Abstract
Individual neurons express receptors for several different growth factors that influence the survival, growth, neurotransmitter phenotype, and other properties of the cell. Although there has been considerable progress in elucidating the molecular signal transduction pathways and physiological responses of neurons and other cells to individual growth factors, little is known about if and how signals from different growth factors are integrated within a neuron. In this study, we determined the interactive effects of nerve growth factor, insulin-like growth factor 1, and epidermal growth factor on the activation status of downstream kinase cascades and transcription factors, cell survival, and neurotransmitter production in neural cells that express receptors for all three growth factors. We document considerable differences in the quality and quantity of intracellular signaling and eventual phenotypic responses that are dependent on whether cells are exposed to a single or multiple growth factors. Dual stimulations that generated the greatest antagonistic or synergistic actions, compared with a theoretically neutral summation of their two activities, yielded the largest eventual change of neuronal phenotype indicated by the ability of the cell to produce norepinephrine or resist oxidative stress. Combined activation of insulin-like growth factor 1 and epidermal growth factor receptors was particularly notable for antagonistic interactions at some levels of signal transduction and norepinephrine production, but potentiation at other levels of signaling and cytoprotection. Our findings suggest that in true physiological settings where multiple growth factors are present, activation of one receptor type may result in molecular and phenotypic responses that are different from that observed in typical experimental paradigms in which cells are exposed to only a single growth factor at a time.
Numerous growth factors have been identified that influence the survival and plasticity of neurons, including the neurotrophins (NGF,2 brain-derived neurotrophic factor, NT-3, and NT-4), basic fibroblast growth factor, insulin-like growth factor 1 (IGF-1), and epidermal growth factor (EGF). These different factors activate receptors with intrinsic tyrosine kinase activity, which then engage downstream kinase cascades, resulting in the activation of transcription factors. For example, NGF activates the TrkA receptor that engages the Raf-mitogen-activated protein kinase kinase (MEK)-extracellular signal-regulated kinase (ERK) pathway resulting in activation of transcription factors, including AP1 (1-3); IGF-1 activates the IGF-1 receptor that is coupled to phosphatidylinositol trisphosphate kinase, Akt kinase, and forkhead transcription factors of the FOXO family (4, 5); and EGF receptors engage the RafMEK-ERK pathway (6, 7). Many neural cells express receptors for multiple growth factors, each of which induce similar effects on the survival, differentiation, and function of the cells. Although there has been considerable progress in elucidating the molecular signal transduction pathways and physiological responses of neural cells to individual growth factors, it is not known how signals from different growth factors are integrated within an individual cell. Indeed, much of our understanding of cellular neurophysiology has come from studies that employed in vitro scenarios in which the signal transduction activity of a single receptor signaling pathway is studied largely in isolation of other inputs. This approach provides an understanding of the linear pathways of cell signaling, but does not provide sufficiently reliable information concerning how multiple inputs integrate to generate a physiological response. The inherent complexity of cellular signaling networks and their importance to a wide range of cellular functions necessitates the development of modeling methods that can be applied toward making predictions and highlighting the appropriate experiments to test our understanding of how these systems are designed and function.
Presumably, the molecular integration of distinct multiple inputs (mediated by specific plasma membrane receptors) occurs at the level of their associated signal transduction cascades. Components of these signal transduction cascades (including kinases, phosphatases, and their substrates) are believed to be pre-assembled into higher order structures by molecular scaffold proteins, including AKAP (protein kinase A anchoring protein) (8), POSH (plenty of SH3 domains) (9), JIP (c-Jun amino-terminal kinase interacting protein) (10), β-arrestins (11), and 14-3-3 proteins (12). These structures compartmentalize signaling pathways in the cell, enhance specificity of target-substrate interactions, and improve the speed and efficiency of signal transduction. Thus, there is a funneling of the complex and diverse signaling inputs from ligands and their specific receptors at the plasma membrane into the higher order multiprotein signaling scaffolds attached either to cytoskeletal proteins or the plasma membrane itself. The high receptor diversity allows specific receptor signaling patterns at the plasma membrane surface, but the signaling proteins that are activated by these receptors are often common among different receptor-activated transduction pathways. Thus the signaling traffic is likely to converge at cytoplasmic nexi, represented by the scaffolding proteins. Clustering of signaling molecules in multiprotein signaling complexes eliminates delays that would otherwise occur as a result of random diffusion in the cytoplasm. An understanding of how multiple inputs are integrated will provide a better appreciation of how multiple neurotrophic factors actually mediate intracellular signaling events in the physiological setting.
One cell type that has provided a valuable model for the elucidation of neurotrophic factor signal transduction mechanisms is PC12 pheochromocytoma cells generated by Greene and Tischler (13). These cells respond to NGF by differentiating into a neuron-like phenotype characterized by cessation of cell division, extension of neurites, acquisition of excitable membrane properties, and the production of the monoamine neurotransmitters dopamine and norepinephrine (13-16). PC12 cells also express receptors for IGF-1 and EGF, and activation of either of these two receptor types promotes cell survival under conditions of serum deprivation or other apoptotic insults (17-19). In this study we investigated how neuronal signaling is controlled by analyzing the signal transduction pathway activation status in PC12 cells stimulated with single growth factors or combinations of growth factors. Our findings reveal distinctly different qualitative and quantitative signaling patterns and phenotypic responses generated by growth factors when they stimulate neural cells in the presence of other growth factors.
EXPERIMENTAL PROCEDURES
Materials—Epidermal growth factor and insulin-like growth factor-1 were both obtained from Sigma, and nerve growth factor was obtained from Invitrogen. Antisera against proline-rich tyrosine kinase 2 (Pyk2) was purchased from BD Transduction Laboratories. Antisera raised against the active auto-tyrosine 418-phosphorylated form of c-Src were obtained from BIOSOURCE. Anti-Shc, -syntaxin, -Hsp40, -calponin, -casein kinase II, -crystallin antisera, and -agarose pre-conjugated anti-PY20 (phosphotyrosine) to agarose beads were all obtained from Santa Cruz Biotechnology, Inc., and antisera against ERK1/2 (extracellular-regulated kinase 1/2), serine-phosphorylated inactive glycogen synthase kinase 3α/β, and protein kinase B/Akt-1 were all obtained from Cell Signaling Technology. Anti-EGF receptor, IGF-1 receptor, and NGF receptor antisera were obtained from Santa Cruz Biotechnology.
Cell Culture—Rat pheochromocytoma cells (PC12) were routinely maintained in RPMI 1640 media supplemented with 10% horse serum, 5% fetal bovine serum, and 1% antibiotic/antimycotic (penicillin/streptomycin/fungizone: Invitrogen) at 37 °C in a 5% CO2 atmosphere. Prior to any cellular stimulations with the growth factors, the cells were serum-deprived for 16 h by removing the horse and bovine serum content of the RPMI growth medium and addition of 10 mm HEPES to compensate for the reduced buffering capacity. For differentiation studies of the PC12 cells, the serum content of the RPMI media was reduced to 1% horse and 1% bovine serum in addition to the 10 mm HEPES application. All PC12 cells were maintained in 100-mm plates coated with poly-d-lysine (Sigma).
Immunoprecipitation and Immunoblotting—After stimulation with EGF (10 ng/ml), NGF (10 ng/ml), or IGF-1 (25 nm), cell monolayers were placed on ice, washed twice in ice-cold Dulbecco's phosphate-buffered saline, and lysed in Nonidet P-40-based solubilization buffer, as described previously (7). Solubilized lysates were clarified by centrifugation at 15,000 rpm for 15 min and diluted to an approximate concentration of 1 mg/ml total protein. Subsequently, a 50-μl aliquot of clarified whole-cell lysate was mixed with an equal volume of 2× Laemmli sample buffer and resolved by SDS-PAGE for determination of intracellular protein activation by protein immunoblotting. Immunoprecipitation of the phosphotyrosine proteins Pyk2 and Shc was achieved using 20 μl of a 50% slurry of anti-PY20 affinity agarose (Santa Cruz Biotechnology) with agitation for 16 h at 4 °C. Immune complexes were washed three times with ice-cold Nonidet P-40 based lysis buffer and transferred to a clean microcentrifuge tube before addition of 20 μl of 2× Laemmli sample buffer. Immunoprecipitates were resolved by SDS-PAGE and electrotransferred to polyvinylidene difluoride (PerkinElmer Life Sciences) membrane for protein immunoblotting. Polyvinylidene difluoride membranes were blocked in a 4% bovine serum albumin, 50 mm Tris-HCl, pH 7.0, 0.05% Tween 20, and 0.05% Nonidet P-40 solution for 1 h at 37 °C before immunoblotting. Immunoblotting of whole-cell lysates for ERK1/2, GSK-3, Akt, and active c-Src (Tyr-418 autophosphorylated) was performed using specific antisera (see under “Materials”) at a 1:1000 dilution with subsequent addition of a 1:7000 dilution of alkaline phosphatase-conjugated anti-mouse/rabbit IgG as a secondary antibody (Sigma). Immunoblotting of anti-phosphotyrosine immunoprecipitates, run out on SDS-PAGE, for Pyk2 and Shc was performed using a 1:1000 dilution with subsequent addition of a 1:7000 dilution of alkaline phosphatase-conjugated anti-mouse/rabbit IgG as a secondary antibody (Sigma). Receptor tyrosine kinase immunoprecipitations were performed on neurotrophin-stimulated PC12 cell lysates by incubation with 4 μg of the respective anti-receptor sera for 16 h at 4 °C followed by addition of 25 μl of a 30% slurry of protein A/protein G-Plus-agarose (Calbiochem). Immune complexes were prepared as above and resolved by SDS-PAGE. Their degree of tyrosine phosphorylation was assessed by immunoblotting with a 1:1000 dilution of a PY20 anti-phosphotyrosine monoclonal antibody followed by a 1:7000 dilution of an anti-mouse alkaline phosphatase-conjugated secondary antibody. Visualization of alkaline-phosphatase labeled proteins was performed using enzyme-linked chemifluorescence (Amersham Biosciences) and quantified using a Typhoon 9410 PhosphorImager.
RNA Extraction—PC12 cells were plated on poly-d-lysine-coated 100-mm Petri dishes at a density of 4 × 106 cells/dish. The cells were grown for 24 h in RPMI 1640 media supplemented with 10% horse serum, 5% fetal bovine serum, and 1% antibiotic/antimycotic. The following day, the media were changed to low serum-starving RPMI 1640 media supplemented with 1% horse serum and 10 mm HEPES. PC12 cells were stimulated individually or in dual combinations with EGF (10 ng/ml), IGF-1 (25 nm), or NGF (10 ng/ml) for 2-, 4-, and 8-h time points in triplicate. Cells were washed with phosphate-buffered saline, removed from the plate with a cell scraper, and isolated by centrifugation at 4 °C. The cell pellets were snapfrozen on dry ice and stored temporarily at -80 °C. A mouse 17K cDNA microarray was utilized to analyze gene expression in the cell pellets. The cell pellets were processed using a Bead Beater (Bio-Spec, Bartlesville, OK) followed by RNA purification using the RNeasy mini kit (Qiagen, CA) according to the manufacturers' instructions. The RNA was examined for quantity and quality using an Agilent Bioanalyzer 2100 (Agilent Technologies, CA).
Radioactive cDNA Probe Preparation and Microarray Hybridization—cDNA probe preparation and microarray hybridization were performed as described previously (20). Briefly, 5 μg of total RNA was reverse-transcribed in a reaction mixture containing 8 μl of 5× first strand RT buffer, 1 μl of 1 μg/μl 12-18-mer poly(dT) primer, 4 μl of 2 mm dNTPs (-dCTP), 4 μl of 0.1 m dithiothreitol, 1 μl (40 units) of RNase-OUT, 6 μl of 3000 Ci/mmol [α-33P]dCTP, and diethyl pyrocarbonate/water to a final volume of 40 μl. The RT mixture was first heated at 65 °C for 10 min, followed by incubation on ice for 2 min. Two microliters of Superscript II reverse transcriptase (Invitrogen) was then added followed by incubation at 42 °C for 35 min. One additional microliter of reverse transcriptase was added, followed by another 35-min incubation. At the end of the incubation, 5 μl of 0.5 m EDTA was added to chelate divalent cations. After addition of 10 μl of 1.0 m NaOH, the samples were incubated at 65 °C for 30 min to hydrolyze the remaining RNA. Following the addition of 25 μl of 1 m Tris, pH 8.0, the samples were purified using Bio-Rad 6 purification columns. cDNA microarrays were pre-hybridized in a 4-ml hybridization buffer containing 3.2 ml of Microhyb (Research Genetics, AL) and 0.8 ml of 50% dextran sulfate, 10 μl of 10 mg/ml denatured human Cot 1 DNA (Invitrogen), and 10 μl of 8 mg/ml denatured poly(dA) (Amersham Biosciences). After at least 4 h of pre-hybridization at 55 °C, ∼106 cpm/ml of heat-denatured cDNA probes were added, followed by 17 h of incubation at 55 °C. Hybridized arrays were washed in 2× SSC and 0.1% SDS once at room temperature followed by two washes in 2× SSC and 0.1% SDS at 65 °C for 15 min each.
Scanning and Quantification—The microarrays were exposed to PhosphorImager screens for 3 days. The screens were then scanned in a STORM PhosphorImager (GE Healthcare) at 50 μm resolution. Quantification of scanned screens was performed with ArrayPro software.
Z-scores and Z-ratios—Raw hybridization intensity data were log transformed and normalized to yield Z-scores, which in turn were used to calculate a Z-ratio value for each gene with respect to the control cells. The Z-ratio was calculated as the difference between the observed gene Z-scores for the experimental and the control comparisons and dividing by the standard deviation associated with the distribution of these differences (21). Z-ratio values ≥ +2.0 or ≤ -2.0 were chosen as cutoff values, defining increased and decreased expression, respectively. This eliminates spurious observations of high fold changes from genes of low intensities comparable with the background. The filtered genes were then tested for p values and Z-ratios. The p values test for repeatability of the intensity of a gene was between replicate arrays. The Z-ratios are a measure of fold change between treatments. We then divided by a standard deviation over all the genes on an array to determine whether a given ratio was statistically significant (p < 0.05), considering the array as a whole.
Gene Clustering Analysis and Gene Pathway Analysis—DIANE software (National Institutes of Health) was used to filter the 17,000 genes. We filtered out genes that did not vary at least 1.25-fold from the log of the mean of the first filter in at least 60% of the genes expressed (p < 0.01). Genes were clustered, and sub-clusters were generated using DIANE software. A complete set of 522 cellular pathways was obtained from the Molecular Signatures Data Base (MSigDB) created by the Broad Institute at the Massachusetts Institute of Technology (22). The complete set was tested for Geneset enrichment using Parametric Analysis of Geneset Enrichment (PAGE) (23). For each pathway a Z-score was computed as described previously (24). For each pathway score, a p value was computed using JMP 6.0 software to test for the significance of the Z-score obtained. These tools were part of DIANE 1.0 (grc.nia.nih.gov).
HPLC Analysis of Norepinephrine—PC12 cells plated on poly-d-lysine-coated plates were maintained in low serum media (0.5% fetal calf serum-containing RPMI) with the supplemented neurotrophins either singly or in dual combination paradigms for 7 days before harvesting in a cell pellet through centrifugation. Cell pellets were snap-frozen and stored at -80 °C. Pellets were individually weighed and ultrasonicated in 10% perchloric acid and centrifuged at 25,000 × g for 12 min. The primary neurotransmitter expressed in the PC12 cells is norepinephrine (NE), and thus we concentrated upon the regulation of this by the neurotrophic factors. NE contents in cell lysates were measured by HPLC with electrochemical detection (25). The analytical column was a Symmetry C18 3.5 μm (4.6 × 150.0 mm) from Waters Corp. (Milford, MA). The mobile phase consisted of 0.01 m sodium dihydrogen phosphate, 0.01 m citric acid, 2 mm sodium EDTA, 1 mm sodium octyl sulfate, 10% methanol, pH 3.5, and was used at flow rate 0.9 ml/min and temperature 25 °C. The installation consisted of a Waters 717 Plus automated injection system, a Waters 1525 binary pump, and Coulochem III detector (ESA, Chelmsford, MA). Waters Breeze system was used for data collection and analysis. The cellular content of NE was calculated initially as picogram/mg of cell weight and then converted to a fold induction over the basal level of NE.
Oxidative Stress Resistance—PC12 cells plated onto poly-d-lysine-coated plates were treated with the various individual and combinations of growth factors for 7 days prior to the application of oxidative stress. PC12 cells (seeded at 2 × 106 cells/100-mm Petri dish) were treated for 60 min with 200 μm hydrogen peroxide and then washed with low serum RPMI growth media. Cells were then allowed to grow for a further 48 h before assessment of cell number by viable cell trypan blue staining.
RESULTS
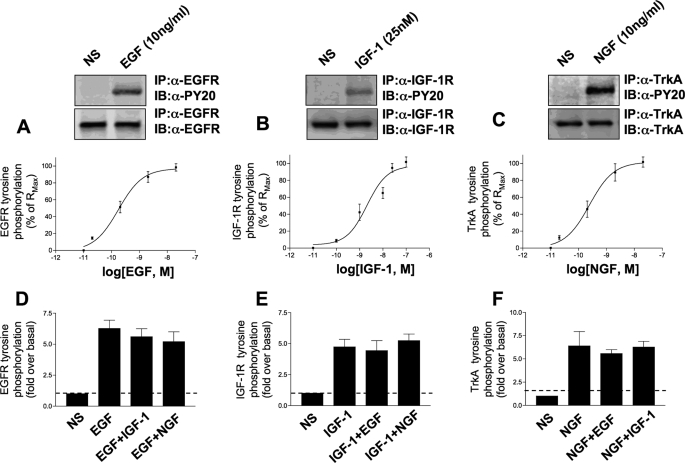
Acute Activation of Cytoplasmic Signaling Factors—Stimulation of PC12 cells with the individual concentrations of EGF, NGF, or IGF-1 resulted in the activation/tyrosine phosphorylation of the multiple cytoplasmic signaling factors (Table 1). The concentrations of the three individual ligands used were considered maximal for this specific cell line. To confirm this, we assessed their ability to induce maximal levels of auto-tyrosine phosphorylation of their cognate receptor tyrosine kinases. The ligand concentration-response relationships for these three receptor tyrosine kinase systems are depicted in Fig. 1. In all subsequent experiments in this study we adhered to the following concentrations of each growth factor: EGF and NGF, 10 ng/ml; IGF-1 25 nm, acknowledging that an extra variable of the growth factor concentration could potentially be considered in further studies.
TABLE 1.
Neurotrophin activation of multiple transduction pathways
The degree of stimulation of the specific output is characterized by the number of asterisks, i.e. high degree of stimulation (2.5-3.0-fold above basal activity) is denoted by three asterisks, two asterisks (1.5-2.5-fold over basal activity), and one asterisk (1-1.5-fold over basal activity) denotes lesser activations. A dash represents no significant activation measured in that immunoblot assay.
|
Ligand
|
Output
|
|||||
|---|---|---|---|---|---|---|
| ERK1/2 | Akt-1 | GSK-3 | Pyk2 tyrosine phosphorylation | Shc-tyrosine phosphorylation | c-Src | |
| EGF | *** | ** | *** | *** | *** | *** |
| IGF-1 | - | *** | * | - | - | - |
| NGF | * | * | - | *** | ** | * |
FIGURE 1.
Maximal stimulation of receptor tyrosine kinase signaling systems. A-C depict log dose-response curves for the ligand-induced tyrosine phosphorylation (expressed as a percentage of Rmax) for the EGF (A), IGF-1 (B), or NGF (C) induction of auto-tyrosine phosphorylation of their cognate receptors. Each experimental point upon the dose-response curves is the mean ± S.E. of three independent experiments. Above the dose-response relationships there are representative Western blots of receptor tyrosine kinase immunoprecipitates probed with an anti-phosphotyrosine sera (PY20) or a specific anti-receptor protein sera (for loading control). The doses used for the Western blots were 10 ng/ml for EGF and NGF and 25 nm for IGF-1, which corresponded to the near maximal level of receptor activation. The effect of additional cell stimulation upon discrete ligand-receptor stimulation is depicted in D-F. The simultaneous activation of the two other receptor tyrosine kinase pathways failed to significantly alter the ability of the individual ligands to fully activate its cognate receptor. The bars in the histograms in D-F represent mean ± S.E. receptor tyrosine phosphorylation fold inductions from three independent experiments. IP, immunoprecipitation; IB, immunoblot.
Differential Signal Output and Activation of Signaling Molecules—If the various signal transduction cascades activated by plasma membrane receptors were independent of each other and possessed their own discrete complement of molecules required to activate their specific cellular processes, one would expect that activation of the pathways would be the same irrespective of additional factors. However, recent data suggest that transduction cascades are not composed of freely available molecules from an inexhaustible cytoplasmic pool, but instead they are pre-assembled often in close proximity to the receptors that activate them. Therefore, as there is probably a high degree of convergence of plasma membrane signals upon these signaling scaffolds, it is more likely that integration of different signals probably occurs at this point. With this concept we investigated how different, both qualitatively and quantitatively, the eventual signal output is with respect to the growth factors when they compete for the activation of the signal transduction scaffolds. Using a neutral hypothesis as a standard comparator (i.e. assuming complete independence of signal transduction pathways), we analyzed the actual resultant output induced by simultaneous ligand stimulation.
It is clear that there is heterogeneity in the ability of the three growth factors to stimulate the various signal transduction pathways. In Fig. 2, the ability of the individual growth factors and the dual combinations (both growth factors applied simultaneously) to activate the ERK1/2 pathway are depicted. Application of EGF by itself resulted in nearly a 3-fold elevation in active ERK1/2 at the 5-min time point, whereas IGF-1 alone resulted in only a minimal activation (1.21 ± 0.02-fold). The theoretical composite of these above basal would therefore be only ∼3-fold, whereas the actual response to the co-stimulation was nearly a 7-fold elevation in active ERK1/2 levels. A similar trend of this activity was also seen at the other experimental time points. The actual ligand combined ERK1/2 stimulation was significantly greater than the theoretical composite at each time point (5 min, p = 0.0026; 10 min; p = 0.00012; 30 min, p = 0.0112). The application of NGF alone also yielded only a modest activation of ERK1/2 (1.65 + 0.03-fold) when compared with EGF at the 5-min time point. In a similar manner, but not as statistically significant, the actual ERK1/2 activation induced by the actual EGF:NGF co-stimulation was greater than the theoretical composite at each of the time points. A similar pattern to this EGF:NGF co-stimulation paradigm was observed when the synergism between IGF and NGF was considered. The degree of ERK1/2 activation induced by the actual co-stimulations was consistently higher (although not statistically significant) than that predicted by the theoretical composites.
FIGURE 2.
Combined hormonal activation of ERK1/2 in PC12 cells. A, representative Western blots depicting ERK1/2 activation upon EGF, IGF, or NGF stimulation for 5, 10, and 30 min duration. Protein loading was controlled by assessing the presence of actin in each lane. The histograms in B depict means ± S.E. ERK activation data from at least three independent experiments per histogram as demonstrated in A. EGF (blue bars), IGF (red bars), and NGF (green bars) indicate responses induced by individual ligand stimulation. Shaded mixed color bars indicate theoretical summation of individual responses compared with the actual resultant response induced by legend co-stimulation (black bars). IB, immunoblot; w.c. lysate, whole-cell lysate; NS, no stimulation.
In Fig. 3, the ability of the individual and mixed growth factors to activate the neuroprotective kinase Akt-1 is shown. In contrast to ERK1/2 activation, IGF-1 in this case appears to be the most effective growth factor. Stimulation with EGF induces a modest (∼2-fold), time-dependent serine 473 phosphorylation of Akt-1. The IGF-1 activation, however, peaks near 3.5-fold at the 10-min time point. Interestingly, when the actual EGF:IGF co-stimulation results are compared with the composite, they are consistently lower than the neutral theoretical composites (5 min, p = 0.048; 10 min, p = 0.042; 30 min, p = 0.00024). Unlike both EGF and IGF-1, the application of NGF does not greatly induce an Akt-1 serine 473 phosphorylation. In addition to this, the co-stimulation of EGF:NGF yields Akt-1 activation results that appear largely similar to the theoretical composites, except at the 5-min time point where the actual co-stimulation results in a greater activation of Akt-1 (p = 0.039). With regard to the general trends of interaction and comparisons of actual co-stimulation results to the theoretical composites, the IGF:NGF interactions with respect to Akt-1 activation show similarity with the EGF:IGF combination. Hence, at each time point the actual IGF:NGF co-stimulation Akt-1 activations are lower than the values expected from the theoretical composites (5 min, p = 0.023; 10 min, p = 0.037; 30 min, p = 0.00065).
FIGURE 3.
Combined hormonal activation of Akt-1 in PC12 cells. A, representative Western blots depicting Akt-1 activation upon EGF, IGF, or NGF stimulation for 5, 10, and 30 min duration. Protein loading was controlled by assessing the presence of actin in each lane. The histograms in B depict means ± S.E. Akt-1 activation data from at least three independent experiments per histogram as demonstrated in A. EGF (blue bars), IGF (red bars), and NGF (green bars) indicate responses induced by individual ligand stimulation. Shaded mixed color bars indicate theoretical summation of individual responses compared with the actual resultant response induced by legend co-stimulation (black bars). IB, immunoblot; w.c. lysate, whole-cell lysate; NS, no stimulation.
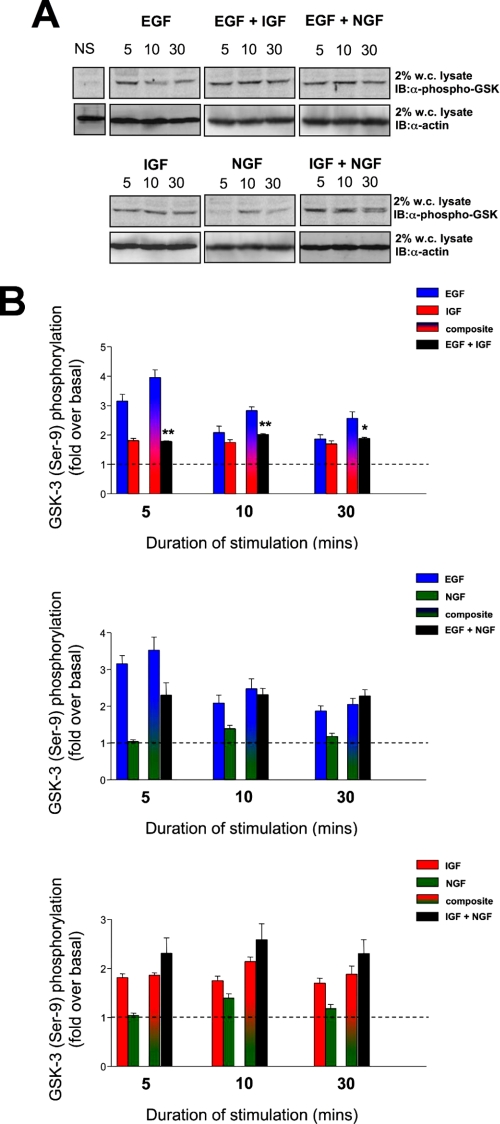
GSK-3 forms the center of many important signaling pathways required for cell survival and resistance to stressors. Interestingly, GSK-3 is typically constitutively catalytically active, and upon stimulation of cell surface receptors it can be phosphorylated by several factors, which results in its inactivation. The inactive form of GSK-3 is measured in Fig. 4. With respect to stimulation with EGF, there appeared to be a strong (3-fold), yet transient inactivation of GSK-3 as indicated by its increase of serine 9 phosphorylation. However, IGF-1 only minimally induced serine 9 GSK-3 phosphorylation. Therefore, the neutral theoretical composite of these responses is a strong induction of serine 9 GSK-3 phosphorylation; however, the actual co-stimulation results demonstrated a significantly smaller GSK-3 serine 9 phosphorylation response (Fig. 4, 5 min, p = 0.023; 10 min, p = 0.019; 30 min, p = 0.047). In a similar manner to the action of IGF-1 upon GSK-3 serine 9 phosphorylation, NGF only induced a minimal stimulation that peaked at the 10-min time point. The theoretical composite GSK-3 serine 9 phosphorylations between EGF:NGF therefore resembled those for EGF:IGF. The actual co-stimulation data did not significantly differ from the theoretical composites for the EGF: NGF combination with respect to GSK-3 serine 9 phosphorylation, yet the co-stimulation result for 5 min was lower than the theoretical composite but was not statistically significantly different. Interestingly, there were also no significant differences between the actual co-stimulation results and the theoretical composites for the IGF:NGF combinations at any of the experimental time points.
FIGURE 4.
Combined hormonal phosphorylation of GSK-3 in PC12 cells. A, representative Western blots depicting GSK-3 phosphorylation upon EGF, IGF, or NGF stimulation for 5, 10, and 30 min duration. Protein loading was controlled by assessing the presence of actin in each lane. The histograms in B depict means ± S.E. GSK-3 phosphorylation data from at least three independent experiments per histogram as demonstrated in A. EGF (blue bars), IGF (red bars), and NGF (green bars) indicate responses induced by individual ligand stimulation. Shaded mixed color bars indicate theoretical summation of individual responses compared with the actual resultant response induced by legend co-stimulation (black bars). IB, immunoblot; w.c. lysate, whole-cell lysate; NS, no stimulation.
A major factor that links the three receptor systems in this study is their ability to induce the activation of tyrosine kinases, both their own intrinsic kinase activity and also associated proteins such as the calcium-dependent tyrosine kinase Pyk2. Pyk2 can act as a molecular scaffold for the generation of large multiprotein signaling complexes through its ability to auto-tyrosine phosphorylate (26). The ability of the three growth factors to induce this activation of Pyk2 is demonstrated in Fig. 5. With respect to individual ligand stimulations, EGF generated the most profound degree of Pyk2 tyrosine phosphorylation (ranging from 2- to 3-fold at 20 min; Fig. 5). NGF, and to a lesser extent IGF-1, generated only minimal levels of Pyk2 tyrosine phosphorylation. The comparison between the theoretical composite Pyk2 tyrosine phosphorylations by EGF:IGF and the actual co-stimulation results demonstrated that only at the later time periods was there a negative interaction between EGF and IGF-1, i.e. the actual co-stimulated level of Pyk2 tyrosine phosphorylation was lower than the theoretical composite (p = 0.036). When the interactions between EGF and NGF were studied, however, there appeared to be a strong and significant negative interaction between EGF and NGF with respect to Pyk2 tyrosine phosphorylation. The actual co-stimulated levels of Pyk2 phosphotyrosine content was profoundly lower (significant at 5 min, p = 0.00039; and 30 min, p = 0.000026) at all the measured time points than the theoretical composite expectation. An opposite interaction type was in evidence when the combination of IGF with NGF was studied with respect to Pyk2 tyrosine phosphorylation. At each time point the actual co-stimulated Pyk2 phosphotyrosine levels were higher than the expected theoretical composites. These differences were statistically significant at the 5- (p = 0.038) and 10-min (p = 0.00021) time points.
FIGURE 5.
Combined hormonal-induced tyrosine phosphorylation of Pyk2 in PC12 cells. A, representative Western blots of anti-phosphotyrosine immunoprecipitates depicting Pyk2 tyrosine phosphorylation upon EGF, IGF, or NGF stimulation for 5, 10, and 30 min duration. Protein expression was controlled by assessing the presence of nonphosphorylated Pyk2 in an anti-Pyk2 immunoprecipitate from the same original cell lysate sample. The histograms in B depict means ± S.E. Pyk2 tyrosine phosphorylation data from at least three independent experiments per histogram as demonstrated in A. EGF (blue bars), IGF (red bars), and NGF (green bars) indicate responses induced by individual ligand stimulation. Shaded mixed color bars indicate theoretical summation of individual responses compared with the actual resultant response induced by legend co-stimulation (black bars). IP, immunoprecipitation; IB, immunoblot; NS, no stimulation.
Another member of the nonreceptor tyrosine kinase superfamily is the Src family kinases. These tyrosine kinases possess both SH2 and SH3 modular interaction domains (27). These kinases are typically held in an inactive state by their tyrosine phosphorylation of a conserved tyrosine residue in their carboxyl terminus by c-Src kinase (Csk) (28). This phosphorylation facilitates the interaction of this phosphotyrosine moiety with the SH2 domain of the protein, thus occluding its own kinase domain and activity. Upon activation (by dephosphorylation of the carboxyl-terminal tyrosine), the first target of the kinase is c-Src itself. Auto-tyrosine phosphorylation at tyrosine 418 by the kinase domain of c-Src potentiates its own catalytic activity and reduces the probability of inactivation by Csk. Therefore, we used the measurement of this initial auto-tyrosine phosphorylation as an indicator of c-Src activation by our three growth factors. Acute application of EGF resulted in a strong activation of c-Src (∼3-fold above basal). which persisted for up to 30 min; IGF-1 induced a smaller and slower responding activation of c-Src that peaked at nearly 2-fold at the 30-min time point. NGF failed to cause a profound activation of c-Src in our experimental paradigm. When we considered the differences between the theoretical composite results and the actual data gained from the co-stimulations, the combination of EGF and IGF produced an interesting biphasic outcome (Fig. 6). At the 5-min time point the actual co-stimulation response was greater (p = 0.029) than the theoretical composite, and at the 30-min time point it was significantly smaller (p = 0.013). The intermediate time point of 10 min represented this transition from potentiation of response to interference of the co-stimulation. With respect to the interactions between EGF and NGF, there was minimal difference between the theoretical composite responses and the actual co-stimulation responses with the exception of the 10-min time point in which the actual co-stimulation response was significantly greater (p = 0.029) than the theoretical composite. A more consistent difference between theoretical composite and actual co-stimulation results was seen with the IGF-1 and NGF interactions and c-Src activation. Hence, at the earlier time points the actual co-stimulated c-Src activation induced by IGF and NGF was significantly greater than the expected composite result (5 min, p = 0.036; 10 min, p = 0.041). At the 30-min time point, however, this positive interaction between these two growth factors was diminished and was no longer significant.
FIGURE 6.
Combined hormone-induced tyrosine phosphorylation of c-Src in PC12 cells. A, representative Western blots of anti-phosphotyrosine immunoprecipitates depicting c-Src Tyr-418 autophosphorylation upon EGF, IGF, or NGF stimulation for 5, 10, and 30 min duration. Protein expression was controlled by assessing the presence of nonphosphorylated c-Src in an anti-Src immunoprecipitate from the same original cell lysate sample. The histograms in B depict means ± S.E. Shc tyrosine phosphorylation data from at least three independent experiments per histogram as demonstrated in A. EGF (blue bars), IGF (red bars), and NGF (green bars) indicate responses induced by individual ligand stimulation. Shaded mixed color bars indicate theoretical summation of individual responses compared with the actual resultant response induced by legend co-stimulation (black bars). IP, immunoprecipitation; IB, immunoblot; NS, no stimulation.
The formation of multiprotein signaling cascades, especially those induced by growth factor ligands, often involve the SH2-containing adapter Shc. This protein contains two functional SH2 domains that allow it to bind with high affinity to proteins containing phosphotyrosine moieties (29). Shc itself is a substrate for tyrosine kinases, and thus we investigated the ability of the three growth factors to induce Shc tyrosine phosphorylation (Fig. 7). Interestingly, and similarly to Pyk2 activation, out of the three growth factors only EGF (2-3-fold above basal) and NGF (2-fold) caused a profound increase in the phosphotyrosine content of Shc (Fig. 7). IGF-1 stimulation resulted in only a minimal stimulation (1.2-fold over basal) at 30 min. Despite causing a minimal stimulation of Shc tyrosine phosphorylation when combined with EGF, the actual co-stimulated levels of Shc tyrosine phosphorylation were significantly lower than the expected theoretical composites at each time point (5 min, p = 0.00018; 10 min, p = 0.000018; 30 min, p = 0.000015). This profound and significant negative interaction was also observed to occur with the co-stimulations of EGF-NGF and IGF-NGF (Fig. 7). This negative interaction was the least profound, however, with the EGF-NGF co-stimulation, although this was statistically significantly different at the later time points (10 min, p = 0.036; 30 min, p = 0.029). The difference between IGF-NGF co-stimulated levels of Shc tyrosine phosphorylation was significantly lower than that expected by the theoretical composites at each time point (5 min, p = 0.037; 10 min, p = 0.00014; 30 min, p = 0.00012). Therefore, despite IGF only mediating a small increase in Shc tyrosine phosphorylation alone when in the presence of co-stimulating EGF or NGF, it greatly antagonized the increase of Shc phosphotyrosine content.
FIGURE 7.
Combined hormone-induced tyrosine phosphorylation of Shc in PC12 cells. A, representative Western blots of anti-phosphotyrosine immunoprecipitates depicting Shc tyrosine phosphorylation upon EGF, IGF, or NGF stimulation for 5, 10, and 30 min duration. Protein expression was controlled by assessing the presence of nonphosphorylated Shc in an anti-Shc immunoprecipitate from the same original cell lysate sample. The histograms in B depict means ± S.E. Shc tyrosine phosphorylation data from at least three independent experiments per histogram as demonstrated in A. EGF (blue bars), IGF (red bars), and NGF (green bars) indicate responses induced by individual ligand stimulation. Shaded mixed color bars indicate theoretical summation of individual responses compared with the actual resultant response induced by legend co-stimulation (black bars). IP, immunoprecipitation; IB, immunoblot.
In contrast to this diverse activity with respect to effects of simultaneous stimulation with tyrosine kinase activity, the actions of the growth factors on Shc tyrosine phosphorylation were surprisingly consistent. Hence all of the combinations of growth factors resulted in an antagonism of Shc tyrosine phosphorylation (Fig. 7). Perhaps the more ubiquitous role of this relatively passive scaffolding protein underlies an important nexus role because a commonality of signaling effects was seen with respect to Shc, making it unlike all the other signaling proteins investigated.
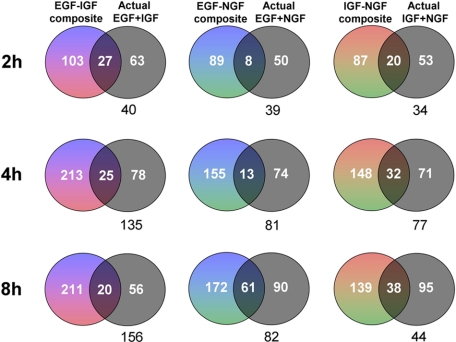
Interaction of Growth Factor Signaling at the Transcriptional Level—Because of the interactive effects of different growth factors at the level of early responsive signaling factors (5-30 min), we determined whether distinctions between individual versus simultaneous stimulations propagated into alterations in gene expression. The activation of genes in the PC12 cells with 2, 4, or 8 h of stimulation with the growth factors individually or in combination was assessed using a mouse 17K gene array. Each experimental growth factor treatment was performed in triplicate with a single array run for each replicate. Significantly regulated (increased or decreased) genes (p < 0.05) from mouse 17K gene arrays are displayed in the form of Venn diagrams (full gene lists are displayed in supplemental Table 1). Individually regulated gene lists (up and down-regulated) for the single ligand-stimulated paradigms were added to each other to create the composite (multicolor) groups whereas the significantly regulated gene lists are denoted in black (Fig. 8). The regulated genes were assessed from predicted composite and actual mixed ligand experiments. Any intersection of regulated genes (irrespective of regulation direction) is denoted by the overlap of the regulated groups. Below each of the Venn diagrams is the numerical gene difference of the actual dual-ligand stimulated paradigm compared with the theoretical composite output. It is clear from Fig. 8 that at each time period of growth factor treatment there is the greatest divergence in genes regulated by the ligands with the actual EGF-IGF co-stimulation compared with the theoretical summation of the two individual gene lists. This therefore suggests that this ligand combination induces the greatest degree of interaction/antagonism compared with the other ligand combinations. The largest divergences occurred between the 4- and 8-h time points.
FIGURE 8.
Regulation of PC12 gene transcription by individual and combined stimulations. Venn diagrams demonstrate the numbers of significantly up- or down-regulated genes (p < 0.05) by composites of individual ligand stimulation (mixed colors, EGF = blue; IGF = red; NGF = green) or the actual results of combined ligand mixing (black) at three time points (2, 4, and 8 h) compared with the control gene levels. Commonalities between regulated genes are shown by the inclusion in the Venn diagram intersections. The difference of the gene numbers between composite and actual is shown in black numerals below each Venn diagram.
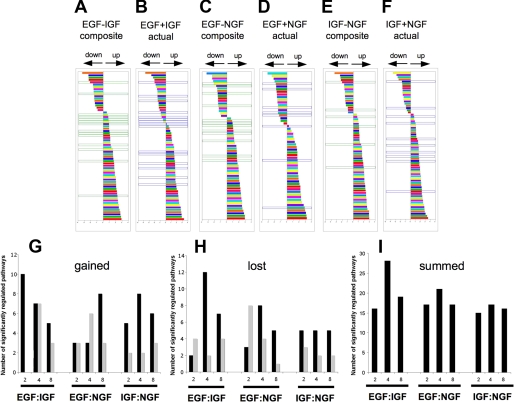
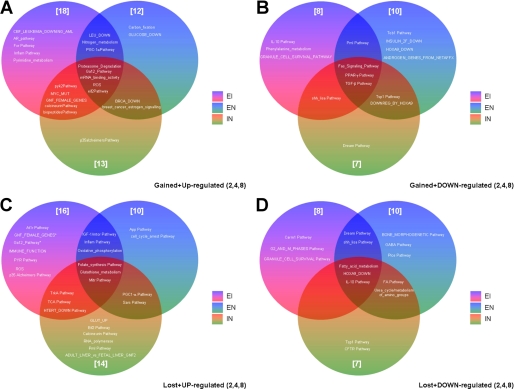
Coordinated Physiological Nature of Pathways Regulated by Growth Factor Interactions—Our data suggest that at the level of individual gene regulation, the pattern of signaling interaction seen at the level of early cytoplasmic protein signal transduction cascades is retained. We next evaluated the divergence of the potential physiological pathways stimulated by the growth factors between the theoretical composites and the actual results generated through dual ligand stimulation. Composite modulated gene lists were created by simple addition and then compared with the actual co-stimulated gene lists. These two lists were then both subjected to phenotypic pathway PAGE analysis (23). The results of the signaling pathway significance analysis (PAGE) are displayed in supplemental Table 2. These results revealed that the EGF:IGF combinations demonstrated the greatest degree of signaling interaction/antagonism between the theoretical composite lists and the actual co-stimulation lists (Fig. 9). The PAGE pathways at the 4-h time point are shown for the EGF:IGF combination (Fig. 9, A, theoretical composite, and B, actual co-stimulation), the EGF:NGF combination (Fig. 9, C, composite, and D, actual), and the IGF:NGF combination (Fig. 9, E, composite, and F, actual). The pathways outlined in green in Fig. 9, E and F, were present only in the theoretical composites, and those in blue were only present in the actual co-stimulation paradigms. The greatest number of gained/lost significantly regulated pathways between the theoretical and actual co-stimulations at this time point was seen with the EGF:IGF paradigm.
FIGURE 9.
PAGE pathway analysis for theoretical ligand combinations versus actual ligand combinations. PAGE pathway trees for the 4-h ligand stimulation time point are shown in A-F. Each colored bar represents a specific PAGE signaling pathway: A (output from composite gene list) and B (output from actual co-stimulation gene list) demonstrate PAGE signaling pathway data from the EGF:IGF data sets. Multiple pathways are shown to be both up- or down-regulated in the two gene sets. Analogous data for EGF:NGF and IGF:NGF paradigms are depicted in C and D, and E and F, respectively. PAGE pathways only generated by theoretical gene list output are bordered in green, and those only created using actual co-stimulated gene list output are bordered in blue. G, number of PAGE pathways present only in the output from actual co-stimulation gene lists (gained: blue bordered, A-F) are depicted for each time point and each ligand co-stimulation paradigm. These “gained” pathways that were up-regulated are denoted by a solid black vertical bar, and gained pathways that show a down-regulation are denoted by a gray vertical bar. The numbers of pathways present in theoretical gene list PAGE outputs only (bordered in green, A-F) that were lost from the PAGE output of actual co-stimulation gene lists are depicted for each time point and ligand combination in H. Up-regulated “lost” pathways are depicted in black, and down-regulated lost pathways are depicted in gray. I, demonstrates the sums of both lost and gained pathways between PAGE outputs from theoretical versus actual co-stimulation gene lists.
Over the three time periods, with respect to the total number of significant distinct signaling pathways “gained,” i.e. not present in the composite list but present in the actual co-stimulation list (Fig. 9G), the EGF:IGF combination showed the greatest magnitude, 32, compared with the EGF:NGF (26) or IGF: NGF combinations (26). Taking into account the converse, i.e. loss of significantly regulated phenotypic pathways (Fig. 9H), the EGF:IGF combination again showed the greatest degree of divergence with 31 pathways lost between the theoretical composite list and the actual co-stimulated pathway list compared with the EGF:NGF (29) and IGF:NGF (22) paradigms. In Fig. 9I the total gained and lost pathways between theoretical composite lists and the actual co-stimulation lists are shown (up and down-regulated), demonstrating that at the 4-h time point the greatest distinctions between theoretical and actual results were typically seen, especially for the EGF:IGF combination. It is interesting to note that not only were the greatest theoretical versus actual divergences greatest for individual gene regulations between the EGF:IGF paradigms (Fig. 8), but this also flows into the potential physiological pathways regulated by these genes.
With respect to the predicted phenotypic nature of the output genes activated by the actual ligand co-stimulation only, there are several interesting examples to note (Fig. 10). Cumulated over the three experimental time points, the EGF:IGF co-stimulation paradigm possessed the most gained significantly regulated (up- or down-regulated) pathways compared with the theoretical results. The co-stimulated EGF:IGF paradigm enhanced the inflammatory regulatory pathway and at the same time down-regulated the specific interleukin-10 pathway. The actual EGF:IGF co-stimulation also engendered a specific increase of potential pyrimidine metabolism while decreasing phenylalanine metabolism. The EGF-containing co-stimulation paradigms (EGF:IGF and EGF:NGF) also seemed to foster the creation of new up-regulated pathways related to transcriptional energy-related responses, e.g. peroxisome proliferator-activated receptor-γ regulation through PGC1-α (Fig. 10). The EGF:NGF/IGF:NGF co-stimulation paradigms created fewer novel, uniquely up-regulated pathways (carbon fixation, glucose_down for EGF:NGF and p35-Alzheimer pathway for IGF: NGF) than the EGF:IGF paradigm. The NGF-containing co-stimulation paradigms (EGF:NGF and IGF:NGF) created novel up-regulated pathways tightly involved in breast cancer signaling, i.e. BRCA_down and breast_cancer_signaling. The IGF-containing co-stimulation paradigms (EGF:IGF and IGF:NGF) up-regulated calcium-dependent tyrosine kinase and dephosphorylation processes (Pyk2 pathway and calcineurin).
FIGURE 10.
Cumulated pathway regulation intersections between gained and lost pathways between theoretical and actual co-stimulations. A and B represent Venn diagrams for PAGE pathways gained selectively in the actual co-stimulation paradigms compared with the theoretical composites cumulated over the three experimental time points (2, 4, and 8 h). A depicts pathways gained and up-regulated, and B depicts pathways gained and down-regulated. C and D represent Venn diagrams for PAGE pathways lost selectively from the theoretical composite results to the actual co-stimulations cumulated over the three experimental time points (2, 4, and 8 h). C depicts pathways lost and up-regulated, and D depicts pathways lost and down-regulated.
It is interesting to note that compared with their respective theoretical composite lists, all the actual co-stimulation events appeared to result in up-regulation of pathways that may control protein/mRNA turnover (proteasome degradation, mRNA binding, and eIF2 pathway) and stress responses (reactive oxygen species pathways). The converse of this specific subset, i.e. gained with all of the co-stimulations, is the subset of commonly down-regulated pathways contains pathways controlling cell fate/death and energy metabolism (Fas, peroxisome proliferator-activated receptor-γ, and transforming growth factor-β). Perhaps, therefore, combinations of ligands, which is typically the normal case for healthy cells, interact functionally to maintain cell survival routes. The converse of this postulate, i.e. commonly down-regulated pathways, seems to involve the coherent down-regulation of pathways linked to cell fate/death. Therefore, these results might suggest an encoded mechanism by which multiple impinging signals do not disrupt crucial cell signaling events and “accidentally” block cell maintenance mechanisms.
We have shown that with respect to the divergent pathways between actual co-stimulations and theoretical composites that the EGF:IGF paradigm is the most distinct from what one would expect from mere summation of the individual pathways induced by single ligand stimulation. Despite this result, however, there were many pathway similarities between the theoretical and co-stimulation PAGE data. It is in fact reassuring that this is largely the case for the ligand mixing paradigms. It would be virtually impossible to coherently control cell metabolism if the simultaneous activation of distinct receptors resulted in a dramatic upheaval of output functionality. As there are many common signaling PAGE pathways derived from both the theoretical composite and actual co-stimulation lists for each of the ligand combinations, we decided to see if there were numerical differences between the magnitudes of these common regulated pathways between different ligand combinations. Therefore, we calculated the numerical difference between the PAGE pathway score (sum of gene Z-scores clustered into that signaling pathway) derived from the theoretical composite gene list and the actual co-stimulation gene lists. To allow comparison of all the common pathways, we represented these differences as a modulus score (square of the difference in pathway Z-scores between the theoretical composite subtracted from the actual co-stimulation pathway Z-value). In Fig. 11, this modulus score for the pathway numerical differences (for each pathway found in the theoretical and composite gene list PAGE output) is represented by a single vertical bar in each (A-I). The mean of all the moduli in a single panel is shown above each panel in Fig. 11. In Fig. 11, A-C represent data from the 2-h time point, D-F from the 4-h time point, and G-I from the 8-h time point. As demonstrated in Fig. 11, A, D, and G, the means of the moduli (representing the magnitude of pathway score differences for theoretical lists compared with actual gene lists) for the EGF:IGF scenarios are the greatest at each time point. At the 4-h time point (Fig. 11, D, E, and F), the mean difference modulus of the EGF:IGF paradigm is significantly greater than for the EGF:NGF (p = 0.0492) or IGF:NGF scenarios (p = 0.0002). Hence even when comparing the same pathways created from theoretical compared with actual gene lists from ligand co-stimulations, the EGF:IGF paradigm shows the greatest quantitative differences in gene regulation.
FIGURE 11.
PAGE pathway difference modulus between common pathways derived from both theoretical ligand and actual ligand co-stimulation gene lists. All histogram vertical bars on each panel represent a modulus of the difference between pathway Z-scores comparing the common PAGE pathway scores from outputs of theoretical composite gene lists and gene lists from actual co-stimulation experiments. A-C, D-F, and G-I represent data from the 2-, 4-, and 8-h time points, respectively. A, D, and G display Z-score moduli for the differences between pathways common between theoretical and actual co-stimulation EGF:IGF paradigms. B, E, and H, and C, F, and I display analogous data for the EGF:NGF and IGF:NGF paradigms, respectively. The mean pathway difference modulus for each paradigm and time point is displayed above each panel.
These analyses (Figs. 9 and 11) demonstrate that not only is the strongest qualitative divergence seen with the EGF-IGF paradigm when comparing theoretical versus actual results in gained or lost pathways but also in the degree of modulation of the conserved pathways between theoretical and actual co-stimulations. This suggests that when the receptors were activated clear and discrete signaling microcomplexes (which we shall describe as “encryptons”) may be formed from signal-transducing proteins that are then relatively stable and are able to transmit complex information from the cell surface to the nucleus to regulate transcription. The specific complexes generated by the EGF-IGF interaction are able to transfer their specific identity throughout the various time points identified, i.e. 5 min to 8 h. Whether the initial microcomplexes formed at 5 min are stably retained during this process or whether they spawn further stable microcomplexes is an interesting question for further research to elucidate.
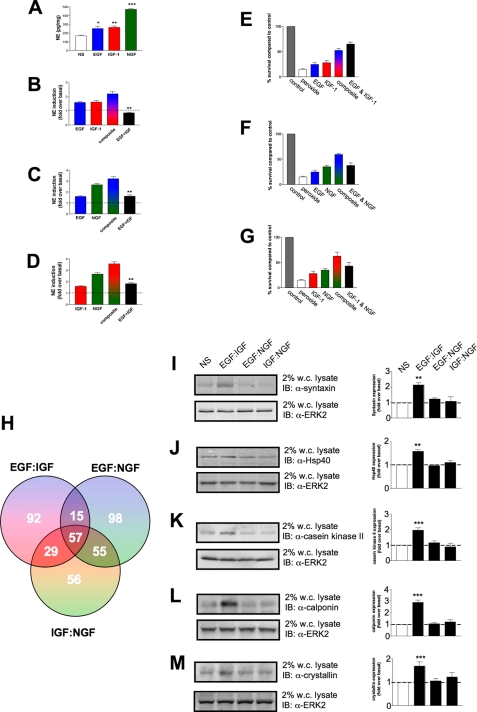
Receptor-mediated Signal Generation Coherently Translates to Discrete Cellular Phenotypes in a Predictable Manner—The results to this point have indicated that the signal transduction machinery of neural cells is configured to respond and encrypt complex plasma membrane messages from multiple ligands to regulate commonly used soluble kinases and the phosphorylation status of protein-protein adapters as well as eventual gene transcription. We next investigated whether these unique enzymatic and genetic changes induced by the ligand stimulations were able to manifest themselves at the level of classically phenotypic neuronal responses. PC12 cells are known to respond to neurotrophic growth factor stimulation by increasing their production of the neurotransmitter NE. To enhance the ability of PC12 cells to generate NE, the cells were maintained for up to 7 days in a low fetal calf serum (0.1%) media supplemented with various growth factors. We investigated whether similar divergences in theoretical to actual ligand co-stimulations occurred when the ability to generate norepinephrine was measured. With 7 days of continuous ligand stimulation, the normally rapidly dividing PC12 cells arrested their cell growth and began to extend neurites within 3-4 days. After 7 days the cells were harvested, and NE levels were quantified by HPLC (Fig. 12). In Fig. 12A, it is clear that all three growth factors increased the generation of NE in the PC12 cells, with NGF being the most potent. Surprisingly, when the results of actual ligand combination upon NE generation are compared with theoretical composites of the individual ligand stimulation data, the EGF-IGF combination reduced the production of NE below the base-line levels observed in cells incubated in low serum media alone. The other two actual ligand combinations also demonstrated a reduced capacity to induce NE generation compared with the theoretical composites, but were still able to elevate the NE levels above base line (Fig. 12). Thus, as with the previous data, it seems that the specificity of EGF:IGF interaction is again maintained in this complex phenotypic output, demonstrating that molecular signaling distinctions created within minutes do propagate and are crucial to determining the eventual nature of the signaling activity of the cells.
FIGURE 12.
Neurotrophin-induced norepinephrine generation and protective actions in PC12 cells. Continuous incubation of PC12 cells with either vehicle control (NS, no stimulation) or the standard doses of EGF, IGF-1, or NGF for 7 days resulted in an elevation in the NE (pg/mg) content of the cells (A). Co-incubation with EGF+IGF (B) resulted in an inhibition of NE-induced cell phenotype, whereas co-incubations between EGF+NGF (C) or IGF-1+NGF (D) resulted in a diminution of the extent of NE production in the cells. In all three cases (B-D) the NE expression in the presence of the combined neurotrophic factors was significantly less than the theoretical composite of the additive effects of each neurotrophin. Bars in each of the histograms represents mean ± S.E. values from three separate experiments. In E-G PC12 cells were treated for 7 days with individual or combinations of the three growth factors, EGF, IGF, or NGF. The viability of the PC12 cells 48 h after an acute (0.2 mm) hydrogen peroxide insult (peroxide) compared with cells not peroxide-insulted (control) was measured by identification of live and dead cells in culture using trypan blue staining. E demonstrates the survival of cells treated with peroxide after EGF, IGF, or combined EGF and IGF incubation. F demonstrates the survival of cells treated with peroxide after EGF, NGF, or combined EGF and NGF incubation. G demonstrates the survival of cells treated with peroxide after IGF, NGF, or combined IGF and NGF incubation. Each histogram in the respective panels demonstrates means ± S.E. survival data from at least three separate experiments. H depicts a Venn diagrammatic representation of the subsets of significantly regulated genes (2-8 h) induced by the various co-stimulating ligand combinations. Protein assessment of five factors related to neuronal transmitter expression and survival were chosen from the EGF:IGF-only regulated group. Their relative expression patterns across the various stimulation patterns is shown in I (syntaxin); J (hsp40); K (casein kinase II); L (calponin), and M (crystallin). Each representative Western blot of lysates from cells treated for 7 days is shown next to quantifications (means ± S.E.) of relative expression levels from three independent stimulation experiments. The cellular levels of ERK2 were employed as an internal protein loading control. IB, immunoblot; w.c. lysate, whole-cell lysate.
In addition to the ability to generate NE, we also assessed whether the ability of the cells to survive external stressors was modified by growth factors in a manner predictable from signaling interaction patterns created at the levels of signal transduction and gene expression established above. After the 7 days of treatment with growth factors alone or in combination, the cells were subjected to an acute hydrogen peroxide stress (0.2 mm), and the number of healthy/live cells present 48 h after this insult was assessed using viable cell stain exclusion (trypan blue). Treatments of the PC12 cells with all of the three growth factors protected the cells to some degree from the hydrogen peroxide insult (Fig. 12). As with the NE production, NGF was the most effective growth factor in promoting cell survival in the face of the oxidative insult. When the theoretical ligand composite effects are compared with the actual co-stimulatory ligand data, the EGF:IGF combination again demonstrates a singular response compared with the other ligand pairs. Hence, when EGF and IGF both co-stimulate the cells, they are more neuroprotective than one would imagine even from the theoretical composite prediction. The other ligand pairs are less neuroprotective than one would predict from the theoretical composite result. To investigate the biological phenotype of these long term effects of ligand incubation, we attempted to isolate the specific nature of the differences between the EGF:IGF combination compared with the other two ligand combinations (EGF:NGF and IGF: NGF). When all significantly regulated genes (2-8 h) were grouped from the three actual ligand co-stimulation combinations using a Venn diagram mechanism of separation, a smaller subset, 92 genes, of EGF:IGF uniquely regulated genes was identified (Fig. 12H; supplemental Table 1). None of the other ligand combinations generated such expression alterations and therefore may, in part, account for the phenotypic alterations noted in Fig. 12 (A-D and E-G). Therefore, the unique signaling quality, observable as early as 5 min after receptor activation, is projected out to 7 days post-stimulation in a manner that appears to control the intrinsic nature of the cells themselves.
DISCUSSION
Signaling pathways once thought to be linear are now known to be highly branched, and transduction modules formerly thought to operate independently participate in a substantial degree of pre-organized signaling routes (41, 42). Our present findings suggest that the complexity and pre-organization of signal transduction scaffolds may be structured into coherent modules that allow the directed flux from multiple growth factor receptors in a discrete and selective manner that specifically allow the temporal and spatial encryption of the original signal input.
We have shown that with full activation of cell surface receptors, there are additional complexities of cell signaling that are only revealed in the presence of other stimulating factors. Extracellular signals, such as hormones, neurotransmitters, and growth factors, regulate a wide variety of cellular activities, including neuronal excitation, cell survival, growth, and differentiation. Intracellular transduction systems receive these signals via receptors and transmit them to the ligand-prescribed cell compartment quickly and precisely, resulting in the amplification of specific biological responses (43, 44). However, cells are often exposed to several stimulating ligands and maintaining the fidelity of the signaling networks is crucial in eliciting the appropriate physiological response. Organizing this requires the accurate selection of effector molecules for regulated activation and deactivation, often by phosphorylation and dephosphorylation events. A principal strategy in achieving this selection specificity is compartmentalization of signaling enzymes (43). Another signaling mechanism that we have recently described (45, 46) is the creation of discrete signaling microcomplexes linked to transmembrane receptors, such as G protein-coupled receptors, that are able to then translocate to different cell compartments after separating from the cell surface receptor and then effectively transmit a unique signal depending upon the nature of the initial receptor stimulation. One can think of these microcomplexes as encryptons as through their physical constitution and protein stoichiometry these signaling microcomplexes of common proteins assemble in a manner that allows functional encryption of the exact nature of the initial input ligands (45). Many cell surface receptors utilize the same types of signaling proteins, so therefore the intrinsic nature of those downstream molecules cannot be specific enough to encrypt the “information” of the stimulating ligand. However, a simple and unique stoichiometric combination of these common proteins would allow the generation of a virtually limitless number of encryptons all able to convey information from a wide range of inputs that impinge upon the cell in a plethora of combinations, temporal sequence, and concentration. We have previously demonstrated that the differential formation of signaling encryptons around the human androgen receptor induced by activation of the gonadotropin-releasing hormone receptor was able to facilitate the generation of nuclear encryptons that possessed opposite and inhibitory effects upon androgen-sensitive gene regulation with subsequent androgen stimulation (45). An encryption capacity is even more important when one considers that in most physiological settings it is likely that any given cell needs to respond coherently to multiple impinging ligands in various sequential combinations. In this case, the ability of cells to form encryptons themselves may differ from times of single compared with times of multiple stimulations, as the signaling complexes from which the encryptons may be created potentially show a high degree of pre-assembly. Therefore, ligand-induced encrypton generation may not only be a function of the ligand, the receptor, and the cell signal transduction machinery but also of the prevailing level of additional cell stimulators.
Besides the formation of the hypothetical encrypton structures, another potential mechanism that could contribute to the distinct signaling and phenotypic response patterns for the different growth factors could be the formation of discrete and specific “signaling endosomes.” The signaling endosome hypothesis envisions that NGF-TrkA complexes are internalized at the axon terminal and retrogradely transported to the cell body. It has been shown following NGF treatment that clathrin-coated vesicles contained NGF bound to TrkA, together with activated signaling proteins for the Ras-MAPK pathway (47). Evidence that these vesicles could actively signal was demonstrated by their ability in vitro to activate Elk, a downstream target of ERK1/2 (47). It has also been shown recently that Trk can generate long lived signaling endosomes (48). This Trk endocytosis can be distinguished from the classical clathrin-based endocytosis of the EGFR. Although Trk and EGFR each stimulate membrane ruffling, only Trk undergoes both selective and specific macroendocytosis at ruffles, which uniquely require the Rho-GTPase, Rac, and the trafficking protein Pincher. This process leads to Trk-signaling endosomes, which are immature multivesicular bodies that retain Rab5. In contrast, EGFR endosomes rapidly exchange Rab5 for Rab7, thereby transiting into late endosomes/lysosomes for degradation. Sustained endosomal signaling between Trk and EGFR has been shown not to reflect intrinsic differences between Trk and EGFR because each elicits long term ERK kinase activation from the cell surface. Thus, a population of stable Trk endosomes, formed by specialized macroendocytosis in neuronal cells, provides a privileged endosome-based system for propagation of signals to the nucleus (48). This endosomal signaling mechanism could also account for some of the distinct signaling and phenotypic response patterns that were obtained for the different growth factors. As with most signaling paradigms, it is likely that multiple and parallel activities may occur to create the full gamut of receptor-mediated responses.
The scenarios represented in this study attempt to approach the complexity of the eventual signaling milieu present in vivo. It is clear from the investigation of the acute activation of soluble kinases that there are discrete and potent effects of multiple ligand stimulation. For example, with respect to the activation of ERK1/2, EGF alone generated a potent activation, whereas IGF-1 failed to do so. Despite this poor activity of IGF-1 alone, when applied simultaneously with EGF, there was a profound and highly significant potentiation of the resultant ERK1/2 activation. This potentiation was far in excess of that which may be expected by mere addition of the magnitudes of the two individual signals. Hence the act of stimulating the IGF-1 receptor, although not inducing a strong ERK1/2 activation alone, appears to significantly potentiate the actions of EGF. Perhaps the stimulation of the IGF-1 receptor relieves some form of repressive activity that the inactive receptor, connected loosely to its signal transduction machinery, exerts upon the ability of EGF to activate ERK1/2. It has been shown previously that ERK1/2 activation is correlated with the cellular growth state of PC12 cells (49) and is responsible for their proliferation and also differentiation (50). Interestingly, these strongly synergistic effects of EGF and IGF-1 show parallels with other cell systems that are responsive to both of these ligands. Hence, Qureshi et al. (51) have also shown that EGF and IGF-1 act synergistically to regulate the growth of human esophageal epithelial cells. Our scenario also shows a significant interaction between these ligands as the EGF:IGF combination also strongly affected cell development as evidenced by its action upon NE production in the PC12 cells. To a much lesser extent this action of IGF-1 also occurs with the NGF-mediated activation of ERK1/2 suggesting perhaps that the inactive IGF-1 can exert a general repressive effect upon agents that compete with it to stimulate ERK1/2. This form of “passive” activity may be indicative of an intrinsic cellular hierarchy of cell signaling that favors certain ligands to control specific downstream signaling factors.
Interestingly, the ligand occupation and activation of the EGFR and NGF TrkA receptors appeared to mediate an analogous and common action against IGF-1R activity with respect to the stimulation of the neuroprotective kinase Akt-1. Thus, activation of EGF or NGF receptors although not strongly activating Akt-1 by themselves significantly attenuated the ability of IGF-1 to stimulate Akt-1 phosphorylation and activation. Activation of additional receptors therefore may disrupt the stoichiometry of the multiprotein complexes required to stimulate one pathway in the cell, demonstrating that there is not a free inexhaustible pool of intracellular signaling molecules available. Additional findings suggest that Shc tyrosine phosphorylation is a significant point of signal convergence for different growth factors in neural cells. In many mammalian tissues, Shc is expressed as two isoforms of 46 and 52 kDa. Shc has no intrinsic kinase domain and transduces signals dependent on the formation and stabilization of protein-protein interactions. Shc physically associates with plasma membrane receptors (such as the growth factor receptors) to engage the receptor-initiated signaling pathways that typically involve the monomeric G protein Ras (52). There are three classical protein-protein interaction domains in Shc; two of these are the phosphotyrosine-binding domain in the amino-terminal region and the SH2 domain in the carboxyl-terminal region. These two domain regions are separated by a proline/glycinerich region called the collagen homology domain (53). Upon receptor tyrosine kinase activation, e.g. EGFR, TrkA, or IGF-1R, Shc binds rapidly to the phosphotyrosine residues on the receptor, generated by ligand-induced tyrosine autophosphorylation, via its phosphotyrosine-binding domains, leading to Shc itself eventually being phosphorylated by the receptor tyrosine kinase (52). The phosphorylated tyrosine residues on the collagen homology domain of Shc provide the docking sites for binding the SH2 domain of Grb2 (growth factor receptor binding 2) and thus recruit Sos (Son of Sevenless), a guanine nucleotide exchange protein (53), leading to the activation of the Ras/Raf/MAPK pathway (54, 55). Consistent with the importance of Shc in mediating the functional integration of multiple ligand stimuli, we found that, compared with individual ligand stimulation, the co-stimulation paradigms all demonstrated an antagonistic behavior of the different growth factors (Fig. 7). Thus, there appears to be a premium on the availability and use of passive adapter molecules that scaffold further signaling factors allowing the activation of downstream pathways. There may therefore be a large number of kinase or phosphatase subtypes in the cell, but a relatively smaller number of molecular scaffolds to which they are attached and thus compartmentalized. Therefore, rather than the kinases/phosphatases being crucial for cell signaling, it may be the passive adapters that coordinate the assembly of these kinases that are the primary molecular regulators of signaling.
From our analysis of the gene expression alterations at various time points after the initial ligand stimulations, it was clear that the output mediated by the simple addition of two ligand signals is distinct from the actual result of simultaneous stimulation (Fig. 8). As with the case at the level of signal transduction kinases and associated proteins, the EGF:IGF-1 co-stimulation showed the greatest distinctions in the genes significantly elevated or diminished compared with unstimulated cells. Thus it appears that the transfer of information from the two different growth factors, initiated within minutes from the receptor stimulation, is accurately conveyed to the complex distal events involved in gene expression. Another interesting finding is that there is a relatively small number of genes regulated by theoretical ligand composite paradigms compared with the actual co-stimulation paradigm (Fig. 8). However, despite the small number of common genes with the more functionally relevant PAGE analysis, a relatively high conservation of functional physiological output was retained.
In addition to considering the individual gene identities, we also applied a functional pathway annotation tool (PAGE analysis) to render a prediction of the integrated outcome of the significantly regulated gene sets. Not only were the co-stimulation gene sets used but PAGE analysis was applied to theoretical additive composite sets. The PAGE analysis clusters the genes into functional signaling pathways and, depending on the number of genes (and regulation direction) filling each cluster, a magnitude value of pathway regulation is assigned. When analysis was performed at each time point of stimulation, there was the greatest functional group diversity between the EGF:IGF-1 co-stimulation and theoretical composite paradigms compared with the other experimental groups (Fig. 9). In essence, the genetic phenotypic output disruption was greatest for the EGF: IGF-1 paradigm, again correlating to the upstream signaling cascades. More physiological pathways were either gained or lost when the actual co-stimulation results were compared with the theoretical composite results for the EGF:IGF-1 paradigms compared with the two other experimental modes. The numbers of significantly regulated functional pathways was most strongly affected at the 2- and 4-h time points. Not only were we interested in the phenotypic output diversity induced by the differences in actual co-stimulation versus the theoretical composites, but we also studied the magnitudes of the changes in pathways between the ligand stimulation paradigms. The results revealed considerable disparity between actual co-stimulation and theoretical composites, but with evidence of retention of the interactive pathway patterns shown at the level of upstream signal transduction. Therefore, with respect to both the predicted physiological nature of the genetic outcome and the relative magnitude of these outcomes, it is clear that the early receptor-transduction pathway interactions are faithfully translated to even complicated genetic response outcomes. Collectively, our findings demonstrate that, despite being mediated by common kinases and adapter proteins, the signal transduction systems are highly constrained, possibly in our hypothetical encrypton structures. It is also likely that these specific signaling structures could also be compartmentalized in specific endosomic populations to enhance their selectivity of action.
As an additional indication of the ability to channel and maintain signaling fidelity, we used the co-stimulation paradigms (always in comparison to the theoretical composites) at even further removed time points. In Fig. 12 the presence of the ligand combinations for 7 days generated a ligand-interactive pattern analogous to that seen at 5 min and the rapid modulation of cytoplasmic signaling molecules. With respect to the ability of the noradrenergic PC12 cells to produce NE, all three ligands alone increased NE production. However, the EGF: IGF-1 combination resulted in a level of NE production below the basal level. On the other hand, when resistance to toxic peroxide stress was evaluated, stimulation with the EGF:IGF-1 combination resulted in the greatest protective effect compared with the two other growth factor combinations (Fig. 12). These findings suggest that co-treatment with EGF and IGF-1 suppressed neuronal differentiation resulting in both decreased production of NE and increased resistance of the cells to oxidative stress. Upon analysis of the factors specifically regulated only by this EGF:IGF paradigm, we demonstrated that in correlation to the ligand co-stimulation gene array results we noted significant up-regulation of proteins linked to support neuroprotective mechanisms and control neurotransmitter levels. In Fig. 12 we noted the significant and EGF:IGF-specific elevation of the cellular levels of the synaptic vesicle protein syntaxin 1A and the molecular chaperone crystallin (Fig. 12), both which have been demonstrated to potently control noradrenergic neurotransmitter expression (30, 37). In addition we noted increases in the levels of actin-binding protein, calponin, the chaperone protein Hsp40, and casein kinase II, all of which have been closely linked to the generation of neuroprotective mechanisms in in vitro and in vivo systems (31-36). In addition to its effects on neurotransmitter levels, the multifunctional chaperone protein crystallin also is linked to neuroprotective cellular activity (38-40). None of these genes or proteins were up-regulated after treatment with the EGF:NGF or IGF:NGF stimulation paradigms demonstrating that distinct ligand combinations can yield very specific phenotypic outcomes.
Compared with the neutral theoretical composites, the EGF: IGF-1 paradigm resulted in the most significant synergism of ERK1/2 activation (Fig. 2), the most significant antagonistic attenuation of Akt-1 activity (Fig. 3), the most significant synergistic activation of GSK-3 (Fig. 4), the most significant antagonistic attenuation of Shc tyrosine phosphorylation (Fig. 7), the greatest disruptions (magnitude and phenotypic type) of genetic output (Figs. 9 and 10), and the most profound eventual phenotype differences (Fig. 12) compared with the EGF:NGF or IGF:NGF paradigms. Thus it seems that there may be preferred or opposed hierarchical interactions between receptor systems that are mediated by the complex signaling network created by the general signaling architecture of the cell. With an increase in the complexity of experimentation, i.e. changes in ligand dose, application order, and changes in cell type (for example, primary neurons compared with glial cells or tumor cells), it may be possible to delineate the actual structure of the matrix that allows the integration and processing of multiple cellular inputs in relation to the eventual effect upon cell biology.
Our current findings illuminate the potential complexities in signaling interactions of growth factors and how they may be configured to allow discrete signal propagation through a hypothetical mechanism (see Fig. 13). Through the creation of signaling entities that link the receptors to the eventual cell effectors, the cell manages and integrates complex multifactorial inputs. As neural cells normally encounter multiple growth factors in vivo, an understanding of how the cell signaling architecture is created and used will be invaluable in assessing how the normal functioning of the nervous system is adversely affected by injury or disease. Previous studies have shown that EGF (56), IGF-1 (57), and NGF (58) can each protect neurons against death induced by oxidative stress. We found that EGF potentiated the cytoprotective effect of IGF-1 against oxidative stress-induced death but did not potentiate the cytoprotective effect of NGF. Our findings therefore suggest the importance of understanding the outcome of growth factor interactions for developing growth factor-based therapeutic interventions.
FIGURE 13.
Signal integration and multiple protein-protein interactions. The ability of single ligand (α) stimulation (left panel) to exert its full effects on cellular physiology is dependent on the ability of its signal proteins to interact with the receptor in a concerted and controlled manner. Signal transduction accessory proteins are initially created in the endoplasmic reticulum (A), before export and loose assembly in Golgi/vesicles (B), and before eventual stable interaction with a cell surface receptor (C). Single ligand stimulation (α) results in the differential association of stable and de novo signaling factors with the receptor at the membrane (D). Eventually, the complex of signaling proteins (encrypton) may dissociate from the receptor (E) and further change certain factors (F) before affecting genetic transcription. With multiple ligand stimulation (α, β), however, the relative apportioning of the stable signaling encryptons (1) with the active receptors (2) may be qualitatively/quantitatively different. Therefore, the nature of the disengaged signaling encrypton may be distinct (3). Because of the differences in encrypton complementation, alterations in transcriptional regulation may occur effecting distinct metabolic cellular outputs (4).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health (Intramural Research Program of the NIA). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains Tables 1-4.
Footnotes
The abbreviations used are: NGF, nerve growth factor; IGF-1, insulin-like growth factor 1; EGF, epidermal growth factor; SH, Src homology; ERK, extracellular signal-regulated kinase; MEK, mitogen-activated protein kinase kinase; HPLC, high pressure liquid chromatography; NE, norepinephrine; GSK-3, glycogen synthase kinase 3; EGFR, EGF receptor; PAGE, Parametric Analysis of Geneset Enrichment.
References
- 1.Vaudry, D., Stork, P. J., Lazarovici, P., and Eiden, L. E. (2002) Science 296 1648-1949 [DOI] [PubMed] [Google Scholar]
- 2.Freeman, R. S., Burch, R. L., Crowder, R. J., Lomb, D. J., Schoell, M. C., Straub, J. A., and Xie, L. (2004) Prog. Brain Res. 146 111-126 [DOI] [PubMed] [Google Scholar]
- 3.Chao, M. V., Rajagopal, R., and Lee, F. S. (2006) Clin. Sci. (Lond.) 110 167-173 [DOI] [PubMed] [Google Scholar]
- 4.Bondy, C. A., and Cheng, C. M. (2004) Eur. J. Pharmacol. 490 25-31 [DOI] [PubMed] [Google Scholar]
- 5.Mattson, M. P., Maudsley, S., and Martin, B. (2004) Ageing Res. Rev. 3 445-464 [DOI] [PubMed] [Google Scholar]
- 6.Groot, M., Boxer, L. M., and Thiel, G. (2000) Eur. J. Cell Biol. 79 924-935 [DOI] [PubMed] [Google Scholar]
- 7.Maudsley, S., Pierce, K. L., Zamah, A. M., Miller, W. E., Ahn, S., Daaka, Y., Lefkowitz, R. J., and Luttrell, L. M. (2000) J. Biol. Chem. 275 9572-9580 [DOI] [PubMed] [Google Scholar]
- 8.Diviani, D., Baisamy, L., and Appert-Collin, A. (2006) Eur. J. Cell Biol. 85 603-610 [DOI] [PubMed] [Google Scholar]
- 9.Tapon, N., Nagata, K., Lamarche, N., and Hall, A. (1998) EMBO J. 17 1395-1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickens, M., Rogers, J. S., Cavanagh, J., Raitano, A., Xia, Z., Halpern, J. R., Greenberg, M. E., Sawyers, C. L., and Davis, R. J. (1997) Science 277 693-696 [DOI] [PubMed] [Google Scholar]
- 11.Willoughby, E. A., and Collins, M. K. (2005) J. Biol. Chem. 280 25651-25658 [DOI] [PubMed] [Google Scholar]
- 12.Shinoda, S., Skradski, S. L., Araki, T., Schindler, C. K., Meller, R., Lan, J. Q., Taki, W., Simon, R. P., and Henshall, D. C. (2003) Eur. J. Neurosci. 17 2065-2076 [DOI] [PubMed] [Google Scholar]
- 13.Greene, L. A., and Tischler, A. S. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 2424-2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tischler, A. S., Greene, L. A., Kwan, P. W., and Slayton, V. W. (1983) Cell Tissue Res. 228 641-648 [DOI] [PubMed] [Google Scholar]
- 15.Garber, S. S., Hoshi, T., and Aldrich, R. W. (1989) J. Neurosci. 9 3976-3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park, S. Y., Avraham, H., and Avraham, S. (2000) J. Biol. Chem. 275 19768-19777 [DOI] [PubMed] [Google Scholar]
- 17.D'Mello, S. R., Borodezt, K., and Soltoff, S. P. (1997) J. Neurosci. 17 1548-1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Párrizas, M., and LeRoith, D. (1997) Endocrinology 138 1355-1358 [DOI] [PubMed] [Google Scholar]
- 19.Yamada, M., Ikeuchi, T., and Hatanaka, H. (1997) Prog. Neurobiol. 51 19-37 [DOI] [PubMed] [Google Scholar]
- 20.Whitney, L. W., Becker, K. G., Tresser, N. J., Caballero-Ramos, C. I., Munson, P. J., Prabhu, V. V., Trent, J. M., McFarland, H. F., and Biddison, W. E. (1999) Ann. Neurol. 46 425-428 [DOI] [PubMed] [Google Scholar]
- 21.Cheadle, C., Cho-Chung, Y. S., Becker, K. G., and Vawter, M. P. (2003) Appl. Bioinformatics 2 209-217 [PubMed] [Google Scholar]
- 22.Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., Paulovich, A., Pomeroy, S. L., Golub, T. R., Lander, E. S., and Mesirov, J. P. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15545-15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, S. Y., and Volsky, D. J. (2005) BMC Bioinformatics. 6 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baur, J. A., Pearson, K. J., Price, N. L., Jamieson, H. A., Lerin, C., Kalra, A., Prabhu, V. V., Allard, J. S., Lopez-Lluch, G., Lewis, K., Pistell, P. J., Poosala, S., Becker, K. G., Boss, O., Gwinn, D., Wang, M., Ramaswamy, S., Fishbein, K. W., Spencer, R. G., Lakatta, E. G., Le Couteur, D., Shaw, R. J., Navas, P., Puigserver, P., Ingram, D. K., de Cabo, R., and Sinclair, D. A. (2006). Nature 444 337-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krasnova, I. N., Cherkas, IuV., Denisenko, T. V., and Kartsova, L. A. (1999) Klin. Lab. Diagn. 7 11-14 [PubMed] [Google Scholar]
- 26.Xiong, W. C., and Mei, L. (2003) Front. Biosci. 8 676-682 [DOI] [PubMed] [Google Scholar]
- 27.Parsons, S. J., and Parsons, J. T. (2004) Oncogene 23 7906-7909 [DOI] [PubMed] [Google Scholar]
- 28.Liu, X., and Pawson, T. (1994) Recent Prog. Horm. Res. 49 149-160 [DOI] [PubMed] [Google Scholar]
- 29.Ravichandran, K. S. (2001) Oncogene 20 6322-6330 [DOI] [PubMed] [Google Scholar]
- 30.Sung, U., Apparsundaram, S., Galli, A., Kahlig, K. M., Savchenko, V., Schroeter, S., Quick, M. W., and Blakely, R. D. (2003) J. Neurosci. 23 1697-1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molteni, R., Ying, Z., and Gomez-Pinilla, F. (2002) Eur. J. Neurosci. 16 1107-1116 [DOI] [PubMed] [Google Scholar]
- 32.Kim, S. A., Chang, S., Yoon, J. H., and Ahn, S. G. (2008) FEBS Lett. 582 734-740 [DOI] [PubMed] [Google Scholar]
- 33.Blanquet, P. R. (1998) Neuroscience 86 739-749 [DOI] [PubMed] [Google Scholar]
- 34.Chao, C. C., Chiang, C. H., Ma, Y. L., and Lee, E. H. Y. (2006) Neurobiol. Aging 27 105-118 [DOI] [PubMed] [Google Scholar]
- 35.Lee, J. H., Kim, K. Y., Park, S. Y., Kim, D., Lee, W. S., Rhim, B. Y., and Hong, K. W. (2004) J. Pharmacol. Exp. Ther. 308 896-903 [DOI] [PubMed] [Google Scholar]
- 36.Natale, J. E., Ahmed, F., Cernak, I., Stoica, B., and Faden, A. I. (2003) J. Neurotrauma 20 907-928 [DOI] [PubMed] [Google Scholar]
- 37.Masilamoni, J. G., Vignesh, S., Kirubaragan, R., Jesudason, E. P., and Jayakumar, R. (2005) Brain Res. Bull. 67 235-241 [DOI] [PubMed] [Google Scholar]
- 38.Che, Y., Piao, C. S., Han, P. L., and Lee, J. K. (2001) J. Neurosci. Res. 65 425-431 [DOI] [PubMed] [Google Scholar]
- 39.Ousman, S. S., Tomooka, B. H., van Noort, J. M., Wawrousek, E. F., O'Connor, K. C., Hafler, D. A., Sobel, R. A., Robinson, W. H., and Steinman, L. (2007) Nature 448 474-481 [DOI] [PubMed] [Google Scholar]
- 40.Ying, X., Zhang, J., Wang, Y., Wu, N., Wang, Y., and Yew, D. T. (2008) J. Mol. Neurosci. 35 253-258 [DOI] [PubMed] [Google Scholar]
- 41.Hunter, T. (2000) Cell 100 113-127 [DOI] [PubMed] [Google Scholar]
- 42.Schlessinger, J. (2000) Cell 103 211-225 [DOI] [PubMed] [Google Scholar]
- 43.Maudsley, S., Martin, B., and Luttrell, L. M. (2007) Curr. Alzheimer Res. 4 3-19 [DOI] [PubMed] [Google Scholar]
- 44.Pawson, T., and Scott, J. D. (1997) Science 278, 2075-2080 [DOI] [PubMed] [Google Scholar]
- 45.Maudsley, S., Davidson, L., Pawson, A. J., Freestone, S. H., Lopez de Maturana, R., Thomson, A. A., and Millar, R. P. (2006) Neuroendocrinology 84 285-300 [DOI] [PubMed] [Google Scholar]
- 46.Maudsley, S., Naor, Z., Bonfil, D., Davidson, L., Pawson, A. J., Larder, R., Pope, C., Nelson, N., Millar, R. P., and Brown, P. (2007) Mol. Endocrinol. 21 1216-1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howe, C. L., Valletta, J. S., Rusnak, A. S., and Mobley, W. C. (2001) Neuron 32 801-814 [DOI] [PubMed] [Google Scholar]
- 48.Valdez, G., Philippidou, P., Rosenbaum, J., Akmentin, W., Shao, Y., and Halegoua, S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12270-12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Traverse, S., Seedorf, K., Paterson, H., Marshall, C. J., Cohen, P., and Ullrich, A. (1994) Curr. Biol. 4 694-701 [DOI] [PubMed] [Google Scholar]
- 50.Robinson, M. J., Stippec, S. A., Goldsmith, E., White, M. A., and Cobb, M. H. (1998) Curr. Biol. 8 1141-1150 [DOI] [PubMed] [Google Scholar]
- 51.Qureshi, F. G., Tchorzewski, M. T., Duncan, M. D., and Harmon, J. W. (1997) J. Surg. Res. 69 354-358 [DOI] [PubMed] [Google Scholar]
- 52.Pelicci, G., Pelicci, G., Dente, L., De Giuseppe, A., Verducci-Galletti, B., Giuli, S., Mele, S., Vetriani, C., Giorgio, M., Pandolfi, P. P., Cesareni, G., and Pelicci, P. G. (1996) Oncogene 13 633-641 [PubMed] [Google Scholar]
- 53.Pelicci, G., Lanfrancone, L., Salcini, A. E., Romano, A., Mele, S., Grazia Borrello, M., Segatto, O., Di Fiore, P. P., and Pelicci, P. G. (1995) Oncogene 11 899-907 [PubMed] [Google Scholar]
- 54.Rozakis-Adcock, M., McGlade, J., Mbamalu, G., Pelicci, G., Daly, R., Li, W., Batzer, A., Thomas, S., Brugge, J., Pelicci, P. G., Schlessinger, J., and Pawson, T. (1992) Nature 360 689-692 [DOI] [PubMed] [Google Scholar]
- 55.Boney, C. M., Gruppuso, P. A., Faris, R. A., and Frackelton, A. R., Jr. (2000) Mol. Endocrinol. 14 805-813 [DOI] [PubMed] [Google Scholar]
- 56.Peng, H., Wen, T. C., Tanaka, J., Maeda, N., Matsuda, S., Desaki, J., Sudo, S., Zhang, B., and Sakanaka, M. (1998) J. Cereb. Blood Flow Metab. 18 349-360 [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y., Tatsuno, T., Carney, J. M., and Mattson, M. P. (1993) J. Cereb. Blood Flow Metab. 13 378-388 [DOI] [PubMed] [Google Scholar]
- 58.Sampath, D., Jackson, G. R., Werrbach-Perez, K., and Perez-Polo, J. R. (1994) J. Neurochem. 62 2476-2479 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.