Abstract
In mammals, xylose is found as the first sugar residue of the tetrasaccharide GlcAβ1-3Galβ1-3Galβ1-4Xylβ1-O-Ser, initiating the formation of the glycosaminoglycans heparin/heparan sulfate and chondroitin/dermatan sulfate. It is also found in the trisaccharide Xylα1-3Xylα1-3Glcβ1-O-Ser on epidermal growth factor repeats of proteins, such as Notch. UDP-xylose synthase (UXS), which catalyzes the formation of the UDP-xylose substrate for the different xylosyltransferases through decarboxylation of UDP-glucuronic acid, resides in the endoplasmic reticulum and/or Golgi lumen. Since xylosylation takes place in these organelles, no obvious requirement exists for membrane transport of UDP-xylose. However, UDP-xylose transport across isolated Golgi membranes has been documented, and we recently succeeded with the cloning of a human UDP-xylose transporter (SLC25B4). Here we provide new evidence for a functional role of UDP-xylose transport by characterization of a new Chinese hamster ovary cell mutant, designated pgsI-208, that lacks UXS activity. The mutant fails to initiate glycosaminoglycan synthesis and is not capable of xylosylating Notch. Complementation was achieved by expression of a cytoplasmic variant of UXS, which proves the existence of a functional Golgi UDP-xylose transporter. A ∼200 fold increase of UDP-glucuronic acid occurred in pgsI-208 cells, demonstrating a lack of UDP-xylose-mediated control of the cytoplasmically localized UDP-glucose dehydrogenase in the mutant. The data presented in this study suggest the bidirectional transport of UDP-xylose across endoplasmic reticulum/Golgi membranes and its role in controlling homeostasis of UDP-glucuronic acid and UDP-xylose production.
Xylose is only known to occur in two different mammalian glycans. First, xylose is the starting sugar residue of the common tetrasaccharide, GlcAβ1,3Galβ1,3Galβ1,4Xylβ1-O-Ser, attached to proteoglycan core proteins to initiate the biosynthesis of glycosaminoglycans (GAGs)2 (1). Second, xylose is found in the trisaccharide Xylα1,3Xylα1,3Glcβ1-O-Ser in epidermal growth factor (EGF)-like repeats of proteins, such as blood coagulation factors VII and IX (2) and Notch (3) (Fig. 1). Two variants of O-xylosyltransferases (XylT1 and XylT2) are responsible for the initiation of glycosaminoglycan biosynthesis, which differ in terms of acceptor specificity and tissue distribution (4-7), and two different enzymatic activities have been identified that catalyze xylosylation of O-glucose residues added to EGF repeats (8-10). On Notch, O-glucose occurs on EGF repeats in a similar fashion as O-fucose, which modifications have been shown to influence ligand-mediated Notch signaling (11-16). Recently, rumi, the gene encoding the Notch O-glucosyltransferase in Drosophila, has been identified, and inactivation of the gene was found to cause a temperature-sensitive Notch phenotype (17). Although this finding clearly demonstrated that O-glucosylation is essential for Notch signaling, the importance of xylosylation for Notch functions remains ambiguous.
FIGURE 1.
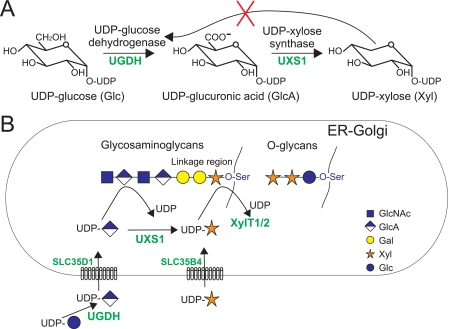
UDP-xylose metabolism in mammalian cells. A, UDP-Xyl is synthesized in two steps from UDP-Glc by the enzymes UGDH, forming UDP-GlcA, and UXS, also referred to as UDP-glucuronic acid decarboxylase. UGDH is inhibited by the product of the second enzyme, UDP-Xyl (42). B, in mammals, UDP-Xyl is synthesized within the lumen of the ER/Golgi, where it is substrate for different xylosyltransferases incorporating xylose in the glycosaminoglycan core (XylT1 and XylT2) or in O-glucose-linked glycans. The nucleotide sugar transporter SLC35D1 (52) has been shown to transport UDP-GlcA over the ER membrane and SLC35B4 (29) to transport UDP-Xyl over the Golgi membrane. The function of this latter transporter is unclear.
Several different Chinese hamster ovary (CHO) cell lines with defects in GAG biosynthesis have been isolated by screening for reduced incorporation of sulfate (18) and reduced binding of fibroblast growth factor 2 (FGF-2) (19, 20) and by direct selection with FGF-2 conjugated to the plant cytotoxin saporin (21). Isolated cells (called pgs, for proteoglycan synthesis mutants) (21) exhibited defects in various stages of GAG biosynthesis, ranging from the initiating xylosyltransferase to specific sulfation reactions (18, 19, 21-25). Mutants that affect overall GAG biosynthesis were shown to have a defect in the assembly of the common core tetrasaccharide. Interestingly, these latter mutants could be separated into clones in which GAG biosynthesis can be restored by the external addition of xylosides as artificial primers and those that cannot (18). The two mutants belonging to the first group are pgsA-745 and pgsB-761. Although pgs-745 is defective in XylT2 (4-6, 18), pgsB-761 exhibits a defect in galactosyltransferase I (B4GalT7), the enzyme that catalyzes the first step in the elongation of the xylosylated protein (25 (see Fig. 1B). Restoration of GAG biosynthesis in the latter mutant presumably occurs through a second β1-4-galactosyltransferase, able to act on xylosides when provided at high concentration but not on the endogenous protein-linked xylose.
Here we describe the isolation of a third CHO cell line (pgsI-208) with the xyloside-correctable phenotype. The mutant is deficient in UDP-xylose synthase (UXS), also known as UDP-glucuronic acid decarboxylase. This enzyme catalyzes the synthesis of UDP-Xyl, the common donor substrate for the different xylosyltransferases, by decarboxylation of UDP-glucuronic acid. Importantly, UXS in the animal cell is localized in the lumen of the ER and/or Golgi (26-28), superseding at first sight the need for the Golgi UDP-xylose transporter, which has been recently cloned and characterized (29). Using this cell variant, experiments were designed that establish the functional significance of UDP-Xyl transport with respect to UDP-glucuronic acid production and xylosylation.
EXPERIMENTAL PROCEDURES
Cell Cultures and Plasmids—Chinese hamster ovary cells (CHO-K1) were obtained from the American Type Culture Collection (CCL-61; Manassas, VA). Mutant pgsA-745 (XylT-deficient) and pgsB-761 (GalTI-deficient) were characterized previously (18, 25), and the mutant line described here was named pgsI-208. Cells were grown under an atmosphere of 5% CO2 in α-minimal essential medium containing ribonucleosides and deoxyribonucleosides (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Biochrom AG, Berlin, Germany).
A cDNA clone of human UXS1 (Hugo Genome Nomenclature Committee ID: 17729) was amplified from prostate gland Marathonready cDNA (Clontech) using primers with EcoRI- and XbaI-flanking start and replacing stop codon, respectively, and cloned in pcDNA3 (Invitrogen) already containing FLAG (N-terminal) and hemagglutinin (C-terminal) tags. XylT2 cDNA (Hugo Genome Nomenclature Committee ID: 15517) was amplified from a human embryonic kidney cDNA library (Invitrogen) using primers with BglII and XbaI flanking the start and stop codon, respectively, and cloned BamHI-XbaI in pcDNA3 containing an N-terminal FLAG tag. A cytoplasmic form of human UXS1 (cytUXS1) was amplified from the full-length clone by primers amplifying the region coding from amino acid 38 to the C terminus and cloned in pcDNA3-FLAG-hemagglutinin like the full-length construct. Arabidopsis thaliana UXS3 (30) (AT5G59290) was amplified by primers covering the complete open reading frame from a cDNA library (31) and cloned into pcDNA3-FLAG-hemagglutinin. Arabidopsis β1,2-xylosyltransferase (AtXylT; AT5G55500) (32) was isolated from the same cDNA library using a sibling selection procedure with PCR primers based on a partial soybean XylT sequence (33).
Complementation Assay—Complementation tests were carried out by cell hybridization as described (34). Approximately 2 × 105 cells of pgsI-208 were added to individual wells of a 24-well plate along with an equal number of pgsA-745 or pgsB-761 cells. After overnight incubation, the mixed cell monolayers were treated for 1 min with 50% (w/w) polyethylene glycol (Mr = 3350) in F-12 medium. The cells were incubated for 1 day, replated in 100-mm diameter tissue culture dishes to obtain ∼300 colonies/dish, and then overlaid with polyester cloth. The replica-plated colonies were incubated with 35SO4 as described (21). Complementation was assessed by the appearance of colonies that regained the capacity to incorporate 35SO4 into acid-precipitable material, as judged by autoradiography.
Complementation by cDNA was carried out by transient transfection using Metafectene (Biontex, Munich, Germany) of pgsI-208 with UXS1 or an empty vector control together with AtXylT. Two days after transfection, cells were fixed in 1.5% glutaraldehyde and stained with polyclonal rabbit anti-horseradish peroxidase (HRP), recognizing xylosylated plant N-glycans (Rockland) followed by alkaline phosphatase-conjugated goat anti-rabbit antibody and developed with Fast-Red substrate (Sigma).
Flow Cytometry—Mutant cells were transfected with plasmids expressing UXS1, cytUXS1, or XylT2 using Metafectene. Cells were selected for stable expression by G418 (Calbiochem), and selected colonies were checked for expression by immunofluorescence using anti-FLAG-M2 monoclonal antibody (Sigma). For flow cytometry, 5 × 105 cells were released with phosphate-buffered saline, 2 mm EDTA and incubated with RB4Ea12, a vesicular stomatitis virus glycoprotein-tagged single chain phage display antibody recognizing heparan sulfate (35, 36), followed by mouse anti-vesicular stomatitis virus glycoprotein tag monoclonal antibody (P5D4; Sigma) and fluorescein isothiocyanate-labeled anti-mouse. Cells (104) were analyzed by flow cytometry (FACScan; BD Biosciences).
Sequence Analysis of CHO UXS1—Chinese hamster UXS1 cDNA was amplified using primers based on conserved parts of human, mouse, and rat sequences covering the sequence from amino acid 37 to 420 (C terminus); GTATGGTACCTTTGTTAACATGAGGTCTAT and GACTTCTAGAGCTGTGGCGCGTCCGGCCCT and cloned BamHI/XbaI (underlined) in pcDNA3 (Invitrogen) for sequencing. Based on this sequence, CHO-specific primers were used to amplify UXS1 mRNA from pgsI-208 in two parts, CAGGAAAATGGTGAACTAAAGT with CGACTCGTTTGCCCTCG to cover amino acids 44-237 and GTCTGCTCCTGGCCTCCACA with TGGGGATGTACTGGTTATTG to cover amino acids 196-406 from a total of 420 amino acids (numbering based on the human sequence). The fragments were cloned in TopoTA (Invitrogen) for sequencing. The genomic fragment around the identified mutation was amplified using AGGTATATGGATCTGGGTCT and ACCTGACATACTGGAACGC and again cloned in TopoTA.
Analyses of Xylosylation of EGF Repeats—A construct encoding EGF repeats 1-5 of the mouse Notch1 extracellular domain in pSecTag (Invitrogen) described previously (37) was transiently transfected in 107 CHO, pgsA-745, and pgsI-208 cells. After 72 h, the fragments were purified by rotating the medium with Ni2+-nitrilotriacetic acid-agarose (30 μl of beads; Qiagen) for 1 h at 4 °C. After extensive washing (five times with 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, and 0.1% SDS), the fragments were eluted with 100 mm EDTA, pH 8.0, and analyzed for the presence of O-glucose glycans by mass spectrometry as described (17, 38). Briefly, samples were reduced and alkylated, separated by SDS-PAGE, and subjected to in-gel trypsin digestion. The resulting tryptic peptides were then analyzed by liquid chromatography-MS/MS on an Agilent XCT ion trap mass spectrometer. O-Glucose-modified peptides were identified by neutral loss searches as described previously (17, 38). A detailed description of the mass spectral analysis of O-glucose glycans on mouse Notch1 is being published separately.3
Nucleotide Sugar Composition—Nucleotide sugars were isolated by formic acid extraction as described for yeast (39). About 8 × 107 cells of wild-type and mutant CHO cells were pelleted and resuspended in 5 ml of 1 m formic acid saturated with butanol. After 1 h of shaking at 4 °C, samples were centrifuged for 5 min at 13,000 × g, and the supernatants were lyophilized and dissolved in 200 μl of water. Insoluble material was sedimented, and 50 μl of the supernatants were separated on a Develosil RPAQUEOUS column (250 × 4.6 mm; Nomura Chemical Co., Ltd., Seto, Japan). The column was equilibrated with 20 mm triethylamine acetate buffer (pH 7.0) at a flow rate of 0.7 ml·min-1. UDP-sugars were detected by UV260 nm absorbance. The peaks of UDP-Glc, UDP-GlcA, and UDP-Xyl were collected based on the retention times of the standards. The combined fractions were reloaded on the same column and run under the same conditions. Identities of peaks were confirmed by mass spectrometry as described (39).
RESULTS
A CHO Cell Mutant Deficient in Xylose Incorporation—A CHO cell clone, designated pgsI-208 according to the nomenclature of previously identified proteoglycan synthesis mutants (21), was selected from a mutagenized population using FGF-2-Saporin, a recombinant chimera consisting of FGF-2 fused to the plant cytotoxin saporin-6 (Selective Genetics, Inc., La Jolla, CA), as described for the isolation of a glucuronyltransferase I mutant (21). Since FGF-2 binds to heparan sulfate, this procedure selects for mutants deficient in heparan sulfate biosynthesis. The addition of naphthol-β-d-xyloside restored the incorporation of 35SO4 in GAGs in pgsI-208 (Table 1) as in the previously isolated mutant pgsA-745, exhibiting a defect in the GAG initiating XylT2 (4-6, 18). This result indicated that all of the downstream enzymes were present, and mutation in an upstream step involved in xylosylation or production of precursors was anticipated.
TABLE 1.
GAG content of mutant pgsl-208
pgsI-208, pgsA-745, and wild-type cells were metabolically labeled with either [6-3H]glucosamine or 35SO4 in the presence or absence of naphthol-β-d-xyloside (Xylβ-O-Np). GAG was isolated as described (21).
|
Strain
|
[6-3H]Glucosamine
|
35SO4
|
|
|---|---|---|---|
| Without Xylβ-O-Np | With Xylβ-O-Np | ||
| cpm/μg cell protein | cpm/μg cell protein | ||
| CHO | 229 | 142 | 251 |
| pgsA-745 | 20 | 3 | 763 |
| pgsI-208 | 38 | 14 | 185 |
The cell line was then fused pairwise to mutants pgsA-745 and pgsB-761, the two previously isolated mutants in which xyloside restores GAG synthesis (18, 25), to test for genetic complementation (34). Fusion of pgsI-208 cells with pgsA-745 and pgsB-761 also restored GAG synthesis (not shown), indicating that the defect was different from these strains.
To test if UDP-Xyl, the substrate for the xylosyltransferases, was available in the Golgi of pgsI-208 cells, AtXylT was expressed by transient transfection. This approach provides a convenient way to measure UDP-Xyl indirectly, since the epitope formed by this enzyme on N-glycans is absent in mammalian cells and can be readily detected on the cell surface using rabbit anti-HRP, which is reactive to plant type N-glycans containing β-linked xylose (40). In wild type CHO cells, transient expression of AtXylT resulted in anti-HRP-positive cells, whereas transfection of mutant pgsI-208 cells did not (Fig. 2A). The simultaneous absence of GAG biosynthesis and xylosylation of N-glycans in the mutant strongly suggested that the substrate UDP-Xyl used by all xylosyltransferases in the ER or Golgi lumen was not available.
FIGURE 2.
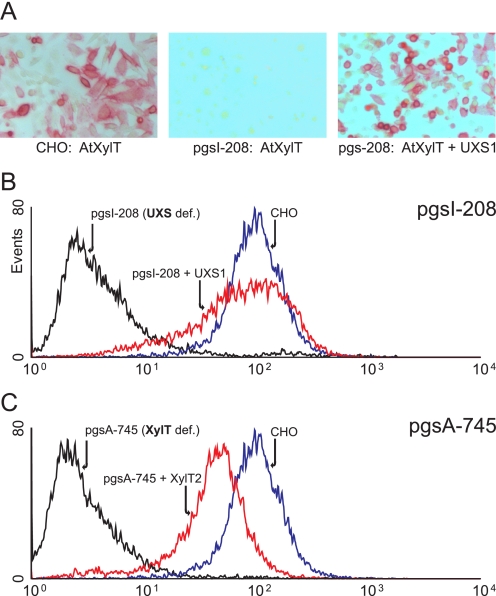
Complementation of pgsI-208 by UXS. A, CHO or pgsI-208 cells transfected with AtXylT. The product of AtXylT, xylosylated N-glycans, was detected on the cell surface using anti-HRP, reactive to xylosylated N-glycans of this protein (40). B and C, pgsI-208 and pgsA-745 cells were stained with antibody RB4Ea12 (35, 36) against heparan sulfate and analyzed by flow cytometry. B, CHO cells (blue) compared with pgsI-208 (black) and pgsI-208 stably transfected with human UXS1 (red). C, CHO cells (blue) compared with xylosyltransferase-deficient mutant pgsA-745 (black) and pgsA-745 stably transfected with human XylT2 (red).
pgsI-208 Exhibits a Defective UDP-xylose Synthase—Current evidence suggests that UDP-xylose is generated inside the ER/Golgi lumen by decarboxylation of UDP-GlcA through the enzyme UXS (26-28). To test if this enzyme was missing from the mutant, a UXS-expressing plasmid was co-transfected with AtXylT. Xylosylation of N-glycans was restored under these conditions, indicating that the absence of UXS was the cause of the observed phenotypes of these cells (Fig. 2A). No salvage pathway for xylose exists in CHO cells, since feeding cells with up to 10 mm xylose did not result in any detectable incorporation of xylose into N-glycans upon expression of AtXylT (not shown).
Reverse transcription-PCR, using two primer pairs covering approximately each half of the coding region of the hamster UXS, showed that mRNA was made. Sequencing of these fragments, however, revealed an exchange of a CAG codon (glutamine 295) for a TAG stop codon. Presumably, truncation of the C-terminal 125 amino acids from the full-length version of the protein (420 amino acids) inactivated the enzyme, suggesting that this point mutation was the cause of the UXS deficiency in the mutant. Sequencing the genomic region confirmed the presence of this mutation but also showed that only one allele was affected. Since only the mutant allele was detected in cDNAs prepared from the mutant, we concluded that the second allele is inactive, a phenomenon not uncommon in CHO cells, which are considered to be partly functionally haploid (41).
pgsI-208 Lacks Both Glycosaminoglycans and Xylosylation of O-Glucose-linked Glycans on EGF Repeats of Notch—Since pgsI-208 was selected for resistance to FGF-2-saporin, the absence of heparan sulfate was anticipated. To examine this possibility, pgsI-208 cells were analyzed by flow cytometry using antibody RB4Ea12 against heparan sulfate (35, 36). Wild-type CHO cells exhibited strong staining by this antibody, whereas pgsI-208 did not and behaved like pgsA-745 cells, defective in XylT2 (Fig. 2). Expression of human UXS1 and XylT2 in pgsI-208 and pgsA-745, respectively, restored antibody binding to a level comparable with wild-type CHO cells (Fig. 2, B and C).
To investigate the presence of xylose in O-glucose-linked glycans, a secreted construct containing the first five EGF-like repeats of mouse Notch1 (37), was expressed in CHO, pgsA-745, and pgsI-208 cells. The C-terminal His tag on this protein allowed its purification from the medium. Tryptic peptides were analyzed for the presence of O-glucose glycans using liquid chromatography-MS/MS (38). We have previously identified a tryptic peptide from EGF 4 bearing an O-glucose trisaccharide when analyzing samples derived from Lec1-CHO cells3 (Fig. 3). In both wild-type cells (not shown) and pgsA-745, the same peptide modified with O-glucose trisaccharide was detected (m/z = 1255.1; Fig. 3A). In contrast, analysis of the sample from pgsI-208 revealed the presence of the peptide modified with a single hexose residue (m/z = 1166.7; Fig. 3B). The peptide modified with a single hexose elutes slightly later (53.7 min; Fig. 3B) than that with the trisaccharide (52.9 min; Fig. 3A), consistent with the predicted effects of two fewer sugars on retention time. Searches of the data showed that the trisaccharide form was present in the sample from pgsA-745 but not pgsI-208 (Fig. 3C), whereas the monosaccharide form was found in pgsI-208 but not pgsA-745 (Fig. 3D). These data suggested that the defect in UXS prevents the addition of xyloses to O-glucose on EGF repeats.
FIGURE 3.
Glycan analysis of Notch EGF1-5 by mass spectrometry. pgsA-745 and pgsI-208 cells were transiently transfected with a construct encoding EGF repeats 1-5 of mouse Notch1. Tryptic peptides of the purified secreted product were analyzed by liquid chromatography-MS/MS for the presence of peptides modified with O-glucose. A, identification of the O-glucose trisaccharide form of a peptide from the pgsA-745 sample. The top panel shows an MS spectrum of material eluting at 52.9 min. The ions labeled [M + 3H]3+ and [M + 4H]4+ match the predicted mass for triply and quadruply charged forms of the O-glucose trisaccharide form of 137SCQQADPCASNPCANGGQCLPFESSYICR165, a tryptic peptide from EGF 4 previously demonstrated to be modified with O-glucose trisaccharide.3 Collision-induced dissociation fragmentation of the triply charged form of this peptide resulted in the MS/MS spectrum shown in the bottom panel. The major product ions at m/z 1210.9, 1167.1, and 1112.9 correspond to sequential losses of a pentose, a second pentose, and a hexose. Numerous sequence fragment ions (y-ions are shown) are observed that confirm the identity of the peptide. Ions selected for fragmentation in the MS spectrum are identified by red diamonds. The position of the parent ion fragmented in the MS/MS spectrum is identified with a blue diamond. B, identification of the O-glucose monosaccharide form of the peptide from the pgsI-208 sample. The top panel shows an MS spectrum of material eluting at 53.7 min, slightly later than the trisaccharide form (consistent with the loss of two hydrophilic xylose residues). The ions labeled [M + 3H]3+ and [M + 4H]4+ match the predicted mass for triply and quadruply charged forms of the glycopeptide. Collision-induced dissociation fragmentation of the triply charged form resulted in the MS/MS spectrum shown in the bottom panel. The major product ion, m/z 1112.8, matches the mass for the unglycosylated peptide. C, the trisaccharide form is present in samples from pgsA-745 but not pgsI-208. The data from both samples was searched for the ion corresponding to the triply charged form of the trisaccharide form, m/z 1255.1 (see A). D, the monosaccharide form is present in samples from psgI-208 but not pgsA-745. The data from both samples was searched for the ion corresponding to the triply charged form of the monosaccharide form, m/z 1166.7 (see B).
The UDP-GlcA Content of pgsI-208 Is Dramatically Increased—The enzyme UDP-glucose dehydrogenase (UGDH) (Fig. 1) is known to be inhibited by UDP-Xyl (42). In Cryptococcus neoformans, the absence of cytosolic UXS results in an drastic increase of the UDP-GlcA concentration due to lack of feedback inhibition of the dehydrogenase by UDP-Xyl (43). To investigate the consequences of reduced UDP-Xyl production in the mammalian cell line where UXS is localized in the ER/Golgi lumen, the composition of the pool of relevant nucleotide sugars was determined in wild-type, XylT2 (pgsA-745), and UXS (pgsI-208) mutants using formic acid extraction and HPLC analyses (39). pgsA-745 and wild-type cells showed the same content of UDP-Glc, UDP-GlcA, and UDP-Xyl (Fig. 4, A and B), indicating that the absence of GAG synthesis had no influence on the ratio of these nucleotide sugars. Conversely, UDP-Xyl was completely absent in pgsI-208, whereas the concentration of UDP-GlcA was dramatically increased to a level about 200 times higher than the level in wild-type cells (Fig. 4). This finding strongly suggested that the luminally produced UDP-Xyl is controlling the cytoplasmically produced UDP-GlcA pool.
FIGURE 4.
UDP-Glc, UDP-GlcA, and UDP-Xyl content of CHO cells. Nucleotide sugars of wild-type, pgsA-745, and pgsI-208 cells were separated in a two-step HPLC as described under “Experimental Procedures” (A-C). Fractions of the first HPLC run were collected, and combined fractions of UDP-Glc, UDP-GlcA, and UDP-Xyl (based on the position of standards (D)) were applied to a second run. E shows the pgsI-208 profile on a decreased scale to demonstrate the dramatic increase of UDP-GlcA compared with UDP-Glc. UDP-GlcA in pgsI-208 is estimated to be increased by a factor of 200 over wild type CHO cell levels based on peak area. *, location of UDP-Xyl as confirmed by mass spectrometry as described (not shown) (39).
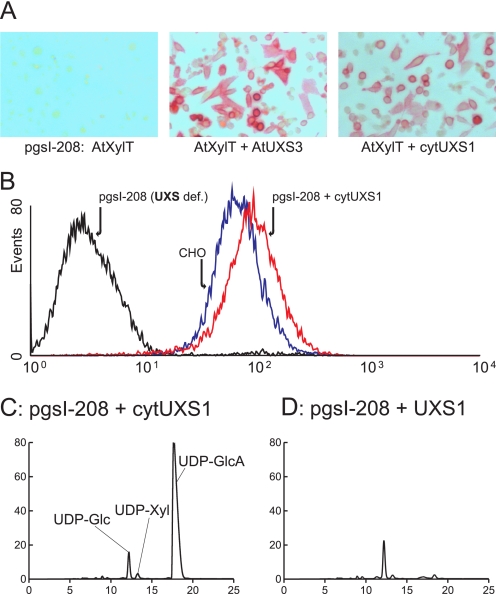
Xylose Incorporation in Glycans Is Restored by Cytoplasmic Expression of UXS—Previous in vitro experiments have shown that UDP-Xyl can be transported across the Golgi membrane (27, 44), and a Golgi-localized UDP-Xyl transporter has been identified (29). The in vivo significance of these findings is not known. To investigate if the transporter can function in vivo, two UXS expressing plasmids, cytoplasmic Arabidopsis UXS3 (30) and a human UXS expressed without the putative N-terminal transmembrane domain (cytUXS1), were transiently expressed together with AtXylT in pgsI-208 cells. Cytoplasmic localization of the UXS proteins was confirmed by immunofluorescence (not shown). Cytoplasmic UXS rendered cells positive for rabbit anti-HRP, which recognizes xylosylated N-glycans (Fig. 5A). More quantitative data were provided by stable expression of cytUXS in pgsI-208 and analyses of the cells with the anti-heparan sulfate antibody by flow cytometry (Fig. 5B). GAG biosynthesis was fully restored in these cells, indicating that sufficient UDP-Xyl could be provided to the lumen of the ER/Golgi from UDP-Xyl produced in the cytoplasm.
FIGURE 5.
Complementation of pgsI-208 by cytoplasmic expression of UXS. A, pgsI-208 cells were transfected with AtXylT or cotransfected with AtUXS3 or cytUXS1 and stained with anti-HRP as in Fig. 2A. B, cells were stained with the anti-heparan sulfate antibody as in Fig. 2B and analyzed by flow cytometry. CHO cells (blue) compared with pgsI-208 (black) and pgsI-208 stably transfected with human UXS1 expressed in the cytoplasm (red). C and D, nucleotide sugar composition analysis (as in Fig. 4) of pgsI-208 expressing human cytUXS1 (C) or UXS1 (D). Identity of the UDP-Xyl peak has been confirmed by mass spectrometry.
As expected, expression of cytoplasmic UXS decreased UDP-GlcA production, but the extent of inhibition was not as great as when UXS was expressed in the ER/Golgi. As shown in Fig. 5C, UDP-Xyl was detectable, but the UDP-GlcA level was only reduced by a factor of ∼4 compared with pgsI-208 cells. In contrast, recombinant expression of UXS in the compartmental lumen normalized nucleotide sugar levels to that of wild-type cells (Fig. 5D).
DISCUSSION
In this report, we describe a new CHO mutant, named here pgsI-208, defective in UXS, the enzyme generating UDP-Xyl. Lack of UDP-Xyl results in absence of both GAG biosynthesis and the addition of xylose to O-glucose glycans of EGF repeats present in Notch. Since CHO cells make both heparan sulfate and chondroitin sulfate, both types of GAGs are anticipated to be lacking in this cell line. Notch is the only protein carrying the Xylα1-3Xylα1-3Glcβ-O-Ser trisaccharide tested in the system (3), but presumably other proteins bearing this sequence would be affected as well. The mutant also has the interesting phenotype of deregulated production of UDP-GlcA and led to the discovery that when provided with a cytosolic source of UDP-Xyl, this nucleotide sugar can be imported into Golgi and/or ER.
UXS was originally cloned from the pathogenic fungus C. neoformans (45) and later from other eukaryotes (28, 30, 46). Mutants in the enzyme have been isolated from C. neoformans (47), where it is one of the capsule structure mutant strains (Cas2), and in Caenorhabditis elegans (26) as one of the squashed vulva (sqv-1) mutants affecting chondroitin and heparan sulfate biosynthesis. In CHO cells, like in C. neoformans (43), inactivation of UXS resulted in an increase of cellular UDP-GlcA. In vitro experiments have shown that UGDH, which generates UDP-GlcA from UDP-Glc, is inhibited by UDP-Xyl (42). Inhibition evidently occurs in vivo as well, since the absence of UDP-xylose resulted in a dramatic increase in UDP-GlcA. The accumulation of UDP-GlcA was most likely due to deregulated synthesis as opposed to a lack of consumption of UDP-GlcA. This conclusion is based on the observation that in pgsA-745, which, like pgsI-208, cannot incorporate UDP-GlcA into glycosaminoglycans, the ratio of UDP-Glc, UDP-GlcA, and UDP-Xyl was unchanged compared with wild-type CHO cells. In yeast, a system that does not utilize UDP-GlcA and UDP-Xyl, expression of UGDH can convert more than half of the UDP-Glc pool into UDP-GlcA (39). After additional expression of UXS, UDP-GlcA was no longer detectable due to substoichiometric accumulation of UDP-Xyl.
In contrast to fungi, where the enzyme is cytoplasmic (43), UXS of mammals (28) and C. elegans (26) and two of the three plant enzymes (30) are localized in the ER/Golgi lumen. Theoretically, physical separation of UDP-Xyl production from UDP-GlcA production provides a mechanism for cells to avoid inhibition of UGDH by UDP-Xyl. However, our current data do not support this hypothesis, because they clearly show that UDP-GlcA production is regulated by UDP-Xyl in mammalian cells. This conclusion was supported by the fact that cytoplasmically expressed UXS inhibited UDP-GlcA production, but less efficiently than the luminally expressed enzyme. The lower inhibition of cytUXS might be explained by differences in the rate of production of UDP-xylose or by other mechanisms. More data are needed for a conclusive interpretation of this interesting finding.
Although, we cannot exclude the possibility that a small portion of UDP-Xyl is produced in the cytoplasm, it seems more likely that UDP-Xyl generated in the Golgi/ER is transported to the cytoplasm to inhibit UGDH. This assumption is supported by observations of Bossuyt and Blanckaert (48) that show that luminal UDP-Xyl stimulates uptake of UDP-GlcA into ER vesicles, possibly by providing an antiport substrate, suggesting the existence of a UDP-GlcA/UDP-Xyl antiporter in the ER membrane. Transport of UDP-Xyl in the other direction, into the ER/Golgi lumen, has also been shown by transport of UDP-Xyl into isolated microsomes (44) or by incorporation of xylose from UDP-[14C]Xyl in permeabilized cells (27). Such a function could be provided by SLC35B4, a recently identified UDP-Xyl transporter of the Golgi (29). According to our results, UDP-Xyl is indeed transported into the Golgi/ER network, since cytosolic UXS allowed the formation of GAGs and xylosylation of N-glycans in the mutant. Thus, UDP-Xyl appears to be transported in both directions across the ER/Golgi membrane.
The exact localization of all proteins in this pathway is not completely resolved. The proteoglycan xylosyltransferases have been shown to be Golgi-localized (49), but evidence exists for co-translational xylosylation (50). Although evidence has been presented for UXS in the Golgi (28), our tagged constructs appear only in the ER.4 The ER and Golgi are also not uniform structures, and production and consumption of UDP-Xyl can take place at different subcellular locations. Whether most of the luminally produced UDP-Xyl is directly used in xylosylation reactions or is transported out of the ER or Golgi and back into the Golgi lumen remains unsolved. Methods to decrease the expression of different potential transporters in this process could provide insight into this problem.
Although a salvage pathway for the regeneration of xylose is anticipated in plants (51), no indication for its occurrence exists in mammalian cells. Feeding of CHO cells with xylose did not restore the mutant phenotype of pgsI-208. Furthermore, auxotrophic mutants dependent on exogenous xylose have not been identified in screening mutagenized cells. No ortholog of the plant enzyme (51) that is able to biosynthesize UDP-Xyl from xylose-1-phosphate is found in mammalian genomes. The pathway via UDP-GlcA, catalyzed by UXS, therefore, appears to be the only way to generate UDP-Xyl in CHO cells and probably all mammals.
Acknowledgments
We thank Gerdy ten Dam and T. van Kuppevelt (University of Nijmegen, The Netherlands) for kindly providing antibody RB4Ea12 and Detlef Neumann (Medizinische Hochschule Hannover, Germany) for help with flow cytometry.
This work was supported, in whole or in part, by National Institutes of Health Grants GM33063 (to J. D. E.) and GM61126 (to R. S. H.). This work was also supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) for the Junior Research Group “Glycomics” established under the roof of REBIRTH, a DFG cluster of Excellence (to R. G.-S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GAG, glycosaminoglycan; CHO, Chinese hamster ovary; EGF, epidermal growth factor; FGF, fibroblast growth factor; HRP, horseradish peroxidase; UGDH, UDP-glucose dehydrogenase; UXS, UDP-xylose synthase; XylT, xylosyltransferase; AtXylT, A. thaliana β1,2-xylosyltransferase; ER, endoplasmic reticulum; MS, mass spectrometry; HPLC, high pressure liquid chromatography.
A. Nita-Lazar, N. A. Rana, H. Takeuchi, R. Orhue, M. P. Myers, K. B. Luther, and R. S. Haltiwanger, submitted for publication.
H. Bakker, T. Oka, A. Ashikov, A. Yadav, M. Berger, N. A. Rana, X. Bai, Y. Jigami, R. S. Haltiwanger, J. D. Esko, and R. Gerardy-Schahn, unpublished results.
References
- 1.Roden, L., and Armand, G. (1966) J. Biol. Chem. 241 65-70 [PubMed] [Google Scholar]
- 2.Hase, S., Kawabata, S., Nishimura, H., Takeya, H., Sueyoshi, T., Miyata, T., Iwanaga, S., Takao, T., Shimonishi, Y., and Ikenaka, T. (1988) J. Biochem. (Tokyo) 104 867-868 [DOI] [PubMed] [Google Scholar]
- 3.Moloney, D. J., Shair, L. H., Lu, F. M., Xia, J., Locke, R., Matta, K. L., and Haltiwanger, R. S. (2000) J. Biol. Chem. 275 9604-9611 [DOI] [PubMed] [Google Scholar]
- 4.Ponighaus, C., Ambrosius, M., Casanova, J. C., Prante, C., Kuhn, J., Esko, J. D., Kleesiek, K., and Götting, C. (2007) J. Biol. Chem. 282 5201-5206 [DOI] [PubMed] [Google Scholar]
- 5.Voglmeir, J., Voglauer, R., and Wilson, I. B. (2007) J. Biol. Chem. 282 5984-5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuellar, K., Chuong, H., Hubbell, S. M., and Hinsdale, M. E. (2007) J. Biol. Chem. 282 5195-5200 [DOI] [PubMed] [Google Scholar]
- 7.Götting, C., Kuhn, J., Zahn, R., Brinkmann, T., and Kleesiek, K. (2000) J. Mol. Biol. 304 517-528 [DOI] [PubMed] [Google Scholar]
- 8.Minamida, S., Aoki, K., Natsuka, S., Omichi, K., Fukase, K., Kusumoto, S., and Hase, S. (1996) J. Biochem. (Tokyo) 120 1002-1006 [DOI] [PubMed] [Google Scholar]
- 9.Ishimizu, T., Sano, K., Uchida, T., Teshima, H., Omichi, K., Hojo, H., Nakahara, Y., and Hase, S. (2007) J. Biochem. (Tokyo) 141 593-600 [DOI] [PubMed] [Google Scholar]
- 10.Omichi, K., Aoki, K., Minamida, S., and Hase, S. (1997) Eur. J. Biochem. 245 143-146 [DOI] [PubMed] [Google Scholar]
- 11.Bruckner, K., Perez, L., Clausen, H., and Cohen, S. (2000) Nature 406 411-415 [DOI] [PubMed] [Google Scholar]
- 12.Xu, A., Haines, N., Dlugosz, M., Rana, N. A., Takeuchi, H., Haltiwanger, R. S., and Irvine, K. D. (2007) J. Biol. Chem. 282 35153-35162 [DOI] [PubMed] [Google Scholar]
- 13.Hicks, C., Johnston, S. H., diSibio, G., Collazo, A., Vogt, T. F., and Weinmaster, G. (2000) Nat. Cell Biol. 2 515-520 [DOI] [PubMed] [Google Scholar]
- 14.Moloney, D. J., Panin, V. M., Johnston, S. H., Chen, J., Shao, L., Wilson, R., Wang, Y., Stanley, P., Irvine, K. D., Haltiwanger, R. S., and Vogt, T. F. (2000) Nature 406 369-375 [DOI] [PubMed] [Google Scholar]
- 15.Shi, S., and Stanley, P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 5234-5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okajima, T., and Irvine, K. D. (2002) Cell 111 893-904 [DOI] [PubMed] [Google Scholar]
- 17.Acar, M., Jafar-Nejad, H., Takeuchi, H., Rajan, A., Ibrani, D., Rana, N. A., Pan, H., Haltiwanger, R. S., and Bellen, H. J. (2008) Cell 132 247-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esko, J. D., Stewart, T. E., and Taylor, W. H. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 3197-3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai, X., and Esko, J. D. (1996) J. Biol. Chem. 271 17711-17717 [DOI] [PubMed] [Google Scholar]
- 20.Zhang, L., Beeler, D. L., Lawrence, R., Lech, M., Liu, J., Davis, J. C., Shriver, Z., Sasisekharan, R., and Rosenberg, R. D. (2001) J. Biol. Chem. 276 42311-42321 [DOI] [PubMed] [Google Scholar]
- 21.Bai, X., Wei, G., Sinha, A., and Esko, J. D. (1999) J. Biol. Chem. 274 13017-13024 [DOI] [PubMed] [Google Scholar]
- 22.Esko, J. D. (1992) Adv. Exp. Med. Biol. 313 97-106 [DOI] [PubMed] [Google Scholar]
- 23.Lidholt, K., Weinke, J. L., Kiser, C. S., Lugemwa, F. N., Bame, K. J., Cheifetz, S., Massague, J., Lindahl, U., and Esko, J. D. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 2267-2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bame, K. J., and Esko, J. D. (1989) J. Biol. Chem. 264 8059-8065 [PubMed] [Google Scholar]
- 25.Esko, J. D., Weinke, J. L., Taylor, W. H., Ekborg, G., Roden, L., Anantharamaiah, G., and Gawish, A. (1987) J. Biol. Chem. 262 12189-12195 [PubMed] [Google Scholar]
- 26.Hwang, H. Y., and Horvitz, H. R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14218-14223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kearns, A. E., Vertel, B. M., and Schwartz, N. B. (1993) J. Biol. Chem. 268 11097-11104 [PubMed] [Google Scholar]
- 28.Moriarity, J. L., Hurt, K. J., Resnick, A. C., Storm, P. B., Laroy, W., Schnaar, R. L., and Snyder, S. H. (2002) J. Biol. Chem. 277 16968-16975 [DOI] [PubMed] [Google Scholar]
- 29.Ashikov, A., Routier, F., Fuhlrott, J., Helmus, Y., Wild, M., Gerardy-Schahn, R., and Bakker, H. (2005) J. Biol. Chem. 280 27230-27235 [DOI] [PubMed] [Google Scholar]
- 30.Harper, A. D., and Bar-Peled, M. (2002) Plant Physiol. 130 2188-2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakker, H., Lommen, A., Jordi, W., Stiekema, W., and Bosch, D. (1999) Biochem. Biophys. Res. Commun. 261 829-832 [DOI] [PubMed] [Google Scholar]
- 32.Strasser, R., Mucha, J., Mach, L., Altmann, F., Wilson, I. B., Glössl, J., and Steinkellner, H. (2000) FEBS Lett. 472 105-108 [DOI] [PubMed] [Google Scholar]
- 33.Elbein, A. D., and Bannon, G. A. (September 4, 2003) U. S. Patent 20,030,166,012
- 34.Esko, J. D. (1989) Methods Cell Biol. 32 387-422 [DOI] [PubMed] [Google Scholar]
- 35.Jenniskens, G. J., Oosterhof, A., Brandwijk, R., Veerkamp, J. H., and van Kuppevelt, T. H. (2000) J. Neurosci. 20 4099-4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ten Dam, G. B., Hafmans, T., Veerkamp, J. H., and van Kuppevelt, T. H. (2003) J. Histochem. Cytochem. 51 727-739 [DOI] [PubMed] [Google Scholar]
- 37.Shao, L., Moloney, D. J., and Haltiwanger, R. (2003) J. Biol. Chem. 278 7775-7782 [DOI] [PubMed] [Google Scholar]
- 38.Nita-Lazar, A., and Haltiwanger, R. S. (2006) Methods Enzymol. 417 93-111 [DOI] [PubMed] [Google Scholar]
- 39.Oka, T., and Jigami, Y. (2006) FEBS J. 273 2645-2657 [DOI] [PubMed] [Google Scholar]
- 40.Faye, L., Gomord, V., Fitchette-Laine, A. C., and Chrispeels, M. J. (1993) Anal. Biochem. 209 104-108 [DOI] [PubMed] [Google Scholar]
- 41.Siminovitch, L. (1976) Cell 7 1-11 [DOI] [PubMed] [Google Scholar]
- 42.Neufeld, E. F., and Hall, C. W. (1965) Biochem. Biophys. Res. Commun. 19 456-461 [DOI] [PubMed] [Google Scholar]
- 43.Griffith, C. L., Klutts, J. S., Zhang, L., Levery, S. B., and Doering, T. L. (2004) J. Biol. Chem. 279 51669-51676 [DOI] [PubMed] [Google Scholar]
- 44.Nuwayhid, N., Glaser, J. H., Johnson, J. C., Conrad, H. E., Hauser, S. C., and Hirschberg, C. B. (1986) J. Biol. Chem. 261 12936-12941 [PubMed] [Google Scholar]
- 45.Bar-Peled, M., Griffith, C. L., and Doering, T. L. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12003-12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi, M., Nakagawa, H., Suda, I., Miyagawa, I., and Matoh, T. (2002) Plant Cell Physiol. 43 1259-1265 [DOI] [PubMed] [Google Scholar]
- 47.Moyrand, F., Klaproth, B., Himmelreich, U., Dromer, F., and Janbon, G. (2002) Mol. Microbiol. 45 837-849 [DOI] [PubMed] [Google Scholar]
- 48.Bossuyt, X., and Blanckaert, N. (2001) J. Hepatol. 34 210-214 [DOI] [PubMed] [Google Scholar]
- 49.Schön, S., Prante, C., Bahr, C., Kuhn, J., Kleesiek, K., and Götting, C. (2006) J. Biol. Chem. 281 14224-14231 [DOI] [PubMed] [Google Scholar]
- 50.Geetha-Habib, M., Campbell, S. C., and Schwartz, N. B. (1984) J. Biol. Chem. 259 7300-7310 [PubMed] [Google Scholar]
- 51.Kotake, T., Yamaguchi, D., Ohzono, H., Hojo, S., Kaneko, S., Ishida, H. K., and Tsumuraya, Y. (2004) J. Biol. Chem. 279 45728-45736 [DOI] [PubMed] [Google Scholar]
- 52.Muraoka, M., Kawakita, M., and Ishida, N. (2001) FEBS Lett. 495 87-93 [DOI] [PubMed] [Google Scholar]