Abstract
Calcium-binding protein 1 (CaBP1), a neuron-specific member of the calmodulin (CaM) superfamily, modulates Ca2+-dependent activity of inositol 1,4,5-trisphosphate receptors (InsP3Rs). Here we present NMR structures of CaBP1 in both Mg2+-bound and Ca2+-bound states and their structural interaction with InsP3Rs. CaBP1 contains four EF-hands in two separate domains. The N-domain consists of EF1 and EF2 in a closed conformation with Mg2+ bound at EF1. The C-domain binds Ca2+ at EF3 and EF4, and exhibits a Ca2+-induced closed to open transition like that of CaM. The Ca2+-bound C-domain contains exposed hydrophobic residues (Leu132, His134, Ile141, Ile144, and Val148) that may account for selective binding to InsP3Rs. Isothermal titration calorimetry analysis reveals a Ca2+-induced binding of the CaBP1 C-domain to the N-terminal region of InsP3R (residues 1-587), whereas CaM and the CaBP1 N-domain did not show appreciable binding. CaBP1 binding to InsP3Rs requires both the suppressor and ligand-binding core domains, but has no effect on InsP3 binding to the receptor. We propose that CaBP1 may regulate Ca2+-dependent activity of InsP3Rs by promoting structural contacts between the suppressor and core domains.
Calcium ion (Ca2+) in the cell functions as an important messenger that controls neurotransmitter release, gene expression, muscle contraction, apoptosis, and disease processes (1). Receptor stimulation in neurons promotes large increases in intracellular Ca2+ levels controlled by Ca2+ release from intracellular stores through InsP3Rs (2). The neuronal type-1 receptor (InsP3R1)2 is positively and negatively regulated by cytosolic Ca2+ (3-6), important for the generation of repetitive Ca2+ transients known as Ca2+ spikes and waves (1). Ca2+-dependent activation of InsP3R1 contributes to the fast rising phase of Ca2+ signaling known as Ca2+-induced Ca2+ release (7). Ca2+-induced inhibition of InsP3R1, triggered at higher cytosolic Ca2+ levels, coordinates the temporal decay of Ca2+ transients (6). The mechanism of Ca2+-dependent regulation of InsP3Rs is complex (8, 9), and involves direct Ca2+ binding sites (5, 10) as well as remote sensing by extrinsic Ca2+-binding proteins such as CaM (11, 12), CaBP1 (13, 14), CIB1 (15), and NCS-1 (16).
Neuronal Ca2+-binding proteins (CaBP1-5 (17)) represent a new sub-branch of the CaM superfamily (18) that regulate various Ca2+ channel targets. Multiple splice variants and isoforms of CaBPs are localized in different neuronal cell types (19-21) and perform specialized roles in signal transduction. CaBP1, also termed caldendrin (22), has been shown to modulate the Ca2+-sensitive activity of InsP3Rs (13, 14). CaBP1 also regulates P/Q-type voltage-gated Ca2+ channels (23), L-type channels (24), and the transient receptor potential channel, TRPC5 (25). CaBP4 regulates Ca2+-dependent inhibition of L-type channels in the retina and may be genetically linked to retinal degeneration (26). Thus, the CaBP proteins are receiving increased attention as a family of Ca2+ sensors that control a variety of Ca2+ channel targets implicated in neuronal degenerative diseases.
CaBP proteins contain four EF-hands, similar in sequence to those found in CaM and troponin C (18) (Fig. 1). By analogy to CaM (27), the four EF-hands are grouped into two domains connected by a central linker that is four residues longer in CaBPs than in CaM. In contrast to CaM, the CaBPs contain non-conserved amino acids within the N-terminal region that may confer target specificity. Another distinguishing property of CaBPs is that the second EF-hand lacks critical residues required for high affinity Ca2+ binding (17). CaBP1 binds Ca2+ only at EF3 and EF4, whereas it binds Mg2+ at EF1 that may serve a functional role (28). Indeed, changes in cytosolic Mg2+ levels have been detected in cortical neurons after treatment with neurotransmitter (29). Other neuronal Ca2+-binding proteins such as DREAM (30), CIB1 (31), and NCS-1 (32) also bind Mg2+ and exhibit Mg2+-induced physiological effects. Mg2+ binding in each of these proteins helps stabilize their Ca2+-free state to interact with signaling targets.
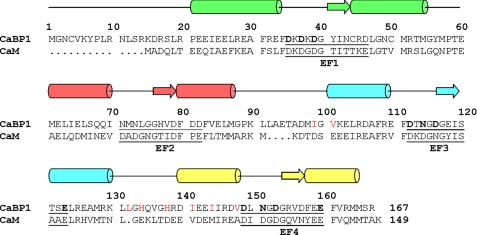
FIGURE 1.
Amino acid sequence alignment of human CaBP1 with CaM. Secondary structural elements (α-helices and β-strands) were derived from NMR analysis. The four EF-hands (EF1, EF2, EF3, and EF4) are highlighted green, red, cyan, and yellow. Residues in the 12-residue Ca2+-binding loops are underlined and chelating residues are highlighted bold. Non-conserved residues in the hydrophobic patch are colored red.
Despite extensive studies on CaBP1, little is known about its structure and target binding properties, and regulation of InsP3Rs by CaBP1 is somewhat controversial and not well understood. Here, we present the NMR solution structures of both Mg2+-bound and Ca2+-bound conformational states of CaBP1 and their structural interactions with InsP3R1. These CaBP1 structures reveal important Ca2+-induced structural changes that control its binding to InsP3R1. Our target binding analysis demonstrates that the C-domain of CaBP1 exhibits Ca2+-induced binding to the N-terminal cytosolic region of InsP3R1. We propose that CaBP1 may regulate Ca2+-dependent channel activity in InsP3Rs by promoting a structural interaction between the N-terminal suppressor and ligand-binding core domains that modulates Ca2+-dependent channel gating (8, 33, 34).
EXPERIMENTAL PROCEDURES
Expression and Purification of CaBP1—CaBP1 has two splice variants expressed in the brain, termed l-CaBP1 and s-CaBP1 (17). Both variants regulate Ca2+ channels with similar efficacy (14) and the extra residues in the long variant can be deleted without affecting CaBP1 binding to InsP3Rs. The short splice variant (19.4 kDa and 167 residues) is more soluble and amenable for NMR structural analysis and was used throughout this study. Recombinant CaBP1 and mutants were expressed and purified from Escherichia coli strain BL21(DE3) as described previously (28).
Construction of CaBP1 N-domain and C-domain Fragments—cDNAs coding for the CaBP1 N-domain (residues 1-91; CaBP1-N) and C-domain (residues 96-167; CaBP1-C) were cloned into protein expression vectors pET-28a(+) and pET-3a(+), respectively. Recombinant CaBP1-C protein was expressed and purified by the same method as full-length CaBP1. The His6-CaBP1-N protein was purified first by nickel-Sepharose (Amersham Biosciences), and then by using Superdex 200 size exclusion chromatography.
Construction of CaBP1 Mutants—The D35A, D37A, D39A, D46A, ΔL132, H134E, and V148A mutants of CaBP1 were generated by using the QuikChange site-directed mutagenesis kit (Stratagene) and the presence of these mutations was confirmed by DNA sequencing. The mutant expression and purification procedures were the same as that for wild type.
Expression and Purification of IP3Rsup-(2-223), IP3Rcore-(224-604), and InsP3R-(1-587)—The recombinant suppressor domain (InsP3Rsup residues, 2-223) and ligand-binding core domain (InsP3Rcore residues, 224-604) containing a GST tag were cloned, expressed, and purified as described by Ref. 35. The GST tag was removed by adding 1 μg of thrombin to the purified GST fusion protein sample that was then applied to a Superdex-75 size exclusion column to remove the GST tag and other impurities. A recombinant dual domain construct containing both InsP3Rsup and InsP3Rcore (InsP3R-(1-587)) was cloned, expressed, and purified as described (35). The recombinant InsP3R-(1-587) protein contained a C-terminal intein-CBD-His9 tag that was first purified with nickel-nitrilotriacetic acid resin (Qiagen) and the CBD-His9 tag was cleaved by treatment with 20 mm dithiothreitol for 24 h. The cleaved protein was released from chitin beads, concentrated, and then chromatographed on a Superdex 200 column.
NMR Spectroscopy—Samples for NMR analyses were prepared by dissolving unlabeled, 15N-labeled, or (15N, 13C)-labeled CaBP1 in 0.3 ml of 95% H2O, 5% [2H]H2O containing 10 mm [2H11]Tris, pH 7.4, 0.1 mm KCl, and 5 mm EDTA (apo-), 5 mm MgCl2 (Mg2+-bound), or 5 mm CaCl2, 5 mm MgCl2 (Ca2+-bound). All NMR experiments were performed at 30 °C on a Bruker Avance 600 MHz spectrometer equipped with triple resonance cryoprobe and z axis gradient. Backbone and side chain assignments were described previously (36, 37). All NMR data were processed and analyzed by using the programs NMRPipe and nmrView.
NMR Structure Calculation—The structures were calculated with XPLOR-NIH (38) that employed the YASAP protocol (39). Distance restraints derived from inter-proton NOEs and dihedral angles (ϕ and ψ) from chemical shift index data are summarized in Table 1. Distance constraints involving Ca2+ bound to loop residues 1, 3, 5, 7, and 12 in EF3 and EF4 (27), and Mg2+ bound to loop residues 1, 3, and 5 in EF1 were introduced as described previously (40). Fifty independent structures were calculated and the 15 lowest energy structures were selected. The final structural statistics are summarized in Table 1 and coordinates were deposited into the RCSB Protein Data bank (accession numbers 2k7b, 2k7c, and 2k7d).
TABLE 1.
Structural statistics for the ensemble structures of CaBP1
| Mg2+-bound N-domain | Mg2+-bound C-domain | Ca2+-bound C-domain | |
|---|---|---|---|
| NOE restraints | 874 | 795 | 814 |
| Intra (|i-j| = 0) | 198 | 284 | 324 |
| Medium (0<|i-j|≤4) | 473 | 338 | 309 |
| Long (|i-j|>4) | 203 | 173 | 181 |
| Hydrogen bonds restraints | 56 | 50 | 50 |
| Dihedral angle restraints (ϕ,ψ) | 96 | 88 | 88 |
| Root mean square deviation from ideal geometry | |||
| Bond length (Å) | 0.0069 ± 0.0001 | 0.0068 ± 0.0003 | 0.0065 ± 0.0001 |
| Bond angle (deg) | 1.87 ± 0.008 | 1.87 ± 0.007 | 1.83 ± 0.0007 |
| Root mean square deviation from average structure (Å) | |||
| Secondary structure (backbone) | 0.58 ± 0.14 | 0.47 ± 0.09 | 0.51 ± 0.10 |
| Secondary structure (heavy) | 1.22 ± 0.10 | 1.33 ± 0.11 | 1.30 ± 0.10 |
| Ramachandran plot (%) | |||
| Most favored region | 73.8 | 77.4 | 79.4 |
| Allowed region | 26.1 | 21.6 | 19.8 |
| Disallowed region | 0.0 | 1.0 | 0.8 |
| Average energy (kcal mol−1) | |||
| Total | 1359.3 | 1217.4 | 1190.8 |
| Distance | 61.5 | 47.7 | 35.3 |
Isothermal Titration Calorimetry—The CaBP1 and InsP3R interactions were measured by a MicroCal VP-ITC microcalorimeter at 30 °C as described previously (28). Proteins were exchanged into buffer containing 15 mm Tris-HCl, pH 7.5, 300 mm NaCl, 3% glycerol, and 1 mm tris(2-carboxyethyl)phosphine with the addition of 2 mm EDTA (apo-state), 5 mm MgCl2 (Mg2+-bound state), or 5 mm MgCl2 and 5 mm Ca2+ (Ca2+-bound state). The InsP3R-(1-587) at a concentration of 50-80 μm was titrated with 1-2 mm CaBP1 in 25 steps of 10 μl. The data were analyzed with a one-binding site model using Micro-Cal Origin 7 for ITC.
Docking Calculation—Structural modeling of the CaBP1-receptor complex (CaBP1-C·InsP3Rsup·InsP3Rcore) was performed using ZDOCK (41). CaBP1-C (PDB 2k7d) was independently docked to either InsP3Rsup (PDB 1xzz) or InsP3Rcore (PDB 1n4k). The top 20 ZDOCK predicted complexes with lowest energy were superimposed. CaBP1-C from each binary complex was structurally aligned using PyMol to generate possible ternary interactions.
RESULTS
CaBP1 Has Two Independent Domains—A critical first step in the NMR structural analysis of CaBP1 was to identify whether the four EF-hands in CaBP1 combine to form two separately folded domains: N-domain (EF1 and EF2) versus C-domain (EF3 and EF4) like what is seen in CaM (42). Alternatively, the four EF-hands might interact to form a single globular domain like what is observed in NCS-1 (43) and CIB1 (44). First, we analyzed the NOESY-HSQC spectra of CaBP1 and were unable to detect NOE-based contacts between the two domains, consistent with this protein having non-interacting domains.
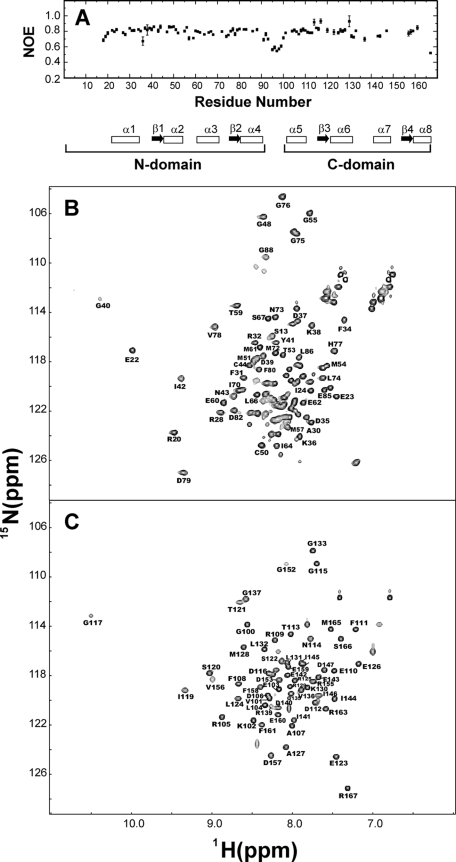
Our second approach was to examine the backbone flexibility of the two domains and the central linker. In Fig. 2A,{1H}-15N NOE measurements indicate relatively low heteronuclear NOE values (∼0.5) for residues in the central linker region (residues 92-98), suggesting that CaBP1 does indeed contain a flexible inter-domain linker. By contrast, much higher heteronuclear NOE values (∼0.8) are found for residues in each domain and indicate the two domains are separately folded.
FIGURE 2.
{1H}-15N NOE data for Mg2+ bound CaBP1 (A) and 15N-1H-HSQC spectra of Mg2+-bound CaBP1-N (B) and CaBP1-C (C). A schematic representation of the secondary structure is shown at the bottom in A with α-helices and β-strands indicated by boxes and arrows, respectively.
A final test for the existence of two independent domains was to analyze NMR spectra of individual domain fragments of CaBP1: N-domain (residues 1-91) and C-domain (96-167). The 1H-15N-HSQC spectra of the domain constructs (Fig. 2, B and C) indicate that each domain is separately folded without having the other domain present. In addition, the backbone amide chemical shifts for each residue in the domain fragments are nearly identical to the corresponding chemical shifts of the full-length protein. Thus, the structures of the isolated domain fragments must remain intact in the full-length protein, consistent with two non-interacting domains.
On the basis of our NMR analyses above, CaBP1 has two independently folded domains (N-domain, EF1 and EF2, and C-domain, EF3 and EF4) separated by a flexible linker. The structures of each domain were analyzed separately below. The C-domain structure was solved by analyzing NMR spectra of a peptide fragment (CaBP1-C, residues, 96-167), whereas the structure of the Mg2+-bound N-domain was solved by analyzing NMR spectra of full-length CaBP1. The Ca2+-bound N-domain was not studied because it does not bind Ca2+ under physiological conditions (28). In summary, we present below three separate NMR solution structures of CaBP1: 1) Mg2+-bound N-domain (PDB 2k7b), 2) Mg2+-bound C-domain (PDB 2k7c), and 3) Ca2+-bound C-domain (PDB 2k7d). The statistics for these structures are summarized in Table 1.
Structure of Mg2+-bound CaBP1—The first 21 N-terminal residues of CaBP1 exhibited weak NMR intensities and could not be accurately analyzed. The remaining residues (22-167) exhibited strong 1H-15N-HSQC peaks and their sequence-specific NMR assignments were analyzed and described previously (36) (BMRB number 15197). The assigned resonances in the HSQC spectrum represent main chain and side chain amide groups that serve as fingerprints of the overall conformation. Three-dimensional protein structures derived from the NMR assignments were calculated on the basis of NOE data, slowly exchanging amide NH groups, chemical shift analysis, and 3JNHα spin-spin coupling constants (see “Experimental Procedures”). The final NMR-derived structures of Mg2+-bound CaBP1 are illustrated in Fig. 3, A and B.
FIGURE 3.
Main chain structures of CaBP1 determined by NMR. Superposition of the 15 lowest energy structures (top) and ribbon representations of the energy-minimized average structure (bottom) are illustrated for: A, Mg2+-bound N-domain (PDB 2k7b); B, Mg2+-bound C-domain (PDB 2k7c); and C, Ca2+-bound C-domain (PDB 2k7d). N-terminal residues (1-21) are unstructured and not shown. EF-hands are highlighted in colors as defined in the legend to Fig. 1. Orange and magenta spheres represent bound Ca2+ and Mg2+.
The Mg2+-bound CaBP1 structure contains a total of eight α-helices and four β-strands: α1 (residues 22-34), α2 (residues 44-54), α3 (residues 61-70), α4 (residues 80-88), α5 (residues 101-111), α6 (121-130), α7 (residues 140-147), α8 (residues 158-165), β1 (residues 41-43), β2 (residues 77-79), β3 (residues 118-120), and β4 (residues 155-157) (Fig. 1). CaBP1 contains two domains comprising four EF-hands (Fig. 3): EF1 (green, residues 26-55) and EF2 (red, residues 62-91)) are linked and form the N-domain; likewise, EF3 (cyan, residues 103-132) and EF4 (yellow, residues 140-169) form the C-domain. The two domains do not interact structurally (Fig. 2). Each EF-hand consists of a helix turn helix structure similar to the closed structure of Ca2+-free EF-hands seen in previous structures of apo-CaM (45-47) and troponin C (48). The interhelical angles for the EF-hands are 126.8° (EF1), 140.2° (EF2), 140.1° (EF3), and 126.2° (EF4) (see Table 2). The overall main chain structures of Mg2+-bound N-domain (Fig. 3A) and C-domain (Fig. 3B) are very similar to those of apo-CaM. The root mean square deviation is 1.5 and 1.3 Å when comparing the main chain atoms of Mg2+-bound CaBP1 with those of apo-CaM in the N-domain and C-domain, respectively.
TABLE 2.
Interhelical angles of the EF-hands in CaM and CaBP1
Residues in the helices are shown below.
|
Interhelical angles
|
|||||
|---|---|---|---|---|---|
| Helix pair | Apo-CaMa | Ca2+-bound CaMb | Mg2+-bound CaBP1c | Ca2+-bound CaBP1c | |
| degree | |||||
| α1-α2 | 130.9 | 103.8 | 126.8 | ||
| α3-α4 | 130.8 | 101.0 | 140.2 | ||
| α5-α6 | 139.5 | 101.0 | 140.1 | 100.6 | |
| α7-α8 | 126.0 | 101.0 | 126.2 | 110.6 | |
Apo-CaM (PDB accession code 1dmo): (a1) 6-18, (a2) 29-38, (a3) 45-55, (a4) 65-75, (a5) 82-90, (a6) 103-112, (a7) 118-127, (a8) 137-143.
Ca2+ bound CaM (PDB accession code 1j7p): (a1) 6-19, (a2) 29-38, (a3) 45-55, (a4) 65-75, (a5) 83-92, (a6) 102-111, (a7) 118-128, (a8) 138-145.
CaBP1: (a1) 22-34, (a2) 45-54, (a3) 61-70, (a4) 80-88, (a5) 101-110, (a6) 121-130, (a7) 141-147, (a8) 158-165.
The NMR structure of Mg2+-bound CaBP1 indicates that Mg2+ is bound at EF1 as evidenced by Mg2+-dependent amide chemical shift changes for residues in the EF1 binding loop (Asp35, Asp37, Asp39, and Gly40). Mg2+-binding caused a large downfield amide proton chemical shift for Gly40 due in part to formation of a strong hydrogen bond between its main chain amide proton and the carboxylate side chain of Asp35. To identify possible chelating interactions with the bound Mg2+, we made the following point mutations (D35A, D37A, D39A, and D46A) to residues in the EF1 loop at positions 1, 3, 5, and 12. Mg2+ binding to each mutant versus wild type was monitored by analyzing the Gly40 amide resonance. The Mg2+-binding analysis revealed that Asp35, Asp37, and Asp39 are each essential for high affinity Mg2+ binding, suggesting that their carboxylate side chains might form coordinate covalent bonds with the bound Mg2+. A similar Mg2+ binding geometry involving acidic side chains from residues at positions 1, 3, and 5 was also observed in the structure of Mg2+-bound calbindin (49). The stereochemical geometry and chelation of the bound Mg2+ at EF1 (magenta sphere, Fig. 3A) was modeled like that described by Ref. 50. The EF2 loop in CaBP1 does not bind Ca2+ or Mg2+ and is structurally distorted by the presence of Gly75 at the fifth position in the binding loop.
Structure of Ca2+-bound CaBP1—The NMR-derived structure of the Ca2+-bound CaBP1-C is shown in Fig. 3C (37) (BMRB number 15623). The secondary structure of Ca2+-bound CaBP1 is nearly identical to that determined above for Mg2+-bound CaBP1 (Fig. 1). By contrast, the overall tertiary structure of Ca2+-bound CaBP1-C (Fig. 3C) is quite different from that of Mg2+-bound CaBP-C (Fig. 3B), reminiscent of the Ca2+-induced closed to open transition seen previously in CaM (45) and troponin C (48). The structures of EF3 and EF4 in Ca2+-bound CaBP1 resemble the familiar “open” conformation of Ca2+ occupied EF-hands in CaM (27) and many other EF-hand proteins. The interhelical angles are 100.6° (EF3) and 110.6° (EF4) for Ca2+-bound CaBP1 (see Table 2). The overall main chain structure of Ca2+-bound CaBP1-C (Fig. 3C) is very similar to that of Ca2+-bound CaM with a root mean square deviation is 1.2 Å when comparing their main chain atoms.
The NMR structure of Ca2+-bound CaBP1 confirms Ca2+ binding at EF3 and EF4, as evidenced by characteristic Ca2+-dependent amide chemical shift changes assigned to Gly117 in EF3 and Gly154 in EF4. Ca2+-binding caused large downfield amide proton chemical shifts for Gly117 and Gly154 due in part to formation of a strong hydrogen bond between its main chain amide proton and the carboxylate side chain of Asp112 (EF3) and Asp149 (EF4), respectively. The geometry of the coordinate covalent bonds formed between chelating amino acid residues in CaBP1 and the bound Ca2+ could not be observed directly in our NMR study. Instead, the stereochemical geometry and chelation of Ca2+ bound at EF3 and EF4 (orange spheres, Fig. 3C) was modeled using structural constraints derived from the x-ray crystal structure of Ca2+-bound CaM (27), which closely resembles the binding site geometry conserved in other EF-hand proteins (51).
Dimerization of CaBP1?—Previous hydrodynamic analyses of CaBP1 suggested a monomer-dimer equilibrium under NMR conditions (28). Indeed, our 15N NMR relaxation analysis (T1 and T2) of CaBP1 in this study suggests an average rotational correlation time of ∼12 ns (consistent with a protein dimer) that decreased somewhat when the protein concentration was lowered 10-fold. But, we did not observe any significant chemical shift changes in NMR spectra recorded as a function of protein concentration (50 μm to 1 mm). Also, intermolecular NOEs could not be detected in 13C-filtered NOESY-HMQC spectra of CaBP1 recorded from a mixed labeled sample. Thus, the CaBP1 monomer-dimer equilibrium must have an exchange rate that is much faster than the chemical shift time scale and the structure of the dimer cannot be resolved by NMR. Such fast exchange kinetics and hence low affinity for dimerization (Kd ∼ 100 μm) is not likely to be physiologically relevant and was not characterized further.
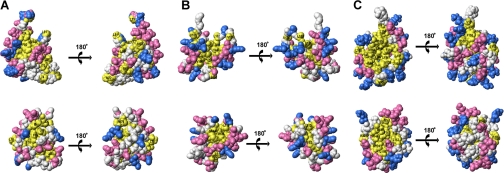
Surface Properties of CaBP1 Versus CaM—Space filling representations of Mg2+-bound and Ca2+-bound CaBP1 are illustrated and compared with those of CaM (Fig. 4). The surface residues of Mg2+-bound CaBP1 are similar to those of apo-CaM (Fig. 4, A and B). The N-domain of Mg2+-bound CaBP1 contains mostly negatively charged residues on the protein surface (highlighted red in Fig. 4A) that remain invariant in CaM, and the overall surface charge is nearly the same between the two. The C-domain surface of Mg2+-bound CaBP1 also looks similar to that of apo-CaM (Fig. 4B). The N-domain has a few exposed hydrophobic residues in Mg2+-bound CaBP1 (Met57, Met61, Met72, and Leu74) not conserved in CaM (Fig. 1) that might serve a functional role in target recognition.
FIGURE 4.
Space-filling representation of CaBP1 (top) and CaM (bottom) illustrating (A) Mg2+-bound N-domain of CaBP1 (PDB 2k7b) and apo-CaM (PDB 1dmo), (B) Mg2+-bound C-domain of CaBP1 (PDB 2k7c) and apo-CaM (PDB 1dmo), and (C) Ca2+-bound C-domain of CaBP1 (PDB 2k7d) and Ca2+-CaM (PDB 1j7p). Acidic residues (Asp and Glu), basic residues (Arg, His, Lys), and hydrophobic residues (Ile, Leu, Phe, Trp, Val) are colored red, blue, and yellow, respectively.
The protein surface of Ca2+-bound CaBP1-C is somewhat different from that of Ca2+-bound CaM (Fig. 4C). The front face of Ca2+-bound CaBP1-C exhibits a striking solvent-exposed hydrophobic surface (highlighted yellow in Fig. 4C) that is wider and more expansive than that of Ca2+-bound CaM. The solvent-exposed hydrophobic patch in Ca2+-bound CaBP1-C contains non-conserved residues located in the loop between EF3 and EF4 (Leu132, His134, Val136, and His138), the helix of EF4 (Ile141, Ile144, and Val148), and the domain linker (Ile99 and Val101). The exposed hydrophobic patch is surrounded by a ring of charged residues that might form electrostatic contacts with target proteins. Basic residues in CaBP1 (Lys102, His134, His138, and Arg139) are replaced by negatively charged residues in CaM that might confer specific electrostatic contacts. We suggest that these nonconserved residues on the surface of Ca2+-bound CaBP1 may form a target binding site and help explain the highly selective binding of CaBP1 to InsP3Rs.
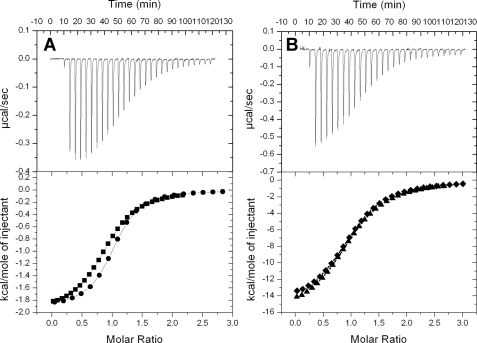
CaBP1 Interaction with N-terminal Cytosolic Residues in InsP3R1—CaBP1 was shown previously to regulate Ca2+-induced channel activity of InsP3Rs and the CaBP1 interaction site was localized to the N-terminal cytosolic region of InsP3R1 (residues 1-604) (13). We performed a series of target binding studies using isothermal titration calorimetry (ITC) and NMR to characterize the structural interaction between CaBP1 and InsP3R1 (Fig. 5). An N-terminal peptide fragment of InsP3R1 (residues, 1-587, called InsP3R-(1-587)), saturated with InsP3, exhibited strongly exothermic binding (ΔH = -1.96 kcal/mol) to Ca2+-bound CaBP1 with a 1:1 stoichiometry and a dissociation constant (Kd) of 3 μm (Fig. 5A). By contrast, InsP3R-(1-587) exhibited about 10-fold weaker binding to Mg2+-bound CaBP1 (ΔH = -1.55 kcal/mol and Kd = 30 μm). InsP3R-(1-587) showed no detectable binding to either Ca2+-free or Ca2+-bound CaM under these same conditions, which was somewhat surprising given that Ca2+-CaM has been suggested to bind InsP3R1 and negatively regulate channel gating (12). Also, apo-InsP3R-(1-587) binds to CaBP1 with approximately the same affinity as the ligand-bound receptor. Thus, ligand-bound InsP3R-(1-587) exhibits Ca2+-induced binding to CaBP1 with high selectivity over CaM.
FIGURE 5.
Isothermal titration microcalorimeteric analysis for the binding of InsP3R1-(1-587) to (A) Ca2+-bound CaBP1 and (B) InsP3. Experimental traces of the calorimetric titration (25 × 10-μl aliquots) are shown in the top panel and the intergrated binding isotherms are shown in the bottom panel. Binding data are overlaid in the bottom panels for the binding of InsP3R-(1-587) to (A) full-length CaBP1 (squares) and CaBP1-C (circles) and (B) InsP3 in the presence (diamonds) or absence (triangles) of CaBP1. The binding isotherms were fit to a one-site model and fitting parameters are reported in Table 3.
The lack of InsP3R-(1-587) binding to CaM was also verified by using NMR in which the 1H-15N-HSQC spectrum of 15N-labeled CaM remained unaffected as a function of adding excess, unlabeled InsP3R-(1-587). The 1H-15N-HSQC spectrum of 15N-labeled CaBP1, by contrast, exhibited striking peak broadening and chemical shift changes upon addition of saturating InsP3R-(1-587), further demonstrating that CaBP1 binds to InsP3R-(1-587). Unfortunately, because all NMR peaks in the HSQC spectrum of CaBP1 are severely broadened and uniformly affected by InsP3R-(1-587) binding, it was not possible to identify any specific binding site residues by chemical shift mapping.
The InsP3R1-binding site on CaBP1 was investigated by performing ITC experiments separately on N-domain and C-domain fragments of CaBP1 (CaBP1-N and CaBP1-C). InsP3R-(1-587) binds to CaBP1-C with nearly the same enthalpy (ΔH = -1.92 kcal/mol), dissociation constant (Kd =∼2 μm), and Ca2+ dependence as seen above for binding to full-length CaBP1 (Fig. 5A). By contrast, InsP3R-(1-587) failed to exhibit any detectable binding to CaBP1-N by ITC. The lack of such binding was verified under NMR conditions wherein the 1H-15N-HSQC spectrum of 15N-labeled CaBP1-N did not change as a function of adding excess InsP3R-(1-587). 1H-15N-HSQC spectra of 15N-labeled CaBP1-C changed dramatically upon adding saturating InsP3R-(1-587) similar to that described above for full-length CaBP1. Thus, InsP3R-(1-587) binds selectively to Ca2+-bound CaBP1-C and does not interact with CaBP1-N.
To probe the CaBP1-binding site within InsP3R-(1-587), ITC studies were performed separately by using the suppressor domain (residues 1-224, InsP3Rsup) and the InsP3 binding core domain (residues 236-604, InsP3Rcore). No heat signal could be detected upon individually adding either the suppressor domain and/or core domain to CaBP1, suggesting either ΔH = 0 or a lack of binding in the micromolar range (Kd ≫ 10-4 m). A lack of binding under NMR conditions was verified by the 1H-15N-HSQC spectrum of 15N-labeled CaBP1 (full-length) that remained unaffected as a function of adding excess suppressor and/or core domain. Thus, CaBP1 does not exhibit high affinity binding to either the suppressor or core domain alone, but rather the two domains must be linked together to have high affinity binding to CaBP1.
The binding of CaBP1 to InsP3R-(1-587) has little or no effect on ligand-binding affinity. The binding of InsP3 to InsP3R-(1-587) is exothermic (ΔH = -16.2 kcal/mol) with a 1:1 stoichiometry and dissociation constant (Kd) of ∼1 μm (Fig. 5B). The apparent Kd measured by ITC is at least 100-fold weaker than the intrinsic ligand binding affinity measured for full-length InsP3R1 (52). The discrepancy could be explained in part by a protein conformational change in InsP3R-(1-587) coupled to InsP3 binding. The intrinsic binding of InsP3 (Ka ∼ 108 m-1) if coupled to an unfavorable conformational change (Keq ∼ 10-2) would yield an overall equilibrium constant of Ktot = Ka × Keq ∼ 106 m-1, consistent with the overall Kd measured by ITC. Thus, InsP3 binding to InsP3R-(1-587) induces a protein conformational change, consistent with predictions from small-angle x-ray scattering analysis (35). The apparent Kd for InsP3 binding to InsP3R-(1-587) is NOT affected by the presence or absence of saturating CaBP1 (Fig. 5B), demonstrating that CaBP1 binding to InsP3R-(1-587) does not block or otherwise influence ligand binding.
Structural Model of the CaBP1·InsP3R-(1-587) Complex—The relatively low solubility of the CaBP1·InsP3R-(1-587) complex has thus far hampered our efforts to directly solve the complex structure by NMR or x-ray crystallography. Instead, we used a computational docking approach that takes into account variables such as shape complementarity, desolvation energetics, and electrostatics to simulate the structure of the protein complex (53). Separate x-ray crystal structures have been solved recently for InsP3Rsup (54) and InsP3Rcore (55). Our ITC analysis indicates that CaBP1-C binds cooperatively to InsP3Rsup and InsP3Rcore only when both domains are connected (Fig. 5). This cooperativity suggests that CaBP1-C might contact both InsP3Rsup and InsP3Rcore in the complex. The first step in the model calculation was to individually dock CaBP1-C to each domain and generate binary complexes: CaBP1-C·InsP3Rsup and CaBP1-C·InsP3Rcore. Structures of the separate binary complexes were then aligned with respect to CaBP1-C to predict the disposition of InsP3Rsup and InsP3Rcore in the ternary complex.
A total of 20 independent docking calculations were performed for each binary complex. A statistical analysis of the CaBP1-InsP3Rsup docked structures revealed a striking tendency for CaBP1-C to bind to an exposed surface on the helical “arm” (residues 66-110) in InsP3Rsup, suggested previously to be functionally important (14, 56). This docking model is also consistent with previous mutagenesis studies, suggesting that the arm residues interact with InsP3Rcore (55). Arm residues (66-81) also form a potential calmodulin binding motif shown previously to inhibit CaBP1 binding to InsP3R1 (14). Finally, it is well known that EF-hand proteins generally bind to helical segments in target proteins (57). Thus, the docking interactions of CaBP1-C with the suppressor helical arm are plausible and well justified experimentally. The family of docked structures of the CaBP1·InsP3Rcore complex revealed a tendency for CaBP1-C to interact with the β-trefoil subdomain (residues 397-420) located on the opposite face from the ligand-binding site. The lowest energy structures of the CaBP1·InsP3Rsup and CaBP1·InsP3Rcore binary complexes were then aligned with respect to CaBP1. Candidate docked structures were selected that minimize any overlap between InsP3Rsup and InsP3Rcore while maintaining a reasonably close distance (<30 Å) between the final residue of InsP3Rsup and initial residue of InsP3Rcore.
A representative structure of the docked CaBP1·InsP3Rsup·InsP3Rcore complex is shown in Fig. 6. CaBP1-C interacts primarily with the arm helix in the suppressor domain (residues 72-94, colored brown in Fig. 6), suggested previously to interact with CaBP1 (8, 13, 14). This CaBP1-binding site on InsP3R1 is located far away from the ligand binding site, consistent with CaBP1 having no effect on the ligand-binding affinity (Fig. 5B). The exposed hydrophobic patch of Ca2+-bound CaBP1-C (Fig. 4C) interacts with both arm helices of InsP3Rsup. The aromatic rings of Phe72 and Trp73 (suppressor domain) contact the side chains of Ile144, Val148, and Met164 of CaBP1. Also, non-conserved CaBP1 residues (Leu132, His134, and Val148 highlighted red in Fig. 1) interact with the C-terminal arm helix that might help explain its highly specific binding to CaBP1 versus CaM. Indeed, the CaBP1 mutants (ΔL132, H134E, V148A) show ∼2-fold weaker binding to InsP3-(1-587) (Table 3). CaBP1 also makes a few contacts with residues in InsP3Rcore (residues 405-409) that are also close to InsP3Rsup helical arm residues, which might explain in part the cooperative interaction. Nearly all exposed residues on the C-terminal arm helix (Leu88, Lys91, His94, Ala95, Leu98, and Thr105) interact with exposed β-trefoil residues from InsP3Rcore, consistent with previous mutagenesis studies (55). The extensive domain interface predicted in Fig. 6 causes InsP3Rsup and InsP3Rcore to interact in a relatively compact arrangement, consistent with previous small-angle x-ray scattering measurements on InsP3R-(1-587) (35). We conclude that CaBP1 binding to the receptor may stabilize a structural interaction between InsP3Rsup and InsP3Rcore that might play a role in channel gating. This cooperative interdomain association appears to reciprocally stabilize the helical arm interaction with CaBP1 and therefore explain why InsP3Rsup and InsP3Rcore are both required for high affinity binding by CaBP1.
FIGURE 6.
Structural model of the docked complex for CaBP1-C·InsP3Rsup/InsP3Rcore. CaBP1-C, InsP3Rsup (residues 2-223), and InsP3Rcore (residues 236-586) are colored red, yellow, and cyan, respectively. Arm helix of InsP3Rcore interacting with CaBP1 is colored brown. The docking calculation was performed using Zdock as described under “Experimental Procedures.”
TABLE 3.
ITC fitting parameters for the binding of InsP3R-(1-587) to various molecules
| Molecule | Kd | ΔH | ΔS |
|---|---|---|---|
| μm | kcal mol−1 | cal mol−1 K−1 | |
| Mg2+-bound CaBP1 | 30 ± 3 | −1.55 ± 0.1 | 15.9 |
| Ca2+-bound CaBP1 | 3 ± 0.3 | −1.96 ± 0.2 | 17.9 |
| Mg2+-bound CaBP1-C | 30 ± 3 | −0.36 ± 0.1 | 21.7 |
| Ca2+-bound CaBP1-C | 2.5 ± 0.3 | −1.92 ± 0.2 | 19.3 |
| Ca2+-bound CaBP1-C (ΔL132) | 3.6 ± 0.3 | −2.22 ± 0.2 | 17.7 |
| Ca2+-bound CaBP1-C (H134E) | 5.6 ± 0.5 | −2.35 ± 0.2 | 16.5 |
| Ca2+-bound CaBP1-C (V148A) | 5.1 ± 0.4 | −2.03 ± 0.2 | 17.3 |
| Mg2+-bound CaBP1-N | -a | - | - |
| Ca2+-bound CaBP1-N | - | - | - |
| Apo-CaM | - | - | - |
| Ca2+-bound CaM | - | - | - |
| InsP3 | 1.9 ± 0.2 | −16 ± 0.4 | −27.5 |
| InsP3 (+CaBP1) | 1.6 ± 0.2 | −15 ± 0.6 | −24.7 |
Indicates no detectable binding.
DISCUSSION
In this study, we determined the NMR solution structures of CaBP1 in both Mg2+-bound and Ca2+-bound states and characterized their structural interaction with InsP3R1. The overall main chain structure of Mg2+-bound CaBP1 (Fig. 3, A and B) is similar to that seen previously in apo-CaM (45) and troponic C (48). One important difference is that Mg2+ is bound tightly at EF1 in CaBP1. The structure of Ca2+-bound CaBP1 is somewhat different from that of CaM (Fig. 4C). At saturating Ca2+ levels, the CaBP1 N-domain does NOT bind Ca2+ but remains in a closed conformation with Mg2+ bound at EF1. The C-domain binds Ca2+ at EF3 and EF4 and adopts the familiar Ca2+-bound open conformation (Fig. 3C) with an exposed hydrophobic patch (Fig. 4C). Many of the exposed hydrophobic residues in CaBP1 (Leu132, His134, Ile144, and Val148) are not conserved in CaM and might play a role in controlling the highly specific and Ca2+-induced binding to InsP3R1 to CaBP1 (Fig. 6). Indeed, the CaBP1 mutants (ΔL132, H134E, and V148A) show noticeably weaker binding to InsP3-(1-587) (Table 3).
Our target binding analysis indicates that Ca2+-bound CaBP1 binds tightly to InsP3R-(1-587) but does not bind to either InsP3Rsup or InsP3Rcore alone. These observations seem somewhat at odds with an earlier report, suggesting that CaBP1 can bind to isolated segments of InsP3Rsup independent of Ca2+ (14). Indeed, such binding of CaBP1 to InsP3Rsup is consistent with our proposed structural model of CaBP1-InsP3R (Fig. 6), showing that CaBP1 forms intimate contacts with the helical arm in InsP3Rsup. However, the affinity of CaBP1 binding to InsP3Rsup alone must be quite low, which would explain why this weak binding escaped detection in our ITC analysis (Fig. 5). Furthermore, we suggest that the affinity of CaBP1 binding to InsP3R1 is significantly enhanced by the cooperative interaction between InsP3Rsup and InsP3Rcore as depicted in Fig. 6. This same interaction also appears to partially block the ligand binding site, which may explain why InsP3 binds with ∼10-fold higher affinity to an isolated fragment of InsP3Rcore than it binds to InsP3R1 (52).
Previous studies have suggested that InsP3R1 binds to both the Ca2+-free and Ca2+-bound forms of CaBP1 (13, 14). In this study, we confirm that InsP3R-(1-587) does indeed bind to both the Mg2+-bound and Ca2+-bound CaBP1. However, our more quantitative ITC analysis reveals that Ca2+-bound CaBP1 binds to InsP3R-(1-587) with ∼10-fold higher affinity compared with that of Mg2+-bound CaBP1. Furthermore, the Mg2+-bound CaBP1 N-domain does NOT bind to InsP3R-(1-587) (Table 3). The lower affinity target binding by Mg2+-bound/Ca2+-free CaBP1 (C-domain) at low, basal Ca2+ levels might represent its binding to IQ-motifs in the receptor (12). Alternatively, we submit that the 10-fold stronger binding by Ca2+-bound CaBP1 may be sufficient to exclude InsP3R1 binding to Ca2+-free CaBP1 under physiological conditions. Thus, CaBP1 would selectively bind to InsP3R1 only when the cell is stimulated (at high cytosolic Ca2+ levels) and modulate Ca2+-dependent channel gating.
InsP3R-(1-587) binds to CaBP1 with at least 100-fold higher affinity than its binding to CaM. The highly selective binding to CaBP1 is explained in part by the large solvent-exposed surface area of the hydrophobic patch in CaBP1-C (Fig. 4C) as well as by a number of non-conserved residues on this surface (Fig. 1). Non-conserved CaBP1 residues (Leu132, His134, Ile144, and Val148) are proposed to make unique hydrophobic contacts with the helical arm of InsP3Rsup (Fig. 6). The highly specific binding of InsP3R-(1-587) to CaBP1 relative to CaM illustrates that CaBP1 is a specialized Ca2+ sensor for regulating InsP3Rs in the brain and retina. This contrasts with CaM that is ubiquitously expressed in all tissues and has a much broader range of target interactions. The specialized target binding by CaBP1 may be augmented by CaBP splice variants and isoforms that exhibit tissue-specific neuronal expression (19-21). We propose that the multiplicity of CaBPs in the central nervous system might play a role in fine tuning their interaction with various InsP3R isoforms and other Ca2+ channel targets.
CaBP1 has been suggested to promote channel opening in the absence of InsP3 (13). CaBP1 binds to InsP3R-(1-587) both in the presence or absence of InsP3, and CaBP1 has little or no effect on InsP3 binding to InsP3R-(1-587) (Fig. 5). Thus, CaBP1 interacts structurally with InsP3R1 even in the absence of InsP3. This is consistent with our docking analysis in which CaBP1-C interacts primarily with the helical arm region of the suppressor domain (Fig. 6), located far from the ligand-binding site. It is also possible that CaBP1 binding to apo-InsP3R1 induces structural interactions between InsP3Rsup and InsP3Rcore that may mimic structural changes caused by ligand-binding and thus explain the observed InsP3-independent channel opening.
Last, our structural studies suggest that the CaBP1 C-domain alone might be sufficient for promoting Ca2+-dependent channel activity because CaBP1-N does not bind to InsP3R-(1-587). However, the current study does not preclude the CaBP1 N-domain from interacting elsewhere in the channel. For example, Ca2+-dependent inactivation of L-type channels was shown recently to require separate binding by both the N-domain and C-domain of CaM (58). A similar bipartite interaction by the two domains of CaBP1 might also be important for regulation of InsP3R1. The CaBP1 N-domain might bind to either the central regulatory domain or the C-terminal cytosolic domain of InsP3R1. In the future, we plan to further investigate the functional interactions between InsP3R1 and CaBP1 by determining the atomic resolution structure of CaBP1/InsP3R-(1-587) and by exploring a possible role for the CaBP1 N-domain.
Acknowledgments
We are grateful to Dr. Jeff de Ropp for help with NMR experiments, Dr. Frits Abildgaard for providing NMR pulse-sequence programs, and Frank Delaglio for writing computer software for NMR data processing and analysis.
The atomic coordinates and structure factors (codes 2k7b, 2k7c, and 2k7d) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants EY012347 and NS045909 (to J. B. A.). This work was also supported by Grant RR11973 to the University of California Davis NMR Facility from the National Institutes of Health, the Canadian Institutes of Health Research and the Canada Foundation for Innovation (to M. I.), and a fellowship from the Canadian Institutes of Health Research (to J. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: InsP3R, inositol 1,4,5-trisphosphate receptor; CaBP1, calcium-binding protein 1; HSQC, heteronuclear single quantum coherence; HMQC, heteronuclear multiple quantum coherence; InsP3, inositol 1,4,5-trisphosphate; NOE, nuclear Overhauser effect; NOESY, NOE spectroscopy; GST, glutathione S-transferase; PDB, Protein Data Bank; ITC, isothermal titration calorimetry.
References
- 1.Berridge, M. J. (2003) Nat. Rev. Mol. Cell. Biol. 4 517-529 [DOI] [PubMed] [Google Scholar]
- 2.Berridge, M. J. (1993) Nature 361 315-325 [DOI] [PubMed] [Google Scholar]
- 3.Iino, M. (1990) J. Gen. Physiol. 95 1103-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor, C. W., and Laude, A. J. (2002) Cell Calcium 32 321-334 [DOI] [PubMed] [Google Scholar]
- 5.Finch, E. A., Turner, T. J., and Goldin, S. M. (1991) Science 252 443-446 [DOI] [PubMed] [Google Scholar]
- 6.Missiaen, L., Parys, J. B., Weidema, A. F., Sipma, H., Vanlingen, S., De Smedt, P., Callewaert, G., and De Smedt, H. (1999) J. Biol. Chem. 274 13748-13751 [DOI] [PubMed] [Google Scholar]
- 7.Berridge, M. J. (2002) Cell Calcium 32 235-249 [DOI] [PubMed] [Google Scholar]
- 8.Devogelaere, B., Verbert, L., Parys, J. B., Missiaen, L., and De Smedt, H. (2008) Cell Calcium 43 17-27 [DOI] [PubMed] [Google Scholar]
- 9.Foskett, J. K., White, C., Cheung, K. H., and Mak, D. O. (2007) Physiol. Rev. 87 593-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sienaert, I., Missiaen, L., De Smedt, H., Parys, J. B., Sipma, H., and Casteels, R. (1997) J. Biol. Chem. 272 25899-25906 [DOI] [PubMed] [Google Scholar]
- 11.Adkins, C. E., Morris, S. A., De Smedt, H., Sienaert, I., Torok, K., and Taylor, C. W. (2000) Biochem. J. 345 357-363 [PMC free article] [PubMed] [Google Scholar]
- 12.Michikawa, T., Hirota, J., Kawano, S., Hiraoka, M., Yamada, M., Furuichi, T., and Mikoshiba, K. (1999) Neuron 23 799-808 [DOI] [PubMed] [Google Scholar]
- 13.Yang, J., McBride, S., Mak, D. O., Vardi, N., Palczewski, K., Haeseleer, F., and Foskett, J. K. (2002) Proc. Natl. Acad. Sci. U. S. A 99 7711-7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasri, N. N., Holmes, A. M., Bultynck, G., Parys, J. B., Bootman, M. D., Rietdorf, K., Missiaen, L., McDonald, F., De Smedt, H., Conway, S. J., Holmes, A. B., Berridge, M. J., and Roderick, H. L. (2004) EMBO J. 23 312-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White, C., Yang, J., Monteiro, M. J., and Foskett, J. K. (2006) J. Biol. Chem. 281 20825-20833 [DOI] [PubMed] [Google Scholar]
- 16.Schlecker, C., Boehmerle, W., Jeromin, A., DeGray, B., Varshney, A., Sharma, Y., and Ehrlich, B. E. (2006) J. Clin. Investig. 116 1668-1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haeseleer, F., Sokal, I., Verlinde, C. L., Erdjument, H., Tempst, P., Pronin, A. N., Benovic, J. L., Fariss, R. N., and Palczewski, K. (2000) J. Biol. Chem. 275 1247-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikura, M. (1996) Trends Biochem. Sci. 21 14-17 [PubMed] [Google Scholar]
- 19.Haynes, L. P., Tepikin, A. V., and Burgoyne, R. D. (2004) J. Biol. Chem. 279 547-555 [DOI] [PubMed] [Google Scholar]
- 20.Menger, N., Seidenbecher, C. I., Gundelfinger, E. D., and Kreutz, M. R. (1999) Cell Tissue Res. 298 21-32 [DOI] [PubMed] [Google Scholar]
- 21.Seidenbecher, C. I., Reissner, C., and Kreutz, M. R. (2002) Adv. Exp. Med. Biol. 514 451-463 [DOI] [PubMed] [Google Scholar]
- 22.Laube, G., Seidenbecher, C. I., Richter, K., Dieterich, D. C., Hoffmann, B., Landwehr, M., Smalla, K. H., Winter, C., Bockers, T. M., Wolf, G., Gundelfinger, E. D., and Kreutz, M. R. (2002) Mol. Cell Neurosci. 19 459-475 [DOI] [PubMed] [Google Scholar]
- 23.Lee, A., Westenbroek, R. E., Haeseleer, F., Palczewski, K., Scheuer, T., and Catterall, W. A. (2002) Nat. Neurosci. 5 210-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou, H., Yu, K., McCoy, K. L., and Lee, A. (2005) J. Biol. Chem. 280 29612-29619 [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita-Kawada, M., Tang, J., Xiao, R., Kaneko, S., Foskett, J. K., and Zhu, M. X. (2005) Pflugers Arch. 450 345-354 [DOI] [PubMed] [Google Scholar]
- 26.Haeseleer, F., Imanishi, Y., Maeda, T., Possin, D. E., Maeda, A., Lee, A., Reike, F., and Palczewski, K. (2004) Nat. Neurosci. 7 1079-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babu, Y. S., Bugg, C. E., and Cook, W. J. (1988) J. Mol. Biol. 204 191-204 [DOI] [PubMed] [Google Scholar]
- 28.Wingard, J. N., Chan, J., Bosanac, I., Haeseleer, F., Palczewski, K., Ikura, M., and Ames, J. B. (2005) J. Biol. Chem. 280 37461-37470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brocard, J. B., Rajdev, S., and Reynolds, I. J. (1993) Neuron 11 751-757 [DOI] [PubMed] [Google Scholar]
- 30.Osawa, M., Dace, A., Tong, K. I., Valiveti, A., Ikura, M., and Ames, J. B. (2005) J. Biol. Chem. 280 18008-18014 [DOI] [PubMed] [Google Scholar]
- 31.Yamniuk, A. P., Nguyen, L. T., Hoang, T. T., and Vogel, H. J. (2004) Biochemistry 43 2558-2568 [DOI] [PubMed] [Google Scholar]
- 32.Cox, J. A., Durussel, I., Comte, M., Nef, S., Nef, P., Lenz, S. E., and Gundelfinger, E. D. (1994) J. Biol. Chem. 269 32807-32813 [PubMed] [Google Scholar]
- 33.Schug, Z. T., and Joseph, S. K. (2006) J. Biol. Chem. 281 24431-24440 [DOI] [PubMed] [Google Scholar]
- 34.Varnai, P., Balla, A., Hunyady, L., and Balla, T. (2005) Proc. Natl. Acad. Sci. U. S. A 102 7859-7864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan, J., Whitten, A. E., Jefferies, C. M., Bosanac, I., Mal, T. K., Ito, J., Porumb, H., Michikawa, T., Mikoshiba, K., Trewhella, J., and Ikura, M. (2007) J. Mol. Biol. 373 1269-1280 [DOI] [PubMed] [Google Scholar]
- 36.Li, C., and Ames, J. B. (2007) Biomol. NMR Assign. 1 77-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, C., and Ames, J. B. (2008) Biomol. NMR Assign. 2 61-63 [DOI] [PubMed] [Google Scholar]
- 38.Schwieters, C. D., Kuszewski, J. J., Tjandra, N., and Clore, G. M. (2003) J. Biol. Chem. 278 37461-37470 [Google Scholar]
- 39.Badger, J., Kumar, R. A., Yip, P., and Szalma, S. (1999) Proteins 35 25-33 [PubMed] [Google Scholar]
- 40.Finley, N. L., Howarth, J. W., and Rosevear, P. R. (2004) Biochemistry 43 11371-11379 [DOI] [PubMed] [Google Scholar]
- 41.Chen, R., Li, L., and Weng, Z. (2003) Protens 52 80-87 [DOI] [PubMed] [Google Scholar]
- 42.Chang, S. L., Szabo, A., and Tjandra, N. (2003) J. Am. Chem. Soc. 125 11379-11384 [DOI] [PubMed] [Google Scholar]
- 43.Bourne, Y., Dannenberg, J., Pollmann, V. V., Marchot, P., and Pongs, O. (2001) J. Biol. Chem. 276 11949-11955 [DOI] [PubMed] [Google Scholar]
- 44.Gentry, H. R., Singer, A. U., Betts, L., Yang, C., Ferrara, J. D., Sondek, J., and Parise, L. V. (2005) J. Biol. Chem. 280 8407-8415 [DOI] [PubMed] [Google Scholar]
- 45.Zhang, M., Tanaka, T., and Ikura, M. (1995) Nat. Struct. Biol. 2 758-767 [DOI] [PubMed] [Google Scholar]
- 46.Kuboniwa, H., Tjandra, N., Grzesiek, S., Ren, H., Klee, C. B., and Bax, A. (1995) Nat. Struct. Biol. 2 768-776 [DOI] [PubMed] [Google Scholar]
- 47.Finn, B. E., Evenas, J., Drakenberg, T., Waltho, J. P., Thulin, E., and Forsen, S. (1995) Nat. Struct. Biol. 2 777-783 [DOI] [PubMed] [Google Scholar]
- 48.Herzberg, O., and James, M. N. (1988) J. Mol. Biol. 203 761-779 [DOI] [PubMed] [Google Scholar]
- 49.Andersson, M., Malmendal, A., Linse, S., Ivarsson, I., Forsen, S., and Svensson, L. A. (1997) Protein Sci. 6 1139-1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malmendal, A., Evenas, J., Thulin, E., Gippert, G. P., Drakenberg, T., and Forsen, S. (1998) J. Biol. Chem. 273 28994-29001 [DOI] [PubMed] [Google Scholar]
- 51.Moncrief, N. D., Kretsinger, R. H., and Goodman, M. (1990) J. Mol. Evol. 30 522-562 [DOI] [PubMed] [Google Scholar]
- 52.Iwai, M., Michikawa, T., Bosanac, I., Ikura, M., and Mikoshiba, K. (2007) J. Biol. Chem. 282 12755-12764 [DOI] [PubMed] [Google Scholar]
- 53.Chen, R., and Weng, Z. (2002) Proteins 47 281-294 [DOI] [PubMed] [Google Scholar]
- 54.Bosanac, I., Alattia, J. R., Mal, T. K., Chan, J., Talarico, S., Tong, F. K., Tong, K. I., Yoshikawa, F., Furuichi, T., Iwai, M., Michikawa, T., Mikoshiba, K., and Ikura, M. (2002) Nature 420 696-700 [DOI] [PubMed] [Google Scholar]
- 55.Bosanac, I., Yamazaki, H., Matsu-Ura, T., Michikawa, T., Mikoshiba, K., and Ikura, M. (2005) Mol. Cell 17 193-203 [DOI] [PubMed] [Google Scholar]
- 56.Patterson, R. L., van Rossum, D. B., Barrow, R. K., and Snyder, S. H. (2004) Proc. Natl. Acad. Sci. U. S. A 101 2328-2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoeflich, K. P., and Ikura, M. (2002) Cell 108 739-742 [DOI] [PubMed] [Google Scholar]
- 58.Dick, I. E., Tadross, M. R., Liang, H., Tay, L. H., Yang, W., and Yue, D. T. (2008) Nature 451 830-834 [DOI] [PMC free article] [PubMed] [Google Scholar]