Abstract
The γ-secretase complex cleaves many transmembrane proteins, including amyloid precursor protein, EphB and ErbB tyrosine kinase receptors, Notch1 receptors, and adhesion factors. Presenilin 1, the catalytic subunit of γ-secretase, associates with the cadherin/catenin cell-cell adhesion/communication system and promotes cadherin processing (Georgakopoulos, A., et al. (1999) Mol. Cell 4, 893–902; Marambaud, P., et al. (2002) EMBO J. 21, 1948–1956), but the mechanism by which γ-secretase and cadherins associate is unclear. Here we report that p120 catenin (p120ctn), a component of the cadherin-catenin complex, recruits γ-secretase to cadherins, thus stimulating their processing while inhibiting production of Aβ peptide and the amyloid precursor protein intracellular domain. This function of p120ctn depends on both p120ctn-cadherin and p120ctn-presenilin 1 binding, indicating that p120ctn is the central factor that bridges γ-secretase and cadherin-catenin complexes. Our data show that p120ctn is a unique positive regulator of the γ-secretase processing of cadherins and a negative regulator of the amyloid precursor protein processing. Furthermore, our data suggest that specific members of the γ-secretase complex may be used to recruit different substrates and that distinct PS1 sequences are required for processing of APP and cadherins.
Presenilin 1 (PS1)6 is a multipass transmembrane protein involved in most cases of early-onset familial Alzheimer disease. Cellular PS1 is cleaved to yield N-terminal fragments (NTFs) and carboxyl-terminal fragments (CTFs) that associate to form functional heterodimers (3). PS1 is an important catalytic component of the γ-secretase proteolytic complex that includes APH-1, PEN-2, and nicastrin (4), the latter acting as a substrate recruiter (5). This complex mediates the amyloidogenic processing of amyloid precursor protein (APP) and production of Aβ peptides (6), the structural components of the amyloid depositions of Alzheimer disease. In addition, the PS1/γ-secretase complex promotes the ε cleavage of cell surface type I transmembrane proteins like APP, receptors, and cell-cell adhesion factors including cadherins. The ε cleavages occur close to the membrane/cytoplasm interface and promote the production of soluble peptides containing the cytoplasmic C-terminal sequence of the cleaved proteins (CTFs). Many of these peptides have been shown to function as transcription factors and cell surface-to-nucleus communication signals (for review, see Ref. 7). In agreement with this function of the γ-secretase proteolytic complex, PS1 is found at the plasma membrane in cell-cell contact sites where it associates with the cadherin-catenin adherens junctions (1). Formation of the PS1-cadherin complexes depends on the juxtamembrane region of cadherins at a site that is also important for the cadherin binding of p120 catenin (p120ctn), an integral component of the cadherin-catenin adherens junctions (8).
Recently, we reported that PS1 promotes the γ-secretase processing and signaling of the cadherin family of proteins, including E- and N-cadherins (2, 9). In addition, using the two-hybrid system analysis, several laboratories reported that PS1 binds directly to members of the p120ctn family of proteins, including δ-catenin and p0071 (10–12). Together, the above reports raise the possibility that these catenin proteins may regulate the γ-secretase processing of cadherins. The p120ctn-catenin family contains 10–12 armadillo repeats (13), and in contrast to β- and γ-catenins that bind the distal sequence of cytosolic cadherins, members of the p120ctn family bind directly to the juxtamembrane sequence of cadherins and regulate their stability (14, 15). p120ctn itself regulates the activity of the Rho family of GTPases (16) and promotes gene expression by binding transcription factor Kaiso and regulating its DNA-binding activity (17, 18). Interestingly, conditional knock-out of mouse hippocampal p120ctn results in abnormalities in synaptic transmission and spine morphology (19, 20). Because cadherins and PS1 are also known to play important roles in neuronal development and function (21, 22), we took advantage of PS1 and cadherin mutants defective in p120ctn binding and employed short hairpin RNA (shRNA) technology to examine the role of p120ctn in the γ-secretase processing of cadherins. Our results indicate that p120ctn plays a central role in the γ-secretase association and processing of cadherins. In contrast, p120ctn suppresses APP processing and Aβ production, possibly by recruiting γ-secretase to cadherins and thus limiting its availability for the processing of APP.
MATERIALS AND METHODS
Antibodies and Reagents—Mouse monoclonal antibody 33B10 against residues 331–350 of the loop domain of PS1 and polyclonal antibody against the PS1 N-terminal fragment (antibody 222) have already been described (1). Polyclonal antibodies against the PS1 C-terminal fragment (S182) and N- and E-cadherin (H63 and H108) were purchased from Sigma and Santa Cruz Biotechnology, respectively. Anti-N-cadherin monoclonal antibody and p120ctn were from BD Biosciences. 33B10 and anti-N-cadherin monoclonal antibody were used only for Western blot analysis. γ-Secretase inhibitor L665,458 was obtained from Calbiochem. Retroviral expression vector expressing pMX/PS1NTF or pMX/PS1CTF was generated by insertion of the corresponding cDNA into HindIII and EcoRI or into EcoRI, respectively. Other PS1 retroviral expression vectors were generated with pMX-IRES-GFP (23) and have been described previously (24). Human N-cadherin was subcloned into pcDNA3.1 with EcoRI and NotI sites. Human E-cadherin was subcloned into the pMX vector with XhoI and NotI sites for viral transduction. The shRNA pSIREN-Retro vector against mouse or human p120ctn was constructed by inserting the sequences reported previously (14). The following oligonucleotides were used for the construction of a retroviral vector for human or mouse p120ctn shRNA: 5′-GATCTCCgccagaagtggtgcgaataTTCAAGAGAtattcgcaccacttctggcTTTTTG-3′, 5′-AATTCAAAAAgccagaagtggtgcgaataTCTCTTGAAtattcgcaccacttctggcGGA-3′, 5′-GATCTCCgccagaggtggttcggataTTCAAGAGAtatccgaaccacctctggcTTTTTG-3′, and 5′-AATTCAAAAAgccagaggtggttcggataTCTCTTGAAtatccgaaccacctctggcGGA-3′. All constructs were verified by sequencing. pcDNA3 vectors encoding VE-cadherin-GFP and Y658E mutant VE-cadherin-GFP were kindly given by Dr. D. Cheresh (25).
Cell Culture, Transfections, and Infections—Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in 5% CO2 at 37 °C. For PS1 expression in PS1-null fibroblasts or E-cadherin expression in SW48 cells, Phoenix α packaging cells were transfected with pMX retroviral vectors (23) and a pVSV-G plasmid (Clontech), and a retroviral gene was transferred into PS1–/– mouse fibroblasts or SW48 cells as described previously (24). The transfection of VE-cadherin-GFP into Chinese hamster ovary cells was performed using Lipofectamine (Invitrogen) according to the manufacturer's protocol. GFP-positive fibroblasts or Chinese hamster ovary cells were sorted by using a MoFlo cell sorter after transduction. For p120ctn shRNA experiments, fibroblasts were infected with control retrovirus or retrovirus expressing p120ctn-specific shRNA, and the cell lysates were subjected to SDS-PAGE and Western blotting after transduction. Clonal p120ctn-depleted HEK293 cell lines were generated by transfection with control pSIREN vector or the vector expressing p120ctn-specific shRNA by Lipofectamine and limited dilution. Human N-cadherin was transfected into p120ctn KD cells by the Lipofectamine Plus™ reagent according to the manufacturer's protocol.
Western Blots (WBs) and Immunoprecipitations—For Western blot analysis, cells were washed with phosphate-buffered saline and solubilized in radioimmune precipitation assay buffer (100 mm HEPES (pH 7.4), 0.1% SDS, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 1 mm EDTA), or HEPES buffer (pH 7.4), 150 mm NaCl, and 1 × complete protease inhibitor mixture containing 0.5% Nonidet P-40 or 1% Triton X-100. For immunoprecipitation, cells were solubilized in HEPES buffer containing 0.5% Nonidet P-40 or 1% digitonin or 10% glycerol and 0.2% dodecyl maltoside. The cell lysates were centrifuged at 16,000 × g for 40 min, and supernatants were pre-cleared with Protein A or Protein G for 2 h at 4 °C. Supernatants were incubated for 2 h or overnight with antibodies and treated for 1–2 h with Protein A or Protein G. Immunoprecipitates were washed with HEPES buffer containing detergent at least four times and analyzed by SDS-PAGE.
In Vitro γ-Secretase Assay—This assay was performed as described previously (9). Briefly, HEK293 cells expressing exogenous N-cadherin were resuspended in hypotonic buffer (10 mm MOPS (pH 7.0), 10 mm KCl, and 1 mm EDTA) and homogenized on ice. A supernatant was prepared by centrifugation at 1,000 × g for 15 min at 4 °C, and crude membranes were isolated from the supernatant by centrifugation at 16,000 × g for 40 min at 4 °C. The membranes were resuspended in 25 μl of assay buffer (150 mm sodium citrate (pH 6.4), completed with protease inhibitor mixture) and incubated for 2 or 12 h at 37 °C. Samples were analyzed on WBs and probed for N-cadherin. For APP intracellular domain (AICD) detection, in vitro assay was conducted in the same way except using SW48 cells.
Sandwich Aβ Enzyme-linked Immunosorbent Assays—Collected conditioned media were analyzed using a BIOSOURCE Aβ40 kit (BIOSOURCE, Camarillo, CA) as described previously (26) or a Wako human Aβ42 kit according to the company's manual. SW48 cells were grown in the growth medium supplemented with 100 μg/ml insulin and 10 μmol/liter phosphoramidon for 24 h (27), and the collected conditioned medium was mixed with Pefablock (1 mm) to minimize degradation of Aβ. Both the collected medium and different amounts of the standards of Aβ40 and Aβ42 were loaded together with the detection antibody supplied by the kits on the coated plates and incubated for 3 h at 23 °C. Data for AICD and Aβ40/42 were statistically analyzed by paired Student's t test.
RESULTS
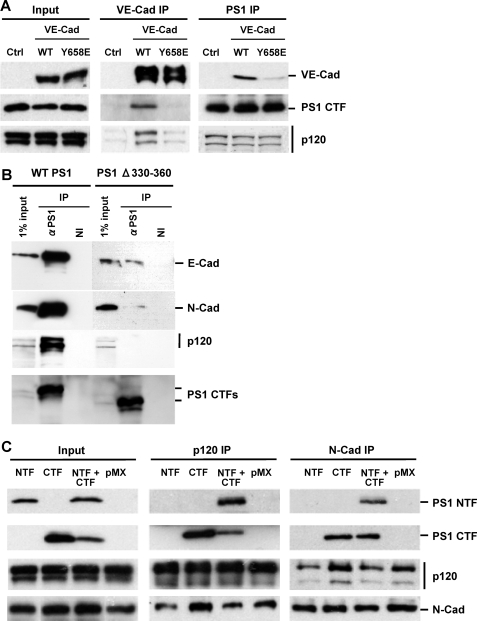
Mutations That Inhibit the p120ctn-Cadherin Binding Also Disrupt the PS1 Association with Cadherins—It has been reported that deletion of E-cadherin amino acid sequence 760–771 (corresponding to residues 604–615 of the mature protein) located at the juxtamembrane region of the protein disrupts its association with both p120ctn and PS1 (8, 28). Furthermore, E-cadherin mutant GGG759–761AAA, which is unable to bind p120ctn, also fails to bind PS1 (2). To further examine the role of p120ctn in the cadherin association of PS1, we used the VE-cadherin mutant Y658E, which is unable to bind p120ctn (25). Fig. 1A shows that this mutant is also unable to bind PS1. Together, these data suggest that sequences critical to p120ctn-cadherin binding (for review, see Ref. 13) are also critical for the association of cadherins with PS1 (1, 2, 8, 9). Additional reports suggested direct binding of PS1 to the p120ctn family of proteins that includes δ-catenin and p0071 (10–12), and we observed that PS1 deletion mutant 330–360, which is unable to associate with E-cadherin (24), also failed to bind p120ctn (Fig. 1B). This last result is in agreement with reports that PS1 sequence 329–362, located in the C-terminal fragment of PS1 (PS1/CTF), promotes PS1 binding to p0071, a member of the p120 catenin family of proteins (10). The observation that sequences critical to p120ctn-cadherin or p120ctn-PS1 binding are also important for the cadherin-PS1 association, combined with evidence that PS1 binds directly to p120ctn, provides support to the theory that p120ctn mediates the PS1-cadherin association instead of competing with PS1 for cadherin (8). This theory is also consistent with our data that PS1 mutant 330–360 binds neither p120ctn nor E-cadherin (Fig. 1B). To verify that PS1/CTF mediates the cadherin-PS1 and p120ctn-PS1 associations, we transduced PS1 knockout (PS1–/–) fibroblasts with the PS1 fragments. Fig. 1C shows that PS1/CTF, when expressed alone, is able to form complexes with both cadherin and p120ctn. In contrast, PS1/NTF co-immunoprecipitates with p120ctn and N-cadherin only in the presence of PS1/CTF (Fig. 1C). Together, these data show that PS1/CTF mediates the PS1 association with both cadherin and p120ctn and that PS1 sequence 330–360 is critical to this association.
FIGURE 1.
Cadherin or PS1 mutants unable to bind p120ctn fail to form PS1-cadherin complexes. A, Chinese hamster ovary cell cultures were transfected with enhanced GFP, WT VE-cadherin-enhanced GFP, or VE-cadherin mutant Y658E-enhanced GFP, which is unable to bind p120ctn. Stable GFP-positive cell populations expressing similar levels of VE-cadherin were sorted and selected. Extracts from all cultures were immunoprecipitated with either anti-VE-cadherin (VE-cad IP)(middle panels) or anti-PS1 (PS1 IP)(right panels) antibodies, and obtained IPs were probed on WBs with antibodies against antigens indicated at the right of the figure as described (1). Input is shown in the left panels. WT VE-cadherin forms complexes with both p120ctn and PS1. In contrast, mutant VE-cadherin binds neither p120ctn nor PS1. As expected, PS1 binds p120ctn in all cultures (lower right panel). B, HEK293 cells transduced with WT PS1 and PS1Δ330–360 were grown to confluency, extracted in Nonidet P-40, and immunoprecipitated with anti-PS1 antibody (Sigma S182 polyclonal antibody) or nonimmune serum (NI). Obtained immunoprecipitated samples were probed on WBs with antibodies against E-cadherin, N-cadherin, p120ctn, and PS1/CTF indicated at the right of the figure. Deletion mutant PS1Δ330–360, which is unable to bind cadherin (24), is also unable to bind p120ctn. C, PS1–/– mouse fibroblasts were transduced with PS1/CTF, NTF, or both. Extract from each culture was prepared in Nonidet P-40 and then immunoprecipitated with either anti-p120ctn (p120ctn IP)(middle panels) or anti-N-cadherin (N-cad IP)(right panels) antibodies. Obtained IPs were probed on WBs with antibodies against antigens indicated at the right of the figure. PS1/CTF binds both p120ctn and N-cadherin, whereas PS1/NTF needs CTF to associate with p120ctn or N-cadherin.
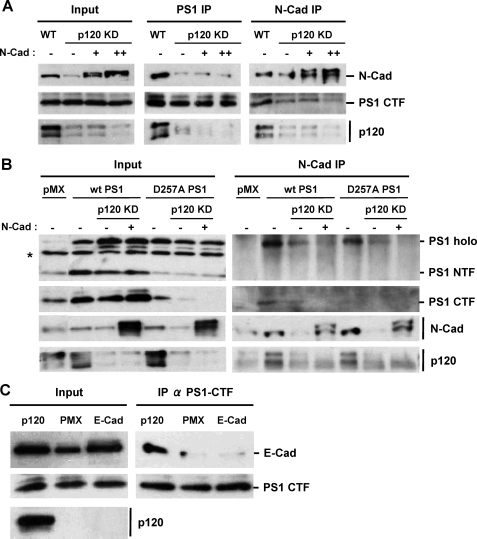
p120ctn Mediates the Association of PS1 and Cadherins—The role of p120ctn in the PS1-cadherin interaction was further examined in p120ctn knockdown (KD) HEK293 cells, which express N-cadherin. Because down-regulation of p120ctn causes a decrease in the cellular levels of cadherins (14, 19, 29), the KD cultures were transfected with N-cadherin so that its levels remained comparable with the levels of the control cultures (Fig. 2A, left panel). Knockdown of p120ctn had no effect on the cellular levels of PS1 fragments (Fig. 2A), and the amounts of the PS1-N-cadherin complexes in wild type (WT) and in p120ctn KD HEK293 cells in the presence or absence of exogenous N-cadherin were analyzed by co-immunoprecipitation. Fig. 2A (middle and right panels) shows that knockdown of p120ctn results in a decrease in the PS1-cadherin complexes regardless of the cellular levels of cadherin, indicating that p120ctn is necessary for PS1 binding to N-cadherin. To examine whether p120ctn is also required for the interaction of full-length (uncleaved) PS1 and N-cadherin, PS1 mutant D257A, which is not processed into fragments (6), was transduced into p120ctn KD HEK293 cells, and uncleaved PS1-cadherin complexes were determined. Fig. 2B shows that knockdown of p120ctn impaired the binding of PS1 mutant D257A to N-cadherin. Similar effects of p120ctn on the complexes between cadherin and full-length WT PS1 were observed when p120ctn KD HEK293 cells were transduced with PS1, a condition that results in excess uncleaved PS1 (Fig. 2B). Together, these data indicate that p120ctn is necessary for the cadherin association of both uncleaved and cleaved PS1. In a different approach, we reintroduced p120ctn into the SW48 colon carcinoma cell line, which expresses no significant amounts of endogenous p120ctn (Fig. 2C) (29). To compensate for reduced levels of cadherin, we transduced these cells with exogenous E-cadherin, and the E-cadherin-PS1 complexes in these cell systems were analyzed by co-immunoprecipitation experiments. Fig. 2C shows that although exogenous p120ctn promoted the PS1-E-cadherin association, exogenous E-cadherin was unable to increase the PS1-E-cadherin association in the absence of p120ctn (Fig. 2C). Together, our data show that p120ctn is a prerequisite for the physical association between PS1 and E-cadherin.
FIGURE 2.
p120ctn is necessary for PS1-cadherin interaction. A, HEK293 cell cultures depleted of p120ctn via shRNA (see “Materials and Methods”) were transiently transfected with zero (–), 0.1 (+), or 0.3 (++) μg of N-cadherin cDNA. A WT culture was used as a control. Extract from each culture was prepared in Nonidet P-40 and then immunoprecipitated with either anti-PS1/CTF (PS1 IP) or anti-N-cadherin (N-cad IP) antibodies. Obtained IPs were probed on WBs with antibodies against antigens indicated at the right of the figure. Note that PS1/CTF levels were not affected by the knockdown of p120ctn or the expression of exogenous N-cadherin. Reintroduction of N-cadherin did not restore the amounts of N-cadherin-PS1 complex in p120ctn knockdown cells. B, control or p120ctn-depleted (p120ctn KD) HEK293 cell cultures were transduced with pMX, WTPS1, or PS1/D257A mutant, and N-cadherin was reintroduced into p120ctn KD cultures as described above. Extracts prepared as described above were immunoprecipitated with anti-N-cadherin antibodies (N-cad IP), and obtained IPs were probed on WBs with antibodies against antigens indicated at the right of the figure. Input is shown in the left panels. Note that knockdown of p120ctn decreases the interaction between uncleaved PS1 and N-cadherin. The double signal of the N-cadherin in the N-cadherin-transfected cultures is due to the presence of unprocessed (immature) protein. Unidentified (artifactual) bands are indicated by an asterisk. C, p120ctn-deficient colon carcinoma SW48 cells were transduced with pMX vector (PMX), E-cadherin (E-cad), or p120ctn. Extracts from the obtained cultures were immunoprecipitated as described above with antibodies against PS1/CTF (IP α PS1/CTF), and IPs were probed on WBs with antibodies against antigens indicated at the right of the figure. Input is shown in the left panels. Results show that p120ctn is needed for E-cadherin/PS1 interaction.
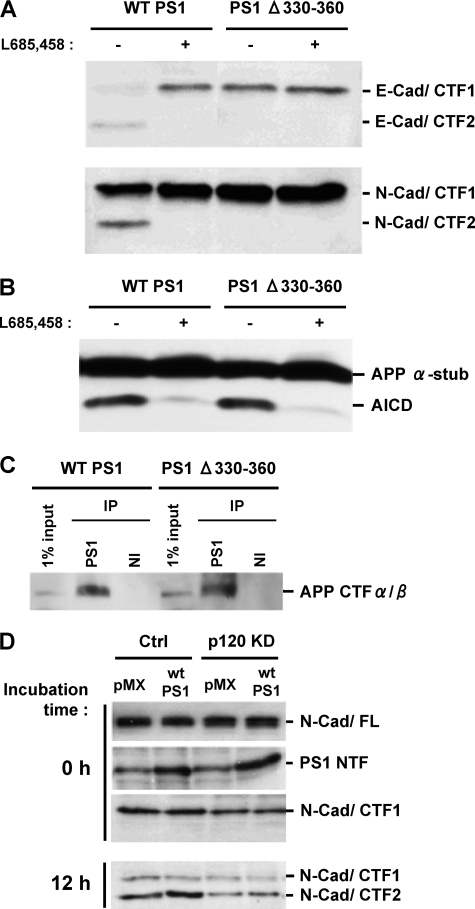
PS1 Deletion Mutant Unable to Bind p120ctn Fails to Process Cadherin Although It Promotes Processing of APP—To investigate the function of p120ctn in the PS1-mediated cadherin cleavage, we took advantage of deletion mutant PS1Δ330–360, which is unable to bind p120ctn (see above and Fig. 1B). Fig. 3A shows that in contrast to WT PS1, this mutant is able to process neither E-cadherin (upper panel) nor N-cadherin (lower panel). Exogenous PS1Δ330–360 replaces endogenous WT PS1 (supplemental figure) (3), and no γ-secretase cleavage products of cadherin are detected in the mutant transfected cells (Fig. 3A). In contrast, this mutant promotes the γ-secretase processing of APP (30). In agreement with this report, we obtained data that deletion mutant PS1Δ330–360 promotes APP processing and production of AICD (Fig. 3B). In addition, Fig. 3C shows that this mutant binds the γ-secretase substrates APP/CTFαβ as efficiently as WT PS1. To further explore the requirement of p120ctn for the γ-secretase processing of cadherins, we examined this processing in p120ctn KD HEK293 cells. Again, the cellular amounts of N-cadherin in the p120ctn KD cultures were adjusted as described above to approximately the same levels as in the control cells (Fig. 3D, upper panel). Fig. 3D shows that knockdown of p120ctn results in decreased production of N-cad/CTF2, the product of the γ-secretase processing of N-cadherin (9). Inhibition of cadherin γ-secretase processing may result in increased levels of N-cad/CTF1 (2). The lack of increased N-cad/CTF1 in Fig. 3D may be due to alternative proteolytic pathways in this cell line as is the case of APP/CTFαβ degradation through pathways other than γ-secretase (31, 32). Together, our results show that binding of PS1 to p120ctn is necessary for the γ-secretase processing of the cadherin substrates. Furthermore, it seems that PS1 sequence 330–360 specifically promotes cadherin processing, but is not necessary for APP processing. Thus, distinct PS1 sequences are important for the γ-secretase processing of APP and cadherins (33, 34).
FIGURE 3.
PS1 mutant unable to bind p120ctn fails to promote cadherin processing although it promotes APP processing. A, HEK293 cells transduced with WT PS1 or PS1Δ330–360 were grown to confluency, and membrane fractions were prepared and used to perform the in vitro γ-secretase assay as described (2, 9) at 37 °C for 2 h in the presence (+) or absence (–) of 1 μm γ-secretase inhibitor L665,458. The resulting samples were submitted to SDS-PAGE and probed on WBs with either anti-E- or anti-N-cadherin antibodies to detect the cadherin products indicated at the right of the figure. PS1Δ330–360 defective in p120ctn binding is unable to promote the production of the γ-secretase cleavage products E-cad/CTF2 and N-cad/CTF2. B, in vitro assay for the detection of AICD was performed as described for A using anti-APP/CTF antibody R1 APP α-stub, (APP CTFα/β). C, HEK293 cells transduced with WT PS1 or PS1Δ330–360 were grown to confluency, extracted in Nonidet P-40, and then immunoprecipitated with an anti-PS1 antibody (S182) or nonimmune serum (NI). Obtained samples were submitted to SDS-PAGE, and the immunoblots were probed with anti-APP antibody R1. PS1Δ330–360 binds APP/CTF as efficiently as WT PS1. D, knockdown of p120ctn decreases the γ-secretase processing of cadherins. Control and p120ctn knockdown HEK293 cell cultures were transduced with either vector (pMX) or PS1 (WT PS1) expression constructs. N-cadherin levels in all cultures were adjusted as described in the legend Fig. 2. Membrane fractions from each culture were used for the in vitro γ-secretase assay at 37 °C for 12 h as described for A. The resulting samples were submitted to SDS-PAGE and immunoblotted with anti-N-cadherin or anti-PS1/NTF antibodies to detect the products indicated at the right of the figure. Knockdown of p120ctn reduced production of the γ-secretase product N-cad/CTF2 even in the presence of increased PS1 levels.
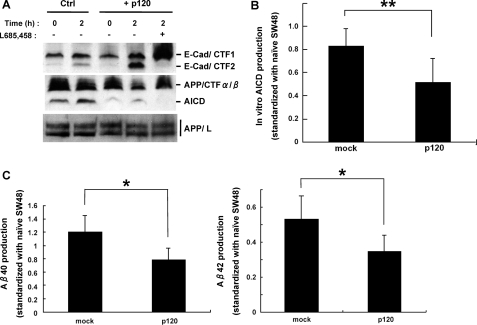
p120ctn Enhances PS1-mediated E-cadherin Processing but Reduces Secreted Aβ Levels—That p120ctn binds PS1 and promotes the γ-secretase processing of cadherins prompted us to ask whether p120ctn may also affect the processing of APP and the production of Aβ. To test this hypothesis, we used the p120ctn-deficient SW48 cell line (29) and the in vitro assay of γ-secretase activity (9). Fig. 4A (upper panel) shows that transduction of p120ctn into these cells stimulated the in vitro production of the E-cadherin γ-secretase product E-cad/CTF2. Furthermore, inhibition of γ-secretase activity resulted in the disappearance of E-cad/CTF2 and the increase of E-cad/CTF1, the precursor of E-cad/CTF2 (2). Interestingly, the cells transduced with p120ctn generated less AICD compared with mock-transduced cells, although the total levels of APP remained unchanged (Fig. 4A, lower panels). The levels of AICD were further quantified and analyzed for statistical significance (Fig. 4B). Secreted Aβx-40 and Aβx-42 levels in conditioned medium from p120ctn-transduced cell cultures were significantly reduced compared to those from mock-transduced cells (Fig. 4C). These results show that p120ctn enhances the γ-secretase processing of cadherins while decreasing the γ-secretase processing of APP indicated by the reduced production of both AICD and Aβ peptides, the products of the ε and γ cleavages of APP, respectively.
FIGURE 4.
p120ctn enhances E-cadherin but attenuates APP processing catalyzed by γ-secretase. A, membranes prepared from SW48 cell cultures transduced with either vector or p120ctn were prepared and used in in vitro γ-secretase assay as described above in the presence or absence of γ-secretase inhibitor L665,458. The upper panel was probed with anti-E-cadherin antibodies and the two lower panels with anti-APP R1 antibodies. The detected products are indicated at the right of the figure. APP/L, full-length APP. Expression of exogenous p120ctn increases the γ-secretase products of E-cadherin but decreases AICD, a γ-secretase product of APP. B, shown are the results from in vitro AICD assay in SW48 cells. AICD was detected in WBs with C-terminal anti-APP antibodies, and signals were quantified by densitometry using ImageJ software. In vitro production of AICD was estimated as the difference between the samples at 2 and 0 h. The effect of p120 on the in vitro production of AICD is statistically significant. Amounts of AICD are indicated in arbitrary units based on the subtracted value of the signal intensity at 0 and 2 h in both mock and p120ctn-transduced cells. The data represent the mean ± S.D. of five experiments. **, p < 0.005. C, secreted Aβ40 and Aβ42 from mock- and p120ctn-expressing SW48 cell cultures were analyzed by an Aβ sandwich enzyme-linked immunosorbent assay. The data represent the mean ± S.E. of five experiments. *, p < 0.05.
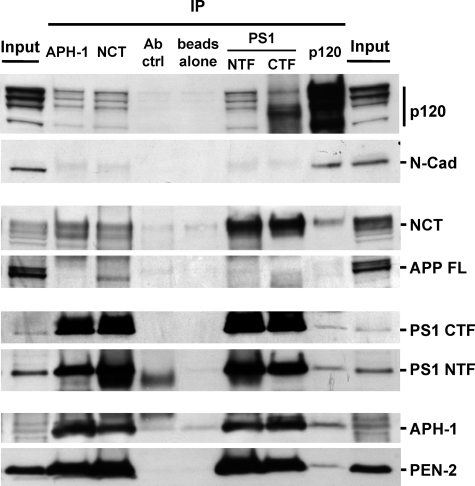
p120ctn Associates with γ-Secretase and Links It to Cadherins—That p120ctn binds both PS1 and cadherins and promotes the γ-secretase processing of cadherins suggests that p120ctn may associate with the γ-secretase complex and thus may facilitate its interaction with the cadherin substrates. To investigate whether p120ctn forms complexes with the γ-secretase holoenzyme, we immunoprecipitated the components of the γ-secretase complex and probed the samples for the presence of p120ctn. Fig. 5 shows that p120ctn co-immunoprecipitates not only with PS1 but also with nicastrin (NCT) and APH-1, both components of the γ-secretase holoenzyme. Inversely, all known components of γ-secretase, including PS1, NCT, APH-1, and PEN-2 (4), co-immunoprecipitate with p120ctn. Together, these data indicate that p120ctn interacts with the γ-secretase complex. It has been reported that NCT interacts directly with APP, and this interaction serves to bring APP to the γ-secretase complex for processing (Fig. 5) (34). In contrast, cadherins bind to p120ctn but not to nicastrin (Fig. 5), and we failed to detect any p120ctn complexes with APP (data not shown). These observations suggest that cadherin substrates bind to the γ-secretase complex via p120ctn, whereas APP substrate is brought to the complex via nicastrin. Thus, APP and cadherins form distinct γ-secretase complexes mediated by distinct factors. By promoting cadherin recruitment, p120ctn may reduce the availability of the holoenzyme to other substrates including APP and thus decrease its processing.
FIGURE 5.
p120ctn associates with γ-secretase and links it to cadherins. Extracts prepared in 0.2% dodecyl maltoside from HEK293 cell cultures were immunoprecipitated with antibodies against APH-1, NCT, PS1-CTF, PS1/NTF, and p120ctn. Obtained IPs were separated by SDS-PAGE and probed on WBs for antigens indicated at the right of the figure. Note that each γ-secretase component co-immunoprecipitates with p120ctn. Conversely, this catenin co-immunoprecipitates with each γ-secretase component probed. Mature full-length APP (APP FL)interacts specifically with nicastrin, whereas N-cadherin interacts with p120ctn. Nonimmune serum (Ab ctrl) and beads alone were used as controls.
DISCUSSION
Classic cadherins, including E-, N-, and VE-cadherins, constitute a large family of cell-cell adhesion/communication factors necessary in both the development and function of mature organisms (35). Cadherins function as central components of the adherens junctions, which include catenins, proteins that bind the intracellular (cytosolic) sequence of cadherins. p120ctn, an armadillo repeat protein member of the catenin family, binds the juxtamembrane cytoplasmic cadherin sequence and regulates cadherin stability. In addition, p120ctn plays important roles in development and gene expression (13, 17). Recent work from our laboratory showed that PS1 localizes at the plasma membrane where it forms complexes with the cadherin-catenin adherens junctions (1) and promotes the γ-secretase processing of cadherins, releasing α- and β-catenins to the cytosol and producing peptides that function in signal transduction (2, 9, 18). Additional in vitro studies indicated that members of the p120ctn family of proteins, including δ-catenin and p0071, bind directly to the large loop of PS1/CTF independently of cadherins (10–12), but the physiological significance of the p120ctn-PS1 binding remained elusive. In this study, we have presented evidence that p120ctn plays a central role in the γ-secretase processing of cadherins. Our data show that p120ctn is necessary for the association of PS1 and cadherin and that mutations that interrupt either the p120ctn-cadherin or p120ctn-PS1 binding also abolish the PS1-cadherin association. Furthermore, PS1 deletion mutant 330–360, which is unable to associate with cadherin (24), also fails to bind p120ctn. The deleted sequence of this mutant is part of the large cytoplasmic loop of PS1/CTF. In agreement with the critical role of sequence 330–360 in p120ctn-PS1 binding, our data show that the PS1/CTF fragment mediates the binding of PS1 to both p120ctn and cadherin. The inability of this PS1 mutant to bind p120ctn is also in agreement with reports that PS1 sequence 329–362 is critical for the binding of PS1 to p0071ctn, another member of the p120ctn family (10). It is not surprising that full-length PS1 binds p120 because amino acid sequence 330–360 in PS1/CTF that binds p120 is also present in full-length PS1. At this point, it is unclear what the function of the p120-full-length PS1 complex is, but it is an interesting question whether full-length PS1 in a γ-secretase complex is able to promote cadherin processing.
To further explore the role of p120ctn in the PS1 association with cadherins, we down-regulated this protein using shRNA technology and employed SW48 cells, which express no significant amounts of endogenous p120ctn (29). These experimental approaches indicated that decreased cellular p120ctn results in decreased levels of PS1-cadherin complexes and that reintroduction of p120ctn in SW48 cells stimulates the PS1-E-cadherin association. In contrast, overexpression of E-cadherin is unable to increase the PS1-E-cadherin association in the absence of p120ctn. Combined with reports that p120ctn binds directly to cytoplasmic cadherin (36), these observations indicate that p120ctn mediates the PS1-cadherin association by binding both proteins and that p120ctn is a prerequisite for the physical association of PS1 and cadherin. However, this conclusion seems at variance with our previous report that overexpression of p120ctn may down-regulate the cadherin-PS1 complexes (8). At this point, we have no clear explanation for the contradiction. It is possible that the previous report resulted from technical problems stemming from multiple co-transfections of the cell line used in those experiments.
We next addressed the physiological significance of the p120ctn-PS1-cadherin complex, focusing on the role of p120ctn in the γ-secretase processing of cadherins and APP. Down-regulation of p120ctn resulted in decreased γ-secretase processing of cadherins, indicated by the reduced production of the cadherin fragment cad/CTF2 (2, 9), whereas introduction of p120ctn in the p120ctn-deficient SW48 cells stimulated production of cad/CTF2. These data show that p120ctn stimulates the γ-secretase processing of cadherins. Interestingly, exogenous p120ctn caused a significant inhibition of the γ-secretase processing of APP in SW48 cells indicated by an inhibition of both Aβ peptides and AICD. These results are consistent with the hypothesis that by promoting the cadherin association with γ-secretase, p120ctn redirects limited amounts of cellular γ-secretase activity from APP processing to cadherin processing. Alternatively, there is a possibility that the decrease in APP products in p120-transduced cells may be due to factors other than the p120-γ-secretase association like an increased degradation of these products, although presently there is no evidence for such a mechanism.
Our data indicate that PS1 sequence 330–360 is necessary for PS1 binding to p120ctn and that this binding links the γ-secretase complex to cadherins, promoting their processing. In contrast, recent literature shows that NCT, another γ-secretase component, links APP and Notch to γ-secretase (34). Thus, APP and cadherins form distinct γ-secretase complexes mediated by distinct factors, suggesting that specific γ-secretase component proteins may be used to link specific substrates toγ-secretase for processing. That deletion mutant PS1Δ330–360 promotes the γ-secretase processing of APP and Notch but not cadherins is consistent with the conclusion that PS1 amino acid sequence 330–360 functions as a specific linker between cadherins and γ-secretase and shows that specific PS1 sequence may be used for the γ-secretase processing of distinct substrates. It is now clear that the γ-secretase complex binds to and processes a large number of substrates (7). That APP and cadherins bind this complex through distinct mechanisms suggests the existence of diverse pathways linking different substrates to γ-secretase.
The absence of p120ctn has been associated with abnormalities in spine development and synaptic transmission (19, 20). Our data raise the possibility that at least some of these abnormalities may be due to the role of p120ctn in the function of γ-secretase. Furthermore, because cadherins, PS1, and p120ctn family members have all been localized at the synapse, some of the effects of PS1 familial Alzheimer disease mutants on synaptic function may be related to abnormalities in the function of the trimeric PS1-p120ctn-cadherin complex.
Supplementary Material
Acknowledgments
We thank Dr. T. Kitamura for providing retroviral vector pMX-IRES-GFP. We also thank Dr. D. Cheresh for GFP fusion VE-cadherin constructs and Dr. A. Reynolds for SW48 cells transduced with p120ctn.
This work was supported, in whole or part, by National Institutes of Health Grants AG-17926, NS047229, and AG-08200. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure.
Footnotes
The abbreviations used are: PS1, presenilin 1; APP, amyloid precursor protein; AICD, APP intracellular domain; GFP, green fluorescent protein; p120ctn, p120 catenin; CTF, carboxyl-terminal fragment; NCT, nicastrin; NTF, N-terminal fragment; WT, wild type; IP, immunoprecipitate; shRNA, short hairpin RNA; WBs, Western blots; MOPS, 3-(N-morpholino)propanesulfonic acid; KD, knockdown.
References
- 1.Georgakopoulos, A., Marambaud, P., Efthimiopoulos, S., Shioi, J., Cui, W., Li, H. C., Schutte, M., Gordon, R., Holstein, G. R., Martinelli, G., Mehta, P., Friedrich, V. L., Jr., and Robakis, N. K. (1999) Mol. Cell 4 893–902 [DOI] [PubMed] [Google Scholar]
- 2.Marambaud, P., Shioi, J., Serban, G., Georgakopoulos, A., Sarner, S., Nagy, V., Baki, L., Wen, P., Efthimiopoulos, S., Shao, Z., Wisniewski, T., and Robakis, N. K. (2002) EMBO J. 21 1948–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thinakaran, G., Borchelt, D. R., Lee, M. K., Slunt, H. H., Spitzer, L., Kim, G., Ratovitsky, T., Davenport, F., Nordstedt, C., Seeger, M., Hardy, J., Levey, A. I., Gandy, S. E., Jenkins, N. A., Copeland, N. G., Price, D. L., and Sisodia, S. S. (1996) Neuron 17 181–190 [DOI] [PubMed] [Google Scholar]
- 4.Takasugi, N., Tomita, T., Hayashi, I., Tsuruoka, M., Niimura, M., Takahashi, Y., Thinakaran, G., and Iwatsubo, T. (2003) Nature 422 438–441 [DOI] [PubMed] [Google Scholar]
- 5.Shah, S., Lee, S. F., Tabuchi, K., Hao, Y. H., Yu, C., LaPlant, Q., Ball, H., Dann, C. E., III, Sudhof, T., and Yu, G. (2005) Cell 122 435–447 [DOI] [PubMed] [Google Scholar]
- 6.Wolfe, M. S., Xia, W., Ostaszewski, B. L., Diehl, T. S., Kimberly, W. T., and Selkoe, D. J. (1999) Nature 398 513–517 [DOI] [PubMed] [Google Scholar]
- 7.Marambaud, P., and Robakis, N. K. (2005) Genes Brain Behav. 4 134–146 [DOI] [PubMed] [Google Scholar]
- 8.Baki, L., Marambaud, P., Efthimiopoulos, S., Georgakopoulos, A., Wen, P., Cui, W., Shioi, J., Koo, E., Ozawa, M., Friedrich, V. L., Jr., and Robakis, N. K. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 2381–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marambaud, P., Wen, P. H., Dutt, A., Shioi, J., Takashima, A., Siman, R., and Robakis, N. K. (2003) Cell 114 635–645 [DOI] [PubMed] [Google Scholar]
- 10.Stahl, B., Diehlmann, A., and Sudhof, T. C. (1999) J. Biol. Chem. 274 9141–9148 [DOI] [PubMed] [Google Scholar]
- 11.Tanahashi, H., and Tabira, T. (1999) Neuroreport 10 563–568 [DOI] [PubMed] [Google Scholar]
- 12.Zhou, J., Liyanage, U., Medina, M., Ho, C., Simmons, A. D., Lovett, M., and Kosik, K. S. (1997) Neuroreport 8 2085–2090 [DOI] [PubMed] [Google Scholar]
- 13.McCrea, P. D., and Park, J. I. (2007) Biochim. Biophys. Acta 1773 17–33 [DOI] [PubMed] [Google Scholar]
- 14.Davis, M. A., Ireton, R. C., and Reynolds, A. B. (2003) J. Cell Biol. 163 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao, K., Allison, D. F., Buckley, K. M., Kottke, M. D., Vincent, P. A., Faundez, V., and Kowalczyk, A. P. (2003) J. Cell Biol. 163 535–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wildenberg, G. A., Dohn, M. R., Carnahan, R. H., Davis, M. A., Lobdell, N. A., Settleman, J., and Reynolds, A. B. (2006) Cell 127 1027–1039 [DOI] [PubMed] [Google Scholar]
- 17.Daniel, J. M. (2007) Biochim. Biophys. Acta 1773 59–68 [DOI] [PubMed] [Google Scholar]
- 18.Ferber, E. C., Kajita, M., Wadlow, A., Tobiansky, L., Niessen, C., Ariga, H., Daniel, J., and Fujita, Y. (2008) J. Biol. Chem. 283 12691–12700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elia, L. P., Yamamoto, M., Zang, K., and Reichardt, L. F. (2006) Neuron 51 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Israely, I., Costa, R. M., Xie, C. W., Silva, A. J., Kosik, K. S., and Liu, X. (2004) Curr. Biol. 14 1657–1663 [DOI] [PubMed] [Google Scholar]
- 21.Feng, R., Wang, H., Wang, J., Shrom, D., Zeng, X., and Tsien, J. Z. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 8162–8167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saura, C. A., Choi, S. Y., Beglopoulos, V., Malkani, S., Zhang, D., Shankaranarayana Rao, B. S., Chattarji, S., Kelleher, R. J., III, Kandel, E. R., Duff, K., Kirkwood, A., and Shen, J. (2004) Neuron 42 23–36 [DOI] [PubMed] [Google Scholar]
- 23.Onishi, M., Kinoshita, S., Morikawa, Y., Shibuya, A., Phillips, J., Lanier, L. L., Gorman, D. M., Nolan, G. P., Miyajima, A., and Kitamura, T. (1996) Exp. Hematol. 24 324–329 [PubMed] [Google Scholar]
- 24.Serban, G., Kouchi, Z., Baki, L., Georgakopoulos, A., Litterst, C. M., Shioi, J., and Robakis, N. K. (2005) J. Biol. Chem. 280 36007–36012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potter, M. D., Barbero, S., and Cheresh, D. A. (2005) J. Biol. Chem. 280 31906–31912 [DOI] [PubMed] [Google Scholar]
- 26.Shioi, J., Georgakopoulos, A., Mehta, P., Kouchi, Z., Litterst, C. M., Baki, L., and Robakis, N. K. (2007) J. Neurochem. 101 674–681 [DOI] [PubMed] [Google Scholar]
- 27.Walker, E. S., Martinez, M., Brunkan, A. L., and Goate, A. (2005) J. Neurochem. 92 294–301 [DOI] [PubMed] [Google Scholar]
- 28.Ohkubo, T., and Ozawa, M. (1999) J. Biol. Chem. 274 21409–21415 [DOI] [PubMed] [Google Scholar]
- 29.Ireton, R. C., Davis, M. A., van Hengel, J., Mariner, D. J., Barnes, K., Thoreson, M. A., Anastasiadis, P. Z., Matrisian, L., Bundy, L. M., Sealy, L., Gilbert, B., van Roy, F., and Reynolds, A. B. (2002) J. Cell Biol. 159 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soriano, S., Lu, D. C., Chandra, S., Pietrzik, C. U., and Koo, E. H. (2001) J. Biol. Chem. 276 29045–29050 [DOI] [PubMed] [Google Scholar]
- 31.Nunan, J., Shearman, M. S., Checler, F., Cappai, R., Evin, G., Beyreuther, K., Masters, C. L., and Small, D. H. (2001) Eur. J. Biochem. 268 5329–5336 [DOI] [PubMed] [Google Scholar]
- 32.Mathews, P. M., Jiang, Y., Schmidt, S. D., Grbovic, O. M., Mercken, M., and Nixon, R. A. (2002) J. Biol. Chem. 277 36415–36424 [DOI] [PubMed] [Google Scholar]
- 33.Kaether, C., Capell, A., Edbauer, D., Winkler, E., Novak, B., Steiner, H., and Haass, C. (2004) EMBO J. 23 4738–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, F., Yu, G., Arawaka, S., Nishimura, M., Kawarai, T., Yu, H., Tandon, A., Supala, A., Song, Y. Q., Rogaeva, E., Milman, P., Sato, C., Yu, C., Janus, C., Lee, J., Song, L., Zhang, L., Fraser, P. E., and St. George-Hyslop, P. H. (2001) Nat. Cell Biol. 3 751–754 [DOI] [PubMed] [Google Scholar]
- 35.Arikkath, J., and Reichardt, L. F. (2008) Trends Neurosci. 31 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yap, A. S., Niessen, C. M., and Gumbiner, B. M. (1998) J. Cell Biol. 141 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.