1.Widdowson, E. M. (1976) Proc. Nutr. Soc. 35 175-180 [DOI] [PubMed] [Google Scholar]
- 2.Hoyenga, K. B., and Hoyenga, K. T. (1982) Physiol. Behav. 28 545-563 [DOI] [PubMed] [Google Scholar]
- 3.Hill, J. O., Fried, S. K., and DiGirolamo, M. (1984) Am. J. Physiol. 247 R318-R327 [DOI] [PubMed] [Google Scholar]
- 4.Bakewell, L., Burdge, G. C., and Calder, P. C. (2006) Br. J. Nutr. 96 93-99 [DOI] [PubMed] [Google Scholar]
- 5.Lamont, L. S., McCullough, A. J., and Kalhan, S. C. (2003) J. Appl. Physiol. 95 1259-1265 [DOI] [PubMed] [Google Scholar]
- 6.Niskanen, L. K., Haffner, S., Karhunen, L. J., Turpeinen, A. K., Miettinen, H., and Uusitupa, M. I. (1997) Eur. J. Endocrinol. 137 61-67 [DOI] [PubMed] [Google Scholar]
- 7.Wolf, I., Sadetzki, S., Kanety, H., Kundel, Y., Pariente, C., Epstein, N., Oberman, B., Catane, R., Kaufman, B., and Shimon, I. (2006) Cancer 106 966-973 [DOI] [PubMed] [Google Scholar]
- 8.Neely, A. N., and Mortimore, G. E. (1974) Biochem. Biophys. Res. Commun. 59 680-687 [DOI] [PubMed] [Google Scholar]
- 9.Seglen, P. O., and Gordon, P. B. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 1889-1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohsumi, Y. (2001) Nat. Rev. Mol. Cell. Biol. 2 211-216 [DOI] [PubMed] [Google Scholar]
- 11.Zeng, X., Overmeyer, J. H., and Maltese, W. A. (2006) J. Cell Sci. 119 259-270 [DOI] [PubMed] [Google Scholar]
- 12.Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., Mizushima, N., Tanida, I., Kominami, E., Ohsumi, M., Noda, T., and Ohsumi, Y. (2000) Nature 408 488-492 [DOI] [PubMed] [Google Scholar]
- 13.Basoglu, M., Yetimalar, Y., Gurgor, N., Buyukcatalbas, S., Kurt, T., Secil, Y., and Yeniocak, A. (2006) Eur. J. Neurol. 13 1089-1097 [DOI] [PubMed] [Google Scholar]
- 14.Connan, F., Murphy, F., Connor, S. E., Rich, P., Murphy, T., Bara-Carill, N., Landau, S., Krljes, S., Ng, V., Williams, S., Morris, R. G., Campbell, I. C., and Treasure, J. (2006) Psychiatry Res. 146 117-125 [DOI] [PubMed] [Google Scholar]
- 15.Katzman, D. K., Christensen, B., Young, A. R., and Zipursky, R. B. (2001) Semin. Clin. Neuropsychiatry 6 146-152 [DOI] [PubMed] [Google Scholar]
- 16.Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T., and Ohsumi, Y. (2004) Mol. Biol. Cell 15 1101-1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai, Y., Hickey, R. W., Chen, Y., Bayir, H., Sullivan, M. L., Chu, C. T., Kochanek, P. M., Dixon, C. E., Jenkins, L. W., Graham, S. H., Watkins, S. C., and Clark, R. S. (2008) J. Cereb. Blood Flow Metab., 28, 540-550 [DOI] [PubMed] [Google Scholar]
- 18.Koike, M., Shibata, M., Tadakoshi, M., Gotoh, K., Komatsu, M., Waguri, S., Kawahara, N., Kuida, K., Nagata, S., Kominami, E., Tanaka, K., and Uchiyama, Y. (2008) Am. J. Pathol. 172 454-469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark, R. S., Bayir, H., Chu, C. T., Alber, S. M., Kochanek, P. M., and Watkins, S. C. (2008) Autophagy 4 88-90 [DOI] [PubMed] [Google Scholar]
- 20.Hopkins, R. O., Ely, E. W., and Jackson, J. C. (2007) Curr. Opin. Crit. Care 13 497-502 [DOI] [PubMed] [Google Scholar]
- 21.Du, L., Bayir, H., Lai, Y., Zhang, X., Kochanek, P. M., Watkins, S. C., Graham, S. H., and Clark, R. S. B. (2004) J. Biol. Chem. 279 38563-38570 [DOI] [PubMed] [Google Scholar]
- 22.Brewer, G. J., Torricelli, J. R., Evege, E. K., and Price, P. J. (1993) J. Neurosci. Res. 35 567-576 [DOI] [PubMed] [Google Scholar]
- 23.Gubern, A., Casas, J., Barcelo-Torns, M., Barneda, D., de la Rosa, X., Masgrau, R., Picatoste, F., Balsinde, J., Balboa, M. A., and Claro, E. (2008) J. Biol. Chem. 283 27369-27382 [DOI] [PubMed] [Google Scholar]
- 24.Kouno, T., Mizuguchi, M., Tanida, I., Ueno, T., Kanematsu, T., Mori, Y., Shinoda, H., Hirata, M., Kominami, E., and Kawano, K. (2005) J. Biol. Chem. 280 24610-24617 [DOI] [PubMed] [Google Scholar]
- 25.Tanida, I., Minematsu-Ikeguchi, N., Ueno, T., and Kominami, E. (2005) Autophagy 1 84-91 [DOI] [PubMed] [Google Scholar]
- 26.Mizushima, N. (2004) Intl. J. Biochem. Cell Biol. 36 2491-2502 [DOI] [PubMed] [Google Scholar]
- 27.Caro, L. H., Plomp, P. J., Wolvetang, E. J., Kerkhof, C., and Meijer, A. J. (1988) Eur. J. Biochem. 175 325-329 [DOI] [PubMed] [Google Scholar]
- 28.Fujimoto, T., and Ohsaki, Y. (2006) Ann. N. Y. Acad. Sci. 1086 104-115 [DOI] [PubMed] [Google Scholar]
- 29.McAnoy, A. M., Wu, C. C., and Murphy, R. C. (2005) J. Am. Soc. Mass. Spectrom. 16 1498-1509 [DOI] [PubMed] [Google Scholar]
- 30.Yue, Z., Jin, S., Yang, C., Levine, A. J., and Heintz, N. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 15077-15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komatsu, M., Waguri, S., Chiba, T., Murata, S., Iwata, J., Tanida, I., Ueno, T., Koike, M., Uchiyama, Y., Kominami, E., and Tanaka, K. (2006) Nature 441 880-884 [DOI] [PubMed] [Google Scholar]
- 32.Zhu, J. H., Horbinski, C., Guo, F., Watkins, S., Uchiyama, Y., and Chu, C. T. (2007) Am. J. Pathol. 170 75-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luxon, B. A., and Weisiger, R. A. (1993) Am. J. Physiol. 265 G831-G841 [DOI] [PubMed] [Google Scholar]
- 34.Listenberger, L. L., Han, X., Lewis, S. E., Cases, S., Farese, R. V., Jr., Ory, D. S., and Schaffer, J. E. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3077-3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akbar, M., Calderon, F., Wen, Z., and Kim, H. Y. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10858-10863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, H. Y., Akbar, M., and Lau, A. (2003) Lipids 38 453-457 [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Rodriguez, R. A., Tabernero, A., Velasco, A., Lavado, E. M., and Medina, J. M. (2004) J. Neurochem. 88 1041-1051 [DOI] [PubMed] [Google Scholar]
- 38.Patil, S., and Chan, C. (2005) Neurosci. Lett. 384 288-293 [DOI] [PubMed] [Google Scholar]
- 39.Ulloth, J. E., Casiano, C. A., and De Leon, M. (2003) J. Neurochem. 84 655-668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ballou, L. R., and Cheung, W. Y. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 371-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker, R. R., and Chang, H. Y. (1999) Biochim. Biophys. Acta. 1438 253-263 [DOI] [PubMed] [Google Scholar]
- 42.Liu, C. L., Chen, S., Dietrich, D., and Hu, B. R. (2008) J. Cereb. Blood Flow Metab., 28, 674-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.