Abstract
The T-box gene Eomesodermin (Eomes) is required for early embryonic mesoderm differentiation in mouse, frog (Xenopus laevis), and zebrafish, is important in late cardiac development in Xenopus, and for CD8+ T effector cell function in mouse. Eomes can ectopically activate many mesodermal genes. However, the mechanism by which Eomes activates transcription of these genes is poorly understood. We report that Eomes protein interacts with Smad2 and is capable of working in a non-cell autonomous manner via transfer of Eomes protein between adjacent embryonic cells. Blocking of Eomes protein transfer using a farnesylated red fluorescent protein (CherryF) also prevents Eomes nuclear accumulation. Transfer of Eomes protein between cells is mediated by the Eomes carboxyl terminus (456-692). A carbohydrate binding domain within the Eomes carboxyl-terminal region is sufficient for transfer and important for gene activation. We propose a novel mechanism by which Eomes helps effect a cellular response to a morphogen gradient.
The T-box genes represent a large multigene family of DNA binding transcription factors, conserved throughout metazoan evolution and important during embryonic development and in adult physiology (1-7). Eomes,4 first discovered in Xenopus, is a novel T-box transcription factor of the T-brain subfamily, and important in multiple tissues and germ layers during embryogenesis (8-16). Ectopic overexpression of Eomes in blastula stage animal pole explants (9, 17) (animal caps) is sufficient to activate transcription of a number of mesodermal, mesendodermal, and endodermal genes (3, 9, 17-20). In Xenopus and zebrafish, Eomes is necessary for normal mesendoderm development, because its suppression in these lineages blocked mesoderm differentiation and gastrulation (9, 18, 19), and endoderm induction (20). In mouse, Eomes (12, 14, 21) is required for the formation of mesoderm, including the node and primitive streak (8, 12), and for specification of definitive endoderm (22). The embryonic node in mouse and chick is thought to be the equivalent of Spemann's Organizer region in Xenopus. Homograft transplantation of the dorsal equatorial region (Organizer) to an ectopic ventral location in early amphibian embryos results in formation of a secondary body axis (23). Zebrafish Eomes (zf-eom) requires Nodal signaling for the specification of the organizer/shield (19), but not for endoderm induction (20). Although direct protein transfer between cells has been reported previously (24-26), the present report is the first to ascribe this property to a T-box transcription factor. Our results show Eomes protein is capable of direct transduction via protein transfer between adjacent cells. Protein transduction is mediated by the Eomes carboxyl terminus, within which a carbohydrate binding domain is sufficient to confer the capability of cell to cell protein transfer to several fluorescent protein tags. Preventing the movement of Eomes protein from cell to cell using CherryF also prevents nuclear accumulation of Eomes. We discuss the implications of our results for embryonic cellular response to morphogen gradient signaling.
EXPERIMENTAL PROCEDURES
Construction of cDNA Plasmids—For DNxSmad2 P445H, polymerase chain reaction was performed using cloned Xenopus Smad2 cDNA as a template (27-29). Primers P399 (forward, 5′-GATCAACTTAAGATGTCGTCCATCTTCATCTTC-3′) and P406 (reverse, 5′-CTTTGTCCAACCACTGCAAGTGTCCA-3′) were used to obtain a 5′Smad2 P445H fragment of 1365 bp. Primers P405 (forward, 5′-GAGCTTCACCTGAATGGACACTTGCAGTG-3′) and P402 (reverse, 5′-GATGGTAGGGGGCGGCTACTTATC-3′) generated a 3′Smad2 P445H cDNA fragment of 135 bp. These two overlapping PCR products, containing the P445H mutation in the overlapping region, were then used with P399 plus P405 in a third “splice” PCR to obtain the full-length P445H sequence (1461 bp). The 1461-bp fragment was restriction digested and cloned 5′ AflII-3′ NotI in pKmRN3P (10). The resulting cDNA plasmid, DNxS2P445H/pKmRN3P, was linearized using SfiI and synthetic capped mRNA was synthesized as previously described (9) using a Megascript T3 kit (Ambion). GSM series cDNAs were synthesized in similar splice-PCRs using mutated forward and reverse oligonucleotides in which each 18-base window encoding 6 glycines was flanked 5′ and 3′ with 18 bases of the appropriate wild type Eomes cDNA sequence. Deletion series cDNAs were performed similarly but with the region to be deleted missing between the two 18-base flanks of Eomes cDNA.
DNxS2 D450E cDNA was prepared as for P445H but using primer pairs P407 (forward, 5′-GACCTTTGCAGTGGTTGGAAAAAGTGTTGACACAGATG-3′) plus P402 (reverse, 5′-GATGGTAGGGGGCGGCTACTTATC-3′) to obtain a 3′Smad2 D450H fragment (119 bp); and P399 plus P408 (reverse, 5′-CATCTGTGTCAACACTTTTTCCAACCACTGCAAAGGTC-3′) to obtain a 5′Smad2 D450E fragment (1380 bp).
To synthesize the GR-Eomes cDNA plasmid, the human glucocorticoid ligand binding domain cDNA (GR) was PCR-amplified as a SmaI fragment, using as a template the cDNA plasmid pSP64T Xbra-GR (30) and primer 5′SR-GR (forward, 5′-GCCTACCCCGGGACAGCCACCATGTCTGAAAATCCTGGTAACAAAACAA-3′) plus primer 3′SP-GR (reverse, 5′-GGATTACCCGGGAGGTCCACCACCACCAGGCTTTTGATGAAACAGAAGTTTTTTGA-3′). The GR PCR product was digested overnight with SmaI, and ligated into vector plasmid pBluescript RN3P (9), creating the plasmid p3′HGR (10). p3′HGR contains a Kozak sequence at its 5′ end (ACAGCCACCATG, including initiator methionine; underlined in 5′SR-GR), and a “PG4P hinge” region at its 3′ end, in-frame with the GR cDNA (9, 10) (underlined in 3′SP-GR). The PG4P hinge would break α helices (prolines) between adjacent polypeptide domains and thereby potentially facilitate independent folding of the two fused proteins. The Eomes cDNA (9) (GenBank™ accession number U75996) was PCR-amplified as a 5′ BamHI-3′ NotI fragment (forward primer 5′BEO, 5′-GCCTACGGATCCATGGTGCCTGGCGCCTGG-3′; reverse primer 3′NEO, 5′-GCATACGGCGGCCGCAGAACTAGAGTAGAAAGAGTAATACCCAAGTCC-3′), digested overnight with BamHI and NotI, and cloned into p3′HGR downstream of and in-frame with GR, creating plasmid HGR-Eomes/RN3P. For GR-Eomes synthetic mRNA, HGR-Eomes/RN3P was linearized with SfiI and transcribed using a T3 Megascript kit (Ambion). Inserts of all synthetic cDNA plasmids were sequenced for verification.
Xenopus Embryo Injection and Culture—Xenopus embryos were staged according to Ref. 31. Egg laying, fertilization, and embryo culture were performed as described previously (9), except that culture media contained 5 μg/ml Gentamycin (Invitrogen). After de-jellying in 2% cysteine hydrochloride (pH 8.0; Sigma), fertilized eggs were placed in 1× modified Barth's saline (MBS) containing 2% Ficoll (Sigma) for 6 min prior to microinjection. Synthetic mRNA was injected at the 2- to 8-cell stage into the animal pole of both blastomeres (9.2 nl/embryo; for co-immunoprecipitation (IP), 9.2 ng of Eomes or Eomes mutant mRNA) using a Drummond Nanoject II microinjector, and embryos were harvested at stage 10.5. Two hours after injection, embryos were transferred to 0.1× MBS. For animal cap assays, caps were dissected in 1× MBS at stage 8 and cultured to stage 10.5, frozen in 1× MBS on dry ice, and stored at -80 °C.
RNA Preparation from Xenopus laevis Animal Caps—Total RNA was isolated from whole embryos and animal caps using the phenol/NETS methods of Sambrook et al. (32) as previously described (9) with the following changes. The proteinase K digestion and LiCl precipitation steps were omitted, and the composition of the NETS homogenization buffer was 0.3 m NaCl, 1 m EDTA, 50 mm Tris (pH 7.5), 1% SDS. Cellular DNA was removed by treatment with RQ1 DNase I (Promega) for 1 h at 37 °C followed by phenol extraction.
RNase Protection Assay—RNase protection assays were performed as previously described (9).
cDNA Synthesis and PCR (RT-PCR)—cDNA was synthesized for 1 h at 50 °C using a ThermoScript kit (Invitrogen) according to the manufacturer's instructions using 1-2 μg of total RNA as template and an oligo(dT) primer. The cDNA was diluted 5-fold with TE (pH 7.5) before PCR. PCR was performed using 2 μl of 5-fold diluted cDNA template plus 0.2 μl of Taq DNA polymerase (Qiagen); heated to 95 °C for 3 min (1 cycle); then thermally cycled at 95 °C for 30 s, 55 °C for 1 min, 68 °C for 1 min (30 cycles). Gene-specific primer pairs were designed based on published sequence data (GenBank). Primer sequences are shown in Table 1. All PCR products were sequenced for verification. All experiments were repeated at least three times.
TABLE 1.
PCR primers for gene expression analysis
| Gene | Primer | Sequence 5′-3′ | Amplicon | Location |
|---|---|---|---|---|
| bp | ||||
| Eomes | P551 | TAAAGAAAGGTTAATATGCTG | 505 | cra |
| P552 | GTACATTGGCTTTTTGTGCC | cra | ||
| Gsc | P474 | CCTTCTGTTTATTGTAGTAATTCC | 305 | 3Ub |
| P475 | CAGAAAATGCCCAAAAGTGG | 3U | ||
| Chd | P476 | CCGGACCCACTCAAAATACC | 301 | 3U |
| P477 | CGTCTGATCTGTAGAGTTTCGC | 3U | ||
| Xwnt8 | P478 | GGATGCAATGCAAAGAATG | 202 | 3U |
| P479 | GTGTAGAGATTTCTACACCGCAG | 3U | ||
| Mix.1 | P480 | CACACAGCTGAAGGGTTAAATTC | 300 | 3U |
| P481 | GTGACACCTCCCCAGAGC | 3U | ||
| Sox17β | P486 | TATAGTATAGAAATCCTAGTACATTTA | 205 | 3U |
| P487c | GTGGCGTATAAAGCACTTG | 3U | ||
| Xnr1 | P490 | CAACAAAGCCAAGGCATAAC | 229 | 3U |
| P491 | ATGATTTTACTGGCCTACAAAAC | 3U | ||
| Xnr2 | P492 | CTGAATTAATTGAGATATACGTGC | 192 | 3U |
| P493 | CAATATTTTATTGAGTGC | 3U | ||
| Xnr5 | P498 | GAGTCTTCTACATTAAGCACAACAG | 326 | 3U |
| P499 | GTCCTTCTTTATTGTTAAAGC | 3U | ||
| myoD | P502 | GGAAGCATCCGGAGGATTC | 303 | 3U |
| P503 | GGATTCTGCCATATTTCCAATG | 3U | ||
| Myf5 | P504 | GAAAGAAAACAGCATGTTTATTAACA | 260 | 3U |
| P505d | GTGTTATAAGCCTCTCTTAATTCC | 3U | ||
| Xbra | P531 | CATACTTTCTCTCCCCGT | 300 | 3U |
| P532 | GAAGGTCTGCTTCTTTTTTAC | 3U | ||
| FGF-R | 5FR | CAGTATACCTGCTTGGCC | 500 | 3U |
| 3FR | GTTAGGCTTCTCCTTGTC | 3U |
cr, protein coding region.
3′-Untranslated region.
Including the last 3 codons of the protein coding region.
Including the last codon of the protein coding region.
Anti-Eomes Antisera—The Xenopus Eomesodermin cDNA (GenBank U75996) was PCR-amplified and fused in-frame to six histidines (His) by cloning 5′ BamHI-3′ HindIII into the pQE-31 vector (Qiagen number 32915). His6-tagged Eomes fusion proteins were produced by overexpression in bacteria and purification on nickel-nitrilotriacetic acid resin (Qiagen number R10-22-40-42/43) according to the manufacturer's instructions with modifications (see below). Purified His6-Eomes protein was sonicated in Complete Freund's adjuvant and used to inoculate New Zealand White rabbits. Antisera were tested by Western blotting against bacterially produced His6-Eomes. The antiserum directed against the Eomes NH2 terminus (amino acid residues 1-214) has been described previously (33). Antiserum to the Eomes central DNA-binding region (215-455) and COOH terminus (456-684) were also generated. NH2- and COOH-terminal directed Eomes antibodies were affinity purified commercially from antisera against bacterially produced Eomes antigen (Sigma Genosys).
Recombinant His6-Eomes protein was overexpressed in Escherichia coli strain SG13009 (pREP4): one single colony was inoculated and grown in 20 ml of LB broth containing 100 μg/ml ampicillin and 25 μg/ml kanamycin at 28 °C overnight with agitation. One liter of LB containing ampicillin, kanamycin, and 0.2% glucose, was inoculated 1:50 with the overnight culture and grown 2-3 h at 28 °C to A600 = 0.6. His6-Eomes protein overexpression was induced by addition of isopropyl 1-thio-β-d-galactopyranoside to 2 mm and culture continued at 28 °C for 4 h. Bacteria were harvested by centrifugation, and resuspended in buffer A (100 mm NaH2PO4, 10 mm Tris-HCl, 6 m guanidine hydrochloride, Invitrogen number 15502-016), pH 8.0). Cells were lysed by magnetic stirring for 1 h at room temperature. Protein extract was cleared by centrifugation at 10,000 × g for 30 min at room temperature and applied to nickel-nitrilotriacetic acid resin. The resin was washed twice in Buffer C (100 mm NaH2PO4, 10 mm Tris-HCl, 8 m guanidine hydrochloride, pH 6.3) and His6-Eomes protein was eluted in Buffer E (100 mm NaH2PO4, 10 mm Tris-HCl, 8 m guanidine hydrochloride, pH 4.5).
Immunoprecipitation and Western Blot Analysis—Immunoprecipitation (IP) and Western blot analysis were performed as described previously (33, 34), except that okadaic acid was not used in the IP buffer (34); extracts were pre-cleared 1 h (h) at 4 °C with agitation using 10 μl of empty Protein G-Sepharose beads (GE Healthcare, 17-0618-01); IP antibody (0.8 μg) was allowed to bind overnight with 100 μl of embryo extract at 4 °C with agitation, then 5 μl of pre-washed empty beads were added and allowed to bind 2 h at 4 °C; after boiling beads in Laemmli sample buffer, before loading gels, excess IgG was removed from buffer by incubating with 10 μl of empty beads for 2 h at 25 °C and 1400 rpm in a Thermomixer R (Eppendorf); and the Protein G band was reduced on blots after incubation in protein A-horseradish peroxidase by washing three times for 10 min plus once for 2 h in TBST (140 mm NaCl, 10 mm Tris-HCl, pH 7.5, 0.1% Tween 20 (Bio-Rad)). Antibodies were diluted 1:1000 in TBST containing 5% milk and allowed to bind to polyvinylidene difluoride (Hybond-P, Amersham, RPN303F) Western blot membrane at 4 °C overnight. Other reagents used were: Smad2 antibody (Upstate/Millipore, 07-408), anti-GFP (Sigma, G6539); recombinant protein A conjugated to horseradish peroxidase (Pierce, 32400, 1:5000); ECL plus reagent (GE Healthcare/Amersham, RPN2132).
Pre-adsorption of the Anti-NH2-terminal Eomes Antibody with Eomes Antigen—Eomes antigen-coupled Sepharose 4B affinity columns were prepared, and Eomes antibodies were affinity purified, by Sigma Genosys. A BSA coupling reaction was performed as for the Eomes NH2-terminal antigen to prepare negative control BSA-coupled Sepharose 4B beads. 0.8 g of CNBr-activated Sepharose 4B (GE Healthcare) was hydrated on a sintered glass filter by washing with 1 mm HCl, then coupling buffer (0.1 m NaHCO3, 0.25 m NaCl, pH 8.5). For the coupling reaction, 18 mg of BSA (Sigma Fraction V) was dissolved in coupling buffer, mixed with the gel suspension, and incubated with mixing for 2 h (h) at room temperature. After coupling of BSA to the Sepharose, any remaining active groups were quenched for 2 h at room temperature in 0.2 m glycine (Bio-Rad) (pH 8.1) using 1:10 (v/v) of gel to buffer. The BSA-coupled Sepharose 4B (BSA-Sepharose) was washed four times with coupling buffer, then with 0.1 m sodium acetate (pH 4.3) (HCl) containing 0.5 m NaCl. BSA-Sepharose was pre-equilibrated using 1× phosphate-buffered saline (pH 7.7) prior to addition of the anti-Eomes antibody. One ml of a 50% slurry of BSA-Sepharose, or Eomes-Sepharose (Eomes amino acid residues 1-214; Sigma Genosys) was incubated with 5 ml of the affinity-purified anti-Eomes NH2-terminal antibody (1.2 mg/ml) for 4 h at 4 °C. As a negative control, buffer alone was incubated with Sepharose (no added antibody). Supernatant was recovered after centrifugation of beads at 3000 × g at 4 °C for 2 min and used to probe Western blots.
Confocal Microscopy—Confocal microscopy of Xenopus animal pole explants was performed using a Leica DMIRE2 confocal microscope and Leica confocal software, within the Pathology Core Facility of The Children's Hospital of Philadelphia.
RESULTS
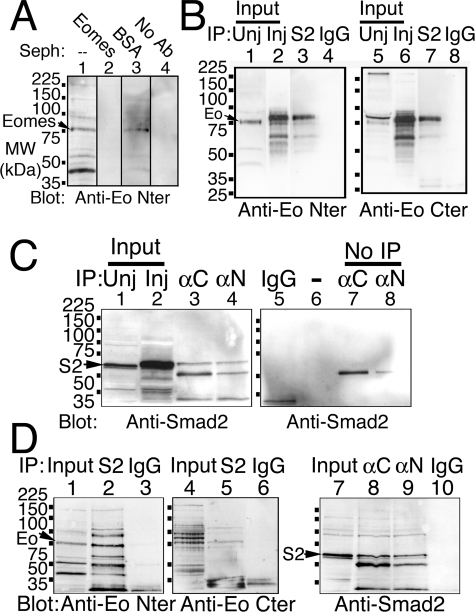
An Anti-Eomes Antibody Recognizes the Eomes Protein—To examine the molecular mechanism by which Eomes activates downstream target genes, we wished to identify candidate protein partners for Eomes. Antisera were raised against the Xenopus Eomes amino (NH2) terminus (33) and in parallel, against its central DNA-binding region and carboxyl (COOH) terminus (this report) by overexpression of histidine (His6-) tagged Eomes protein in bacteria. The NH2- and COOH-terminal antisera were affinity-purified on Eomes-Sepharose 4B columns. Both of the resulting anti-Eomes antibody preparations recognized their respective bacterially produced antigens on Western blots (data not shown); and also recognized an endogenous polypeptide in Xenopus embryo extracts of the expected size for Eomes protein (76 kDa) (33) (Fig. 1A, lane 1). Pre-adsorption of the Eomes anti-NH2-terminal antibody with Eomes NH2-terminal antigen coupled to Sepharose 4B (Eomes-Sepharose) completely prevented its recognition of the 76-kDa Eomes protein band on Western blots (Fig. 1A, lane 2). Pre-adsorption with Eomes-Sepharose also prevented detection of several other polypeptides (Fig. 1A, compare lanes 1 and 2). In contrast, pre-adsorption with BSA-Sepharose failed to block recognition of Eomes but did deplete the other non-Eomes polypeptides (Fig. 1A, lane 3). Protein A-horseradish peroxidase failed to recognize any polypeptides in the absence of added primary antibody (Fig. 1A, lane 4). These data suggest that our purified anti-Eomes antisera specifically recognized the 76-kDa Eomes protein. As detection of other polypeptides was depleted with BSA, these are unrelated to Eomes.
FIGURE 1.
Eomes protein interacts with Smad2. A, the anti-Eomes NH2-terminal antibody is specifically blocked by addition of the Eomes NH2-terminal antigen. Lysates from stage 10.5 embryos were left untreated (lanes 1 and 4), or immunodepleted by incubation with Sepharose that had been covalently cross-linked to the Eomes NH2-terminal antigen (lane 2) or bovine serum albumin (lane 3); immunodepleted lysates were Western blotted with the anti-Eomes NH2-terminal antibody (lanes 1-3) or no antibody (lane 4). B, overexpressed Eomes and Smad2 interact in vivo in Xenopus embryos. Lysates from uninjected stage 10.5 embryos (lanes 1, and 5), or embryos that had been injected with mRNAs for Eomes plus Smad2 (lanes 2 and 6), were immunoprecipitated with antibodies to Smad2 (lanes 3 and 7), or an irrelevant GFP antibody (lanes 4 and 8); immunoprecipitates were Western blotted with the Eomes NH2-terminal (lanes 1-4) or COOH-terminal (lanes 5-8) antibodies. C, overexpressed Smad2 and Eomes interact in vivo in Xenopus embryos. Smad2 and Eomes were coexpressed in Xenopus embryos. Uninjected extract (lane 1) or Eomes antibodies alone (lanes 7 and 8) are shown for comparison; co-injected extracts (lane 2) were immunoprecipitated with anti-Eomes antibodies (anti-COOH terminus, lane 3; anti-NH2 terminus, lane 4), or an irrelevant GFP antibody (lane 5); immunoprecipitates were Western blotted with an anti-Smad2 antibody. (No protein was loaded in lane 6.) D, endogenous Eomes and Smad2 interact in vivo in Xenopus embryos. Extracts (lanes 1, 4, and 7) were immunoprecipitated with anti-Smad2 antibody (lanes 2 and 5), anti-Eomes antibodies (lanes 8 and 9), or a GFP antibody (lanes 3, 6, and 10); immunoprecipitates were Western blotted using anti-Eomes antibodies (lanes 1-6) or anti-Smad2 antibody (lanes 7-10).
Eomes Protein Interacts Physically with Smad2—Eomes required both transforming growth factor-β signaling and phosphorylated Smad2 to activate several of its downstream target genes (data not shown). Thus Eomes protein could have cooperated with Smad2 by physically associating with phosphorylated Smad2 as a transcriptional co-activator. To test this, we overexpressed Xenopus Eomes plus Smad2 in animal caps. IP of Smad2 co-precipitated Eomes protein from caps in which both Eomes and Smad2 had been overexpressed (Fig. 1B, lanes 3 and 7). Overexpressed Eomes protein (Fig. 1B, lanes 2, 3, 6, and 7) is myc-tagged and therefore of slightly greater mass than endogenous Eomes (Fig. 1B, lanes 1 and 5; myc-Eomes migrates more slowly than expected; see below and Fig. 3L). A similar result was obtained using either of our two affinity purified antibodies, one against the Eomes NH2 terminus (Fig. 1B, lane 3), the other against its COOH terminus (Fig. 1B, lane 7). Conversely, immunoprecipitation of Eomes using either of these two antibodies co-precipitated phosphorylated Smad2 in similarly injected caps (Fig. 1C, lanes 3 and 4), whereas a GFP antibody failed to precipitate Eomes (Fig. 1B, lanes 4 and 8) or Smad2 (Fig. 1C, lane 5). These interactions in injected embryo extracts were confirmed in uninjected Xenopus gastrulae, with immunoprecipitation of either Eomes or Smad2 able to co-precipitate the other partner (Fig. 1D, lanes 2, 5, 8, and 9). We conclude that Eomes and Smad2 form a functional transcription factor complex to activate several mesodermal genes during Xenopus embryonic development.
FIGURE 3.
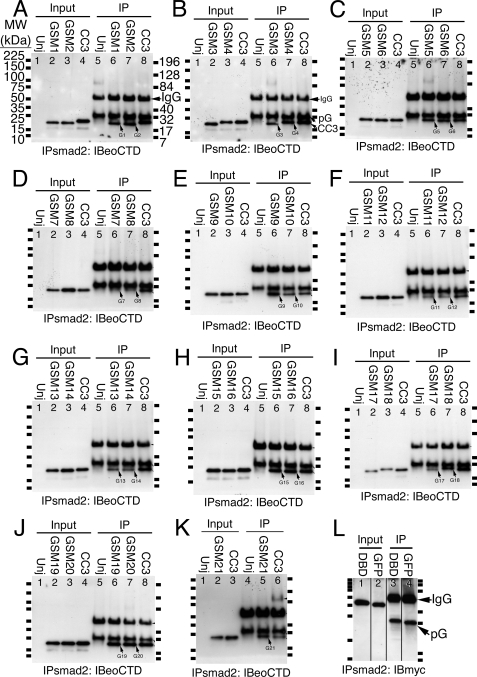
The Eomes carboxyl-terminal region contains a carbohydrate binding domain. A, GSM1 and GSM2 bind to Sepharose beads. Eomes glycine-scanning mutant GSM1, GSM2, or CC3, were overexpressed in Xenopus embryos. Extracts were left untreated (lanes 1-4) or immunoprecipitated (lanes 5-8) with an anti-Smad2 antibody and Western blotted with anti-Eomes COOH-terminal antibodies. Uninjected extracts are shown for comparison (Unj, lanes 1 and 5). (GSM1, -2 lack a myc epitope tag.) B, GSM3 was slightly diminished, GSM4 binds to beads. (GSM3 lacks myc.) C, GSM5, 6 bind beads. D, GSM7, 8 failed to bind beads. E, GSM9 was slightly increased, GSM10 binds beads. F, GSM11, 12 failed to bind beads. G, GSM13, 14 failed to bind beads. H, GSM15, 16 bind beads. I, GSM17 failed to bind beads; GSM18 was diminished. GSM18 migrated slower than expected on gels. J, GSM19, 20 bind beads. K, GSM21 was slightly increased in binding to beads. L, myc-tagged DBD or GFP failed to bind to beads. Eomes DBD or GFP was overexpressed in Xenopus embryos. Extracts were left untreated (lanes 1 and 2) or immunoprecipitated (lanes 3 and 4) with an anti-Smad2 antibody and Western blotted with an anti-myc epitope antibody. (The myc tag caused anomalously slow gel migration of DBD and GFP.)
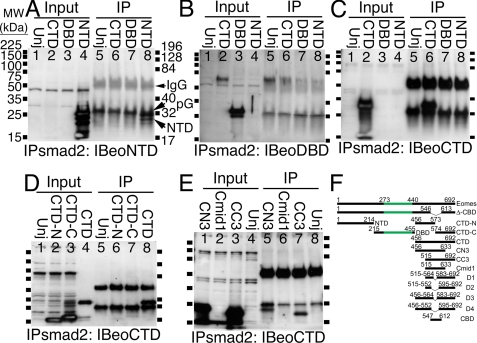
The COOH-terminal Domain of Eomes Binds to Agarose Polysaccharide Beads—To investigate the physical interaction between Eomes and Smad2 in more detail, we attempted to map within Eomes the location of its affinity surface for Smad2. Eomes was divided into three pieces: 1) an NH2-terminal domain (Met1-Tyr214, NTD); 2) a central, DNA binding domain containing the T-box (Ser215-Asp455, DBD); and 3) a COOH-terminal domain (Arg456-Ser692, CTD). mRNA encoding NTD, DBD, or CTD was microinjected into the animal pole of Xenopus embryos, embryos were frozen at stage 10.5 and subjected to co-immunoprecipitation analysis using an anti-Smad2 antibody for the immunoprecipitation, and probing the Western blot for Eomes. We found that anti-Smad2 beads bound poorly to NTD (Fig. 2A, lane 8) and not at all to DBD (Fig. 2B, lane 7). In contrast, the beads bound robustly to CTD (Fig. 2C, lane 6). Dividing CTD in half virtually eliminated binding (Fig. 2D, lanes 6 and 7). Removing a 60-amino acid NH2- or COOH-terminal segment from CTD (called CC3, CN3, respectively) both retained robust binding but eliminated one antibody epitope each (Cmid1 was not recognized by the anti-CTD antibody; Fig. 2, E and F). These results suggested that there existed an affinity site in the middle region of CTD, because binding activity was prevented by halving of CTD between Asp573 and Ala574, but not by removal of 60 amino acids from either end.
FIGURE 2.
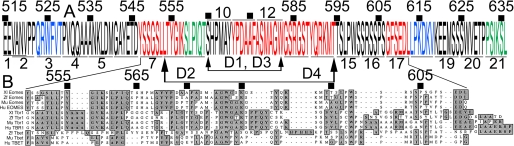
Thecentral region of the Eomes CTD interacts with Sepharose beads. A, the Eomes NTD largely failed to bind to Sepharose. The Eomes NTD, DBD, or CTD were overexpressed in Xenopus embryos. Extracts were left untreated (lanes 1-4) or immunoprecipitated (lanes 5-8) with an anti-Smad2 antibody and Western blotted with anti-Eomes NH2-terminal antibodies. Uninjected extracts are shown for comparison (Unj, lanes 1 and 5). (Central clearing of some input bands was remediated by reducing input.) B, the Eomes DBD did not bind to beads. The blot in A was re-probed with anti-Eomes DBD antibodies. C, the Eomes CTD binds to beads. The blot in A was re-probed with anti-Eomes CTD antibodies. D, the NH2- and COOH-terminal halves of CTD (CTD-N, CTD-C) failed to bind to beads. CTD-N, CTD-C, or CTD were overexpressed in Xenopus embryos. Extracts were left untreated (lanes 1-4) or immunoprecipitated (lanes 5-8) with an anti-Smad2 antibody and Western blotted with anti-Eomes COOH-terminal antibodies. E, both CN3 and CC3 bind to beads. CN3, Cmid1, or CC3 were overexpressed in Xenopus embryos. Extracts were left untreated (lanes 1-4) or immunoprecipitated (lanes 5-8) with an anti-Smad2 antibody and Western blotted with anti-Eomes COOH-terminal antibodies. (The anti-Eomes-CTD antibody failed to recognize Cmid1 (lanes 2 and 6)). F, Eomes constructs used in co-immunoprecipitation analysis. Eomes is 692 amino acids long. The carboxyl-terminal region amino acid residues 547-612 are deleted in Δ-CBD. NTD encodes 1-214; CTD-N, 456-573; DBD, 215-455; CTD-C, 574-692; CTD, 456-692; CN3, 456-633; CC3, 515-692; Cmid1, 515-633; D1, 515-564 fused to 583-692; D2, 515-552 fused to 595-692; D3, 456-564 fused to 583-692; D4, 456-552 fused to 595-692; CBD, 547-612. Eomes T-box (273-440) highlighted green.
We therefore constructed a series of cDNAs, using wild type CC3 as a substrate, in which 21 windows of six Eomes CC3 amino acids were individually mutated. This was accomplished by substituting six glycine residues in place of the natural Eomes 6-amino acid sequence within each such window between Glu515 and Ile633. The resulting “glycine-scanning” mutation clones are called GSM1 through GSM21. Each GSM mRNA was injected individually into Xenopus embryos and subjected to co-immunoprecipitation analysis in direct comparison to wild type CC3. Ten of the 21 GSMs had the same affinity for beads as wild-type CC3 peptide (GSMs 1, 2, 4-6, 10, 15, 16, 19, and 20; Fig. 3, A-C, E, H, and J). Seven GSMs had greatly reduced affinity (GSMs 7, 8, 11-14, and 17; Fig. 3, D, F, G, and I). Two GSMs had slightly reduced (GSMs 3, 18; Fig. 3, B and I), and two slightly increased (GSMs 9, 21; Fig. 3, E and K) affinity for beads. The myc epitope tag, when fused to negative control protein DBD or GFP, failed to bind to beads (Fig. 3L).
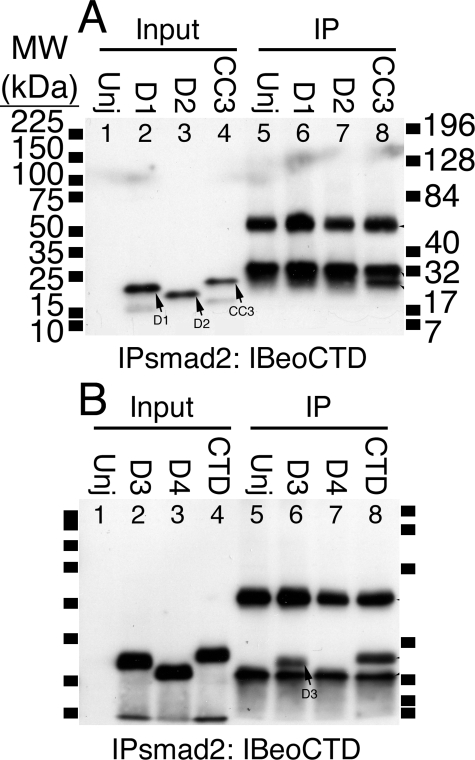
The seven GSMs with reduced affinity are located centrally within CTD, confirming our previous finding that dividing CTD in half eliminated binding (Fig. 2D). To further confirm our GSM results we made two internal deletions, one spanning the region of GSM10-12 (Ser565-Trp582), and one spanning GSM8-14 (Leu553-Thr594). These deletions were made within the context of either CC3 or CTD (Fig. 2F). When made in CC3, both deletions virtually eliminated binding (D1 and D2, Fig. 4A). In CTD, the small deletion (D3) had little effect, whereas the large deletion (D4) eliminated binding (Fig. 4B), potentially implicating Arg456-Asn514 (lacking in CC3; Fig. 2F) in imparting structural stability to CTD. We conclude that the central region within CTD conferred bead binding activity to Eomes (Fig. 5A).
FIGURE 4.
Internal deletions within CTD or CC3 failed to bind to Sepharose beads. A, D1 and D2 failed to bind to beads. Eomes deletion mutants D1, D2, or CC3 were overexpressed in Xenopus embryos. Extracts were left untreated (lanes 1-4) or immunoprecipitated (lanes 5-8) with an anti-Smad2 antibody and Western blotted with anti-Eomes COOH-terminal antibodies. Uninjected extracts are shown for comparison (Unj, lanes 1 and 5). B, deletion mutant D3 in CTD binds to beads; D4 failed to bind to beads.
FIGURE 5.
The Eomes CBD is evolutionarily conserved. A, summary of results from glycine-scanning and deletion mutation of Eomes. Glu515-Leu636 is depicted in which 21 GSMs were made: red letters indicate that binding to Sepharose beads was eliminated within a 6-amino acid window; blue, diminished binding; green, increased binding. Deletions are indicated by arrows; GSMs over- or underlined; and amino acid numbers bulleted above the sequence. B, ClustalW alignment of Eomes Tyr547-Leu612 (CBD) with Tbr1 and Tbet from Xenopus, zebrafish, mouse, human (GenBank accession numbers: xlEomes, P79944; zfEomes NP_57175; muEomes, O54839; huEOMES, NP_005433; xtTbr1, NP_001072587; zfTbr1, NP_001108562; muTbr1, Q64336; huTBR1, NP_006584; zfTbet, XP_001338262; muTbet, AF241242; huTBET, NP_037483.1).
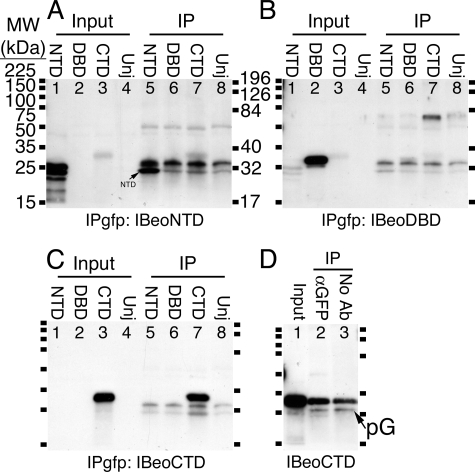
Although full-length Eomes protein bound specifically to anti-Smad2 beads (Fig. 1), we tested the ability of NTD, DBD, and CTD to bind to anti-GFP beads. We found that CTD bound to anti-GFP beads (Fig. 6, C, lane 7, and D, lane 2) or empty protein G-agarose beads (Fig. 6D, lane 3), whereas NTD mostly failed (Fig. 6A, lane 5) and DBD failed to bind to beads (Fig. 6B, lane 6). CTD also bound to unsubstituted agarose (see below). We conclude that the Tyr547-Leu612 region of CTD, spanned by GSMs 7-17 (Fig. 5A) represents a binding domain for polysaccharides.
FIGURE 6.
The Eomes CTD interacts with Sepharose beads. A, the Eomes NTD largely failed to bind to Sepharose. The Eomes NTD, DBD, or CTD were overexpressed in Xenopus embryos. Extracts were left untreated (lanes 1-4) or immunoprecipitated (lanes 5-8) with an anti-GFP antibody and Western blotted with anti-Eomes NH2-terminal antibodies. Uninjected extracts are shown for comparison (Unj, lanes 4 and 8). B, the Eomes DBD did not bind to beads. The blot in A was re-probed with anti-Eomes DBD antibodies. C, the Eomes CTD binds to beads. The blot in A was re-probed with anti-Eomes CTD antibodies. D, CTD binds beads without added antibody. The Eomes CTD was overexpressed in Xenopus embryos. Extracts were left untreated (lane 1) or immunoprecipitated with an anti-GFP antibody (lane 2) or no antibody (lane 3), and Western blotted with anti-Eomes COOH-terminal antibodies.
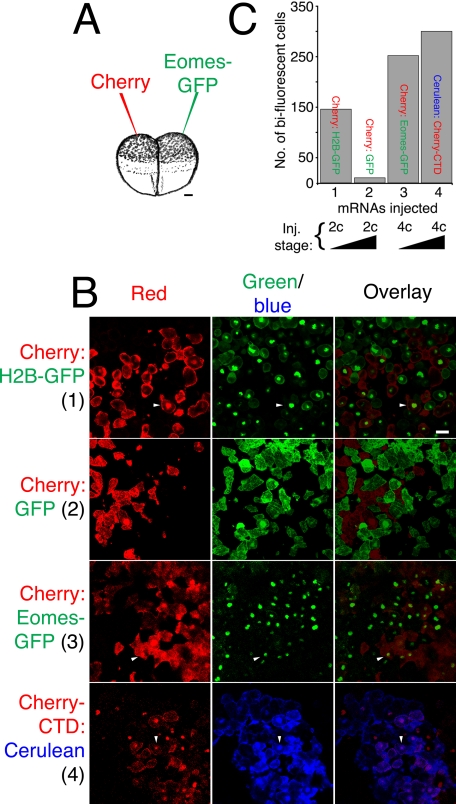
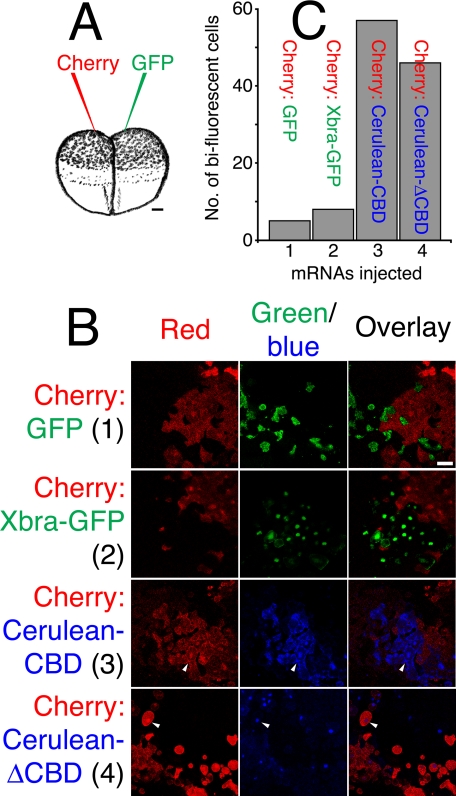
The COOH-terminal Domain of Eomes Mediates Transfer of Eomes Protein between Cells—It was possible that the binding of Eomes to polysaccharide beads reflected an underlying function. For example, beads might mimic the solvent-exposed surface of intra- or extracellular glycosylated protein residues. To test whether Eomes protein is capable of direct protein transfer between cells, we utilized a simple assay (Fig. 7A): we injected two cells of both two- and four-cell stage Xenopus embryos with synthetic mRNA encoding fluorescent proteins of two different colors. Embryos were cultured until mid-blastula stage 8, animal poles explanted (caps), and living caps subjected to confocal microscopy. Injections into two-cell embryos in which cytokinesis has not yet been completed would be expected to yield caps with many cells containing fluorescent proteins of both color. In contrast, for proteins unable to diffuse across or be transported across the cell plasma membrane, later stage injections should yield cells of only one or the other color.
FIGURE 7.
Xenopus embryos can be used to measure intercellular translocation of proteins. A, experimental design. Depiction of two-cell Xenopus embryos, which were injected into the animal pole. One cell was injected with mRNA encoding fluorescent protein of one color (e.g. Cherry), and the other cell with mRNA for another color (e.g. Eomes-GFP). Bar, 100 μm. B, mRNAs can diffuse between cells before completion of cytokinesis in two-cell embryos. One cell of mid-stage two-cell Xenopus embryos was injected with mRNA for Cherry, the other, histone H2B-GFP (top row). Animal poles were excised at stage 8 and subjected to confocal microscopy. Fluorescent images were obtained in the same confocal plane for each fluorescent color (shown individually and overlaid). Late two-cell embryos prevented diffusion of mRNA between cells (Cherry on one side, versus GFP on the other side; second row). Eomes protein is translocated between adjacent cells. Four-cell mid-stage embryos were injected with mRNA for Cherry on one side (left or right) versus Eomes-GFP on the other side (right or left; third row). The Eomes CTD is sufficient to mediate translocation of Eomes between cells. Late four-cell stage embryos were injected with mRNA for Cherry-Eomes-CTD on one side (left or right) versus Cerulean on the other side (right or left; fourth row). Bar, 38 μm. C, quantitation of Eomes protein translocation. The number of bi-fluorescent cells observed in 10 microscopic fields of view were counted. Views were randomly chosen and contained cells of both fluorescent colors. Arrowheads indicate bi-fluorescent cells.
To determine whether this assay was effective, we injected one cell of a two-cell stage Xenopus embryo with mRNA encoding a monomeric red fluorescent protein (Cherry) (35), and the second cell with histone H2B fused to GFP (H2B-GFP), at the two-cell stage, before completion of the first cytokinesis. As expected, in 10 microscopic fields of view chosen at random, many cells were found in which both fluorescent colors were present (Fig. 7B, top row). After completion of cytokinesis, two-cell embryos were injected with Cherry in one cell and GFP in the other cell: very few bi-fluorescent cells were observed (Fig. 7B, second row). In contrast, when four-cell embryos were injected; with one left-(or right-) side cell receiving Cherry, and the other side (right or left) receiving Eomes fused to GFP; many cells contained both fluorescent proteins (Fig. 7B, third row). Similarly, when the Eomes CTD (fused to Cherry) was tested opposite a monomeric form of a blue fluorescent protein (Cerulean) (36), CTD crossed the plasma membrane into adjacent cells (Fig. 7B, bottom row). Quantitation of these results is shown in Fig. 7C. We conclude that Eomes protein was translocated between adjacent cells, and that the Eomes CTD was sufficient to confer this capability.
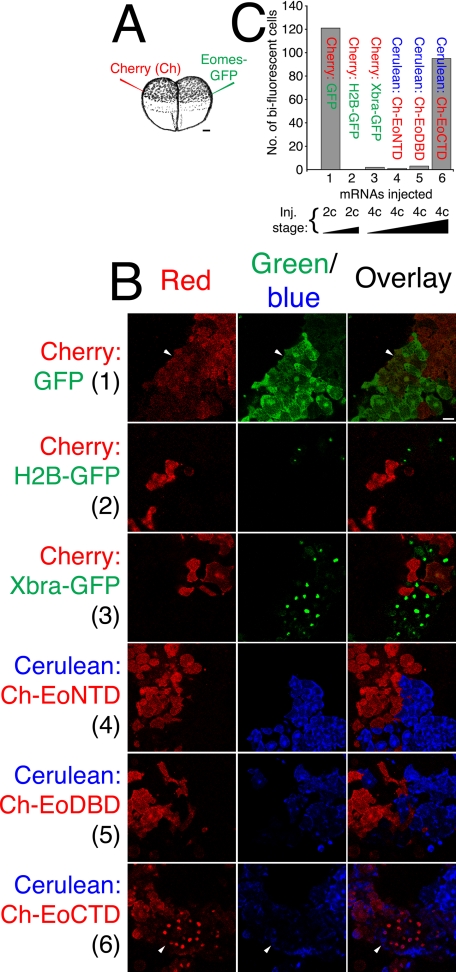
Xbra-GFP, Eomes-NTD, and Eomes-DBD Fail to Cross the Cell Plasma Membrane—To determine whether the ability to be translocated across the plasma membrane was a general property of T-box proteins, we injected four-cell embryos with Cherry in one cell and Xbra-GFP in the other; Xbra-GFP failed to move between adjacent cells (Fig. 8B, third row), as did H2B-GFP when injected at the late two-cell stage (Fig. 8B, second row). As a control, injection of mid-two-cell embryos with one cell receiving Cherry and the other, GFP, yielded many bi-fluorescent cells (Fig. 8B, first row). As a control for mRNA dose effects, in this experiment, embryos were injected distal to the plane of cell division (Fig. 8A). Similarly to Xbra and histone H2B, both Eomes-NTD and Eomes-DBD failed to cross the plasma membrane (Fig. 8B, fourth and fifth rows). As expected, Cherry-CTD crossed the plasma membrane to adjacent Cerulean-injected cells (Fig. 8B, bottom row). Quantitation of these results is shown in Fig. 8C. We conclude that the ability to cross the plasma membrane is not a general property of the T-box transcription factors, and within Eomes, this property specifically mapped to the CTD. Moreover, the Eomes CTD was necessary and sufficient for protein transduction.
FIGURE 8.
Eomes intercellular translocation is mediated by the Eomes carboxyl-terminal region. A, two-cell Xenopus embryos were injected into the animal pole. One cell was injected with mRNA encoding fluorescent protein of one color, and the other cell with mRNA for another color. Injections were targeted at maximal distance from the embryonic midline (plane of first cell division). Bar, 100 μm. B, expression of Cherry in one cell and GFP in the other at the two-cell mid-stage allows diffusion of mRNA between cells (top row). Expression of Cherry versus H2B-GFP in late two-cell embryos prevents mRNA diffusion (second row). Xbra-GFP did not move between cells (versus Cherry, third row). Neither Cherry-Eomes-NTD (versus Cerulean, fourth row) nor Cherry-Eomes-DBD (versus Cerulean, fifth row) moved between cells. Cherry-Eomes-CTD did move between cells (versus Cerulean, bottom row). Bar, 38 μm. C, quantitation of Eomes protein translocation. The number of bi-fluorescent cells observed in 10 microscopic fields of view were counted. Views were randomly chosen and contained cells of both fluorescent colors. Arrowheads indicate bi-fluorescent cells.
The Eomes Carbohydrate Binding Domain (CBD) Is Sufficient for Cell-Cell Protein Transfer—It was possible that the Eomes polysaccharide affinity surface (CBD; 547-612) could mediate Eomes protein transfer across the plasma membrane. When four-cell embryos were injected (Fig. 9A) with Cherry in one animal pole cell (left or right), and Cerulean-CBD on the other side (right or left, Fig. 9A), CBD was found in many cells along with Cherry (Fig. 9B, third row). Eomes lacking CBD (Cerulean-ΔCBD) also crossed the plasma membrane (Fig. 9B, bottom row). Neither Cherry, GFP, nor Xbra-GFP were translocated (Fig. 9B, first and second rows). Quantitation of these results is shown in Fig. 9C. We conclude that the Eomes CBD is sufficient, but not necessary for Eomes protein transduction.
FIGURE 9.
The Eomes CBD is sufficient, but not necessary for translocation of Eomes protein between cells. A, experimental design. Injections were performed at the four-cell stage. Bar, 100 μm. B, Cherry and GFP mRNAs did not diffuse, nor did their proteins move between cells (top row). Xbra failed to move between cells (Cherry versus Xbra-GFP, second row). The Eomes CBD was sufficient to confer intercellular translocation (Cherry versus Cerulean CBD, third row). The Eomes CBD was not necessary for protein translocation (Cherry versus Cerulean-ΔCBD, bottom row). Bar, 38 μm. C, quantitation of Eomes protein translocation. Arrowheads indicate bi-fluorescent cells.
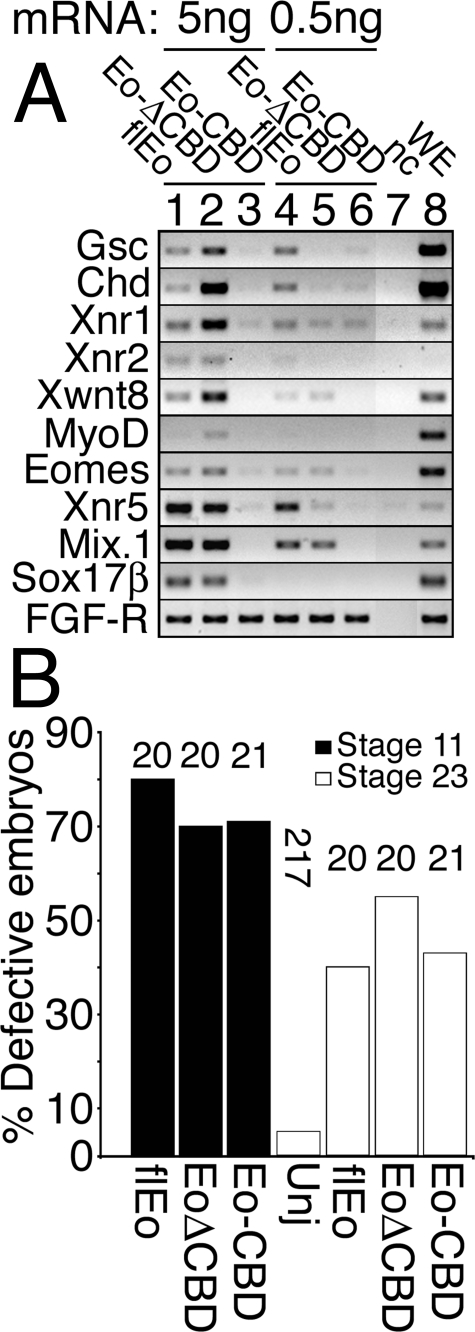
The Eomes CBD Is Important for Gene Activation—The Eomes CTD, containing the CBD, harbors a transcriptional activation domain (3). We therefore tested whether CBD conferred trans-activation capacity on Eomes by overexpressing Eomes-CBD in caps and assaying for early embryonic gene expression by RT-PCR. Although CBD itself failed to activate genes in caps, Eomes lacking CBD (Eomes-ΔCBD) was impaired in its ability to activate genes at moderate dose (0.5 ng of mRNA per cap) but not at high dose (5 ng; Fig. 10A). Both CBD and Eomes-ΔCBD also caused a high frequency of gastrulation defects when injected into the animal pole of intact embryos (Fig. 10B). We conclude that the Eomes CBD is important for Eomes gene activation. As CBD caused gastrulation defects in whole embryos, this suggested CBD interacted and interfered with important protein partners.
FIGURE 10.
The Eomes CBD is important in gene activation. A, Eomes-CBD failed to activate early embryonic genes. Although Eomes-ΔCBD activated genes at high dose (5 ng of mRNA per cap), Eomes-ΔCBD was significantly impaired for gene activation at moderate dose (0.5 ng per cap). Xenopus embryos were injected into the animal pole with mRNA encoding full-length Eomes (lanes 1 and 4), Eomes-ΔCBD (lanes 2 and 5), and CBD (lanes 3 and 6). Injected or uninjected (nc, lane 7) animal poles were explanted (caps) at stage 8 and cultured until stage 10.5; and caps were frozen. Caps and control stage 10.5 whole embryos (WE, lane 8) were analyzed by RT-PCR for early embryonic genes. B, quantitation of embryonic defects in animal pole-injected, and uninjected, embryos. Embryos were scored for normalcy of gastrulation at stage 11, and for defects resulting from earlier inhibition of gastrulation at stage 23. Number of embryos (n) is shown above the histogram.
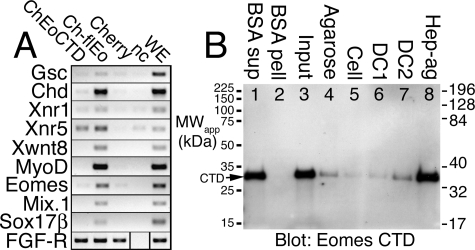
The Eomes CTD Activates Xnr5 but Does Not Bind DNA—Eomes-ΔCBD was capable of access to adjacent cells, which suggested other regions within CTD were also sufficient for cell transfer activity. We therefore tested CTD for its ability to activate gene transcription in caps. We found that Cherry-Eomes-CTD was capable of activating Xnr5 (Fig. 11A). As CTD contains the nuclear localization signal for Eomes (Fig. 8B, bottom row), we asked whether CTD was capable of binding to DNA-cellulose. CTD bound to DNA-cellulose beads (Fig. 11B, lanes 6 and 7) similarly to unsubstituted beads (Fig. 11B, lanes 4 and 5), and failed to bind beads to which BSA had been covalently cross-linked (Fig. 11B, lane 2). All three Eomes peptides, NTD, DBD (data not shown), and CTD, bound to heparin-agarose (positive control; Fig. 11B, lane 8). We conclude that despite its nuclear localization, CTD did not bind to DNA.
FIGURE 11.
The Eomes CTD activates Xnr5 but fails to bind DNA. A, Cherry-Eomes-CTD activated Xnr5. Xenopus embryos were injected into the animal pole with mRNA encoding Cherry-Eomes-CTD, Cherry-f.l.Eomes (full-length), or Cherry alone; injected or uninjected (nc) animal poles were explanted (caps) at stage 8 and cultured until stage 10.5; and caps were frozen. Caps and control stage 10.5 whole embryos (WE) were analyzed by RT-PCR for early embryonic genes. B, the Eomes CTD failed to bind DNA. The Eomes CTD was overexpressed in Xenopus embryos. Extracts (lane 3) were co-precipitated with BSA cross-linked-agarose beads (lane 2), unsubstituted (plain) agarose (lane 4), DNase-treated DNA-cellulose (lane 5), mock-treated DNA-cellulose (lane 6), DNA-cellulose (lane 7), or heparin-agarose (lane 8), and Western blotted with anti-Eomes COOH-terminal antibodies. Supernatant from BSA beads is shown as a control (lane 1).
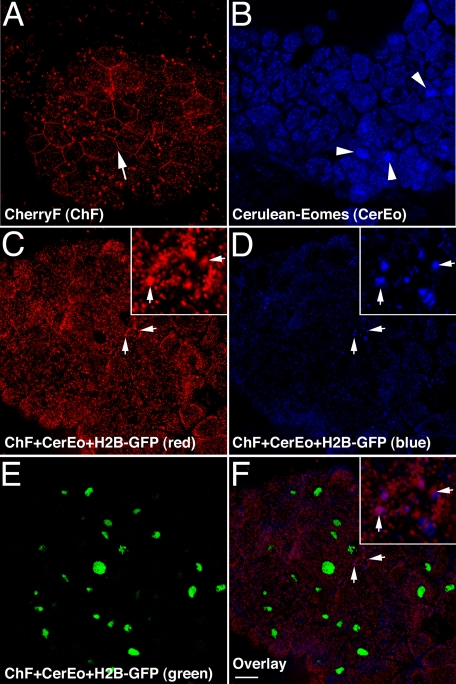
Eomes Protein Translocation Is Required for Its Nuclear Accumulation—As a transcription factor, Eomes must first enter the cell nucleus before activating gene transcription. We asked whether the transfer of Eomes protein between adjacent cells was necessary for its ability to activate genes, as measured by the nuclear accumulation of Eomes. The farnesylation signal sequence from the Xenopus H-ras protein (37) was fused to Cherry (CherryF) and overexpressed in caps. As expected, CherryF localized to the plasma membrane, and also showed some cytoplasmic staining (Fig. 12A). Cerulean-Eomes localized to nuclei (Fig. 12B). When three fluorescently tagged proteins: CherryF (ChF), Cerulean-Eomes (CerEo), and H2B-GFP, were co-expressed in caps, Cerulean-Eomes failed to accumulate in nuclei (Fig. 12D). Instead, in some cells, CherryF and Cerulean-Eomes co-localized to membrane-associated puncta (Fig. 12, C, D, and F). This suggested that attachment of lipidated CherryF to the plasma membrane was responsible for preventing the nuclear accumulation of Cerulean-Eomes. We conclude that CherryF prevented Cerulean-Eomes translocation between cells, and that translocation of Cerulean-Eomes between cells was required for its nuclear localization.
FIGURE 12.
Eomes requires intercellular translocation for nuclear access. A, farnesylated Cherry (CherryF) binds to the plasma membrane (arrow). CherryF was expressed in caps and imaged by confocal microscopy. B, Cerulean-Eomes localizes to cell nuclei in caps (arrowheads). Cerulean-Eomes was expressed in caps and imaged by confocal microscopy. C-F were imaged in the same confocal plane; CherryF prevents Eomes accumulation in nuclei and pauses Cerulean-Eomes translocation at membrane puncta (arrows). CherryF, Cerulean-Eomes, and histone H2B-GFP were co-expressed in caps and imaged by confocal microscopy. Inset shows 4-fold magnification of membrane-localized puncta. C, red fluorescence; D, blue fluorescence; E, green fluorescence; F, overlay of C-E. Bar, 38 μm.
DISCUSSION
Eomes Forms a Complex with Phosphorylated Smad2 to Activate Several Mesodermal Genes—Activin signaling activates mesodermal genes through phosphorylation of Smad2, which then associates with Smad4, translocates to the cell nucleus, and associates with other transcription factors (38, 39). We observed that a truncated dominant negative activin receptor blocks the ability of Eomes to activate several mesodermal genes in caps (data not shown). Furthermore, a point-mutated dominant negative Smad2, which blocks endogenous Smad2 phosphorylation, also prevents Eomes from activating a number of mesodermal genes (data not shown). This suggests that Eomes requires phosphorylated Smad2 to activate these target genes, and that Eomes might activate some genes by associating physically with Smads 2 and 4 in a promoter-bound transcription factor complex. This agrees with work by others that Eomes interacts genetically with nodal in zebrafish and mouse (19, 20, 22) and suggests that the mechanism of this interaction is one of protein complex formation with Smad2.
The Eomes CTD Binds to Polysaccharides—Using co-immunoprecipitation analysis, we found that the Eomes protein associates physically with Smad2 (Fig. 1). To map the Eomes domain for Smad2 binding we divided Eomes into three equal fragments, NTD, DBD, and CTD, and used them in co-immunoprecipitations. Unexpectedly, we found that CTD, unlike full-length Eomes, bound to agarose beads (Fig. 6). The Eomes domain for carbohydrate binding was localized to the middle portion of the COOH-terminal third of Eomes (Fig. 2), and was further delineated by employing a glycine-scanning mutational approach that identified residues within the Eomes COOH-terminal portion between Tyr547 and Leu612 that are critical for binding. These residues, when mutated to glycine (in groups of six), substantially diminished binding of an Eomes COOH-terminal peptide (CC3) to Sepharose beads (Fig. 3). Three segments important for binding were identified: 1) Tyr547-Lys558 (12 amino acids); 2) Tyr571-Thr594 (24 amino acids); and 3) Gly607-Leu612 (6 amino acids). Two additional 6-amino acid blocks had slightly diminished binding: Gln523-Thr528 and Leu613-Val618; and two had increased binding: Ser559-Thr564 and Pro631-Leu636 (Fig. 3). Furthermore, deletion of 42 amino acids (Leu553-Thr594) abolished binding of the Eomes CTD to beads (Fig. 4B). These data are summarized in Fig. 5A.
The entire Eomes CTD (Arg456-Ser692, Xenopus) is highly conserved among vertebrates (70% identical between human and Xenopus; ClustalW comparison, Fig. 5B). The Eomes CBD (Tyr547-Leu612) within the CTD is also highly conserved (65% identical between human and Xenopus Eomes). The equivalent region of human Tbr1 or Tbet is 46 or 22% identical, respectively. Comparing this domain among human, mouse, Xenopus, and zebrafish, a number of blocks of similarity are found which segregate by subfamily member (Eomes, Tbr1, and Tbet; Fig. 5B). These blocks correlate well with our glycine-scanning mutant data, such that almost the entire region spanned by GSM7-9 is conserved among species between Eomes and Tbr1. Region GSM10, dispensable for binding, lies largely within a non-evolutionarily conserved region. GSM11-14, required for binding, is conserved; and with the exception of Trp599 and Ser604, GSM15 is neither required for binding nor highly conserved. Throughout this region Tbet is less well conserved than Eomes and Tbr1, with the exception of region GSM11-13, suggestive of a potential CBD in Tbet. No similarity exists between Eomes and any other carbohydrate binding domains (data not shown), and there is no apparent small, conserved motif located within the Eomes CBD region (Fig. 5B).
Eomes Protein Is Transported between Adjacent Embryonic Cells—The ability of the Eomes CTD peptide to bind polysaccharides suggested that it may normally do so intra- or extracellularly. It is unusual for transcription factors to be trafficked between cells (25). We find that full-length Eomes protein gains access to adjacent embryonic cells by crossing the plasma membrane, a barrier to which other proteins (including Xbra, histone H2B, and GFP) are impermeant (Figs. 7 and 8). The capability for intercellular protein translocation maps to the Eomes CTD (Figs. 7 and 8); and the Eomes CBD within the CTD is sufficient for cell-cell transfer (Fig. 9).
Whereas CBD fails to activate early embryonic genes in caps (Fig. 10A), it is capable of interfering with normal cell movements during gastrulation (Fig. 10B). Although Eomes lacking the CBD (Eomes-ΔCBD) activates genes at high dose, at a moderate dose Eomes-ΔCBD fails to effectively activate early embryonic genes in caps (Fig. 10A). Thus at moderate doses, Eomes-ΔCBD is impaired for gene activation. Eomes-ΔCBD may be similarly impaired for cell translocation, and may only translocate between cells at high doses. CTD activates Xnr5 in caps (Fig. 11A) despite its lack of DNA-binding activity (Fig. 11B).
When direct access of Cerulean-Eomes to the plasma membrane is blocked using a farnesylated red fluorescent protein, nuclear accumulation of Eomes is also blocked (Fig. 12). This suggests that Eomes protein does not simply diffuse across the lipid bilayer (as thought to occur for HIV tat protein) (25), but rather is actively transported from one cell to the next. When CherryF and Cerulean-Eomes are co-expressed, CherryF is partially displaced from the plasma membrane (Fig. 12C), and Cerulean-Eomes is excluded from nuclei, and localizes in some cells to plasma membrane-associated puncta (Fig. 12, C and D). (GFP-Eomes localizes to membrane puncta more readily, perhaps because of the propensity of GFP for aggregation, which may result in a reduced cell-cell transfer rate; data not shown.) This suggests CherryF and Cerulean-Eomes compete for access to the plasma membrane, reducing the rate of Cerulean-Eomes translocation and allowing visualization of Cerulean-Eomes membrane puncta just before, or during, transit. It also suggests that during or after transfer of Eomes protein between cells, Eomes protein is post-translationally modified to subsequently allow it to gain access to the cell nucleus. This putative co-translocational activation appears to regulate the nuclear import of Eomes, which itself is logically required for Eomes to activate its target genes. The Eomes CTD with its carbohydrate binding domain bears no similarity to other known peptides capable of intercellular translocation (ClustalW analysis of peptides in Ref. 25 and BLASTp analysis (40, 41), data not shown (24, 26).
Why should Eomes protein be regulated so as to require translocation between cells before being capable of entering cell nuclei? This novel regulation may represent a molecular mechanism for ensuring a community effect (42, 43) during embryonic development. Cells that first activate the Eomes gene may wait for input from neighboring cells before making a decision to differentiate as mesoderm. In naïve cells Eomes is kept out of the nucleus and thereby prevented from activating its target genes. Cells that activate Eomes before their neighbors could directly signal their tentative decision in favor of mesoderm via translocation of Eomes protein into neighboring cells; prior translocation of Eomes is required for Eomes to activate genes. A critical threshold concentration of Eomes protein may be required within naïve cells before Eomes protein is transferred to its neighbor. Once transferred, Eomes protein would become activated for nuclear import, activate genes, and trigger cell fate commitment. This mechanism would help ensure that mesodermal cells differentiate as groups (42, 43).
Future work will identify the remaining residues within the Eomes CTD that, like CBD, are also capable of protein translocation between cells. It will be important to identify the Eomes transporter, or otherwise elucidate the mechanism for Eomes translocation. Although cell to cell protein translocation is not a general property of the T-box proteins (Figs. 8 and 9), identification of an Eomes transporter could lead to elucidation of other protein substrates, perhaps including other transcription factors currently thought to behave in a cell-autonomous fashion.
Acknowledgments
We thank Jim Smith and Frank Conlon for human glucocorticoid receptor ligand binding domain cDNA; Doug Melton for Xenopus Smad2 cDNA; John Gurdon for antisera raised against Xenopus Eomes; Anthony Gotter, Shelby Blythe, Christine Reid, and Jennifer Skirkanich for technical advice; Robert J. Levy for critical comments.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) U75996.
This work was supported, in whole or in part, by National Institutes of Health Grants HL070168 and HL74731. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Eomes, Eomesodermin; caps, Xenopus animal pole explants; DN, dominant-negative; Xbra, Xenopus brachyury; Xnr, Xenopus nodal-related gene; P445H, mutant DN-xSmad2; D450E, mutant DNxSmad2; GR, human glucocorticoid receptor ligand binding domain; GFP, green fluorescent protein; IP, immunoprecipitation; MBS, modified Barth's saline; RT, reverse transcriptase; BSA, bovine serum albumin; DBD, DNA binding domain; CBD, carbohydrate binding domain.
References
- 1.Ryan, K., and Chin, A. J. (2003) Birth Defects Res. Part C. Embryo Today 69 25-37 [DOI] [PubMed] [Google Scholar]
- 2.Smith, J. (1999) Trends Genet. 15 154-158 [DOI] [PubMed] [Google Scholar]
- 3.Conlon, F. L., Fairclough, L., Price, B. M., Casey, E. S., and Smith, J. C. (2001) Development 128 3749-3758 [DOI] [PubMed] [Google Scholar]
- 4.Showell, C., Binder, O., and Conlon, F. L. (2004) Dev. Dyn. 229 201-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson, V., and Conlon, F. L. (2002) Genome Biol. 3 REVIEWS3008 [DOI] [PMC free article] [PubMed]
- 6.Ataliotis, P., Ivins, S., Mohun, T. J., and Scambler, P. J. (2005) Dev. Dyn. 232 979-991 [DOI] [PubMed] [Google Scholar]
- 7.Glimcher, L. H., Townsend, M. J., Sullivan, B. M., and Lord, G. M. (2004) Nat. Rev. Immunol. 4 900-911 [DOI] [PubMed] [Google Scholar]
- 8.Russ, A. P., Wattler, S., Colledge, W. H., Aparicio, S. A., Carlton, M. B., Pearce, J. J., Barton, S. C., Surani, M. A., Ryan, K., Nehls, M. C., Wilson, V., and Evans, M. J. (2000) Nature 404 95-99 [DOI] [PubMed] [Google Scholar]
- 9.Ryan, K., Garrett, N., Mitchell, A., and Gurdon, J. B. (1996) Cell 87 989-1000 [DOI] [PubMed] [Google Scholar]
- 10.Ryan, K., Russ, A. P., Levy, R. J., Wehr, D. J., You, J., and Easterday, M. C. (2004) Hum. Gene Ther. 15 842-855 [DOI] [PubMed] [Google Scholar]
- 11.Ryan, K., Butler, K., Bellefroid, E., and Gurdon, J. B. (1998) Mech. Dev. 75 155-158 [DOI] [PubMed] [Google Scholar]
- 12.Ciruna, B. G., and Rossant, J. (1999) Mech. Dev. 81 199-203 [DOI] [PubMed] [Google Scholar]
- 13.Hancock, S. N., Agulnik, S. I., Silver, L. M., and Papaioannou, V. E. (1999) Mech. Dev. 81 205-208 [DOI] [PubMed] [Google Scholar]
- 14.Bulfone, A., Martinez, S., Marigo, V., Campanella, M., Basile, A., Quaderi, N., Gattuso, C., Rubenstein, J. L., and Ballabio, A. (1999) Mech. Dev. 84 133-138 [DOI] [PubMed] [Google Scholar]
- 15.Intlekofer, A. M., Takemoto, N., Wherry, E. J., Longworth, S. A., Northrup, J. T., Palanivel, V. R., Mullen, A. C., Gasink, C. R., Kaech, S. M., Miller, J. D., Gapin, L., Ryan, K., Russ, A. P., Lindsten, T., Orange, J. S., Goldrath, A. W., Ahmed, R., and Reiner, S. L. (2005) Nat. Immunol. 6 1236-1244 [DOI] [PubMed] [Google Scholar]
- 16.Pearce, E. L., Mullen, A. C., Martins, G. A., Krawczyk, C. M., Hutchins, A. S., Zediak, V. P., Banica, M., DiCioccio, C. B., Gross, D. A., Mao, C. A., Shen, H., Cereb, N., Yang, S. Y., Lindsten, T., Rossant, J., Hunter, C. A., and Reiner, S. L. (2003) Science 302 1041-1043 [DOI] [PubMed] [Google Scholar]
- 17.Ryan, K., Garrett, N., Bourillot, P., Stennard, F., and Gurdon, J. B. (2000) Mech. Dev. 94 133-146 [DOI] [PubMed] [Google Scholar]
- 18.Bruce, A. E., Howley, C., Dixon Fox, M., and Ho, R. K. (2005) Dev. Dyn. 233 105-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruce, A. E., Howley, C., Zhou, Y., Vickers, S. L., Silver, L. M., King, M. L., and Ho, R. K. (2003) Development 130 5503-5517 [DOI] [PubMed] [Google Scholar]
- 20.Bjornson, C. R., Griffin, K. J., Farr, G. H., 3rd, Terashima, A., Himeda, C., Kikuchi, Y., and Kimelman, D. (2005) Dev. Cell 9 523-533 [DOI] [PubMed] [Google Scholar]
- 21.Strumpf, D., Mao, C. A., Yamanaka, Y., Ralston, A., Chawengsaksophak, K., Beck, F., and Rossant, J. (2005) Development 132 2093-2102 [DOI] [PubMed] [Google Scholar]
- 22.Arnold, S. J., Hofmann, U. K., Bikoff, E. K., and Robertson, E. J. (2008) Development 135 501-511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spemann, H., and Mangold, H. (2001) Int. J. Dev. Biol. 45 13-38 [PubMed] [Google Scholar]
- 24.Cardarelli, F., Serresi, M., Bizzarri, R., Giacca, M., and Beltram, F. (2007) Mol. Ther. 15 1313-1322 [DOI] [PubMed] [Google Scholar]
- 25.Joliot, A., and Prochiantz, A. (2004) Nat. Cell Biol. 6 189-196 [DOI] [PubMed] [Google Scholar]
- 26.Sugiyama, S., Di Nardo, A. A., Aizawa, S., Matsuo, I., Volovitch, M., Prochiantz, A., and Hensch, T. K. (2008) Cell 134 508-520 [DOI] [PubMed] [Google Scholar]
- 27.Derynck, R., Gelbart, W. M., Harland, R. M., Heldin, C. H., Kern, S. E., Massague, J., Melton, D. A., Mlodzik, M., Padgett, R. W., Roberts, A. B., Smith, J., Thomsen, G. H., Vogelstein, B., and Wang, X. F. (1996) Cell 87 173. [DOI] [PubMed] [Google Scholar]
- 28.Graff, J. M., Bansal, A., and Melton, D. A. (1996) Cell 85 479-487 [DOI] [PubMed] [Google Scholar]
- 29.Newfeld, S. J., Chartoff, E. H., Graff, J. M., Melton, D. A., and Gelbart, W. M. (1996) Development 122 2099-2108 [DOI] [PubMed] [Google Scholar]
- 30.Tada, M., O'Reilly, M. A., and Smith, J. C. (1997) Development 124 2225-2234 [DOI] [PubMed] [Google Scholar]
- 31.Nieuwkoop, P. D., and Faber, J. (1994) Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis, 2nd Ed., Garland Publishing, Inc., New York
- 32.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 33.Stennard, F., Zorn, A. M., Ryan, K., Garrett, N., and Gurdon, J. B. (1999) Mech. Dev. 86 87-98 [DOI] [PubMed] [Google Scholar]
- 34.Messenger, N. J., Kabitschke, C., Andrews, R., Grimmer, D., Nunez Miguel, R., Blundell, T. L., Smith, J. C., and Wardle, F. C. (2005) Dev. Cell 8 599-610 [DOI] [PubMed] [Google Scholar]
- 35.Shu, X., Shaner, N. C., Yarbrough, C. A., Tsien, R. Y., and Remington, S. J. (2006) Biochemistry 45 9639-9647 [DOI] [PubMed] [Google Scholar]
- 36.Malo, G. D., Pouwels, L. J., Wang, M., Weichsel, A., Montfort, W. R., Rizzo, M. A., Piston, D. W., and Wachter, R. M. (2007) Biochemistry 46 9865-9873 [DOI] [PubMed] [Google Scholar]
- 37.Apolloni, A., Prior, I. A., Lindsay, M., Parton, R. G., and Hancock, J. F. (2000) Mol. Cell. Biol. 20 2475-2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massague, J., and Gomis, R. R. (2006) FEBS Lett. 580 2811-2820 [DOI] [PubMed] [Google Scholar]
- 39.Massague, J., Seoane, J., and Wotton, D. (2005) Genes Dev. 19 2783-2810 [DOI] [PubMed] [Google Scholar]
- 40.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul, S. F., Wootton, J. C., Gertz, E. M., Agarwala, R., Morgulis, A., Schaffer, A. A., and Yu, Y. K. (2005) FEBS J. 272 5101-5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Standley, H. J., and Gurdon, J. B. (2002) Int. J. Dev. Biol. 46 993-998 [PubMed] [Google Scholar]
- 43.Standley, H. J., Zorn, A. M., and Gurdon, J. B. (2002) Int. J. Dev. Biol. 46 279-283 [PubMed] [Google Scholar]