Abstract
Cyclin-dependent kinase 5 (Cdk5) is a proline-directed serine/threonine kinase. We have previously reported that Cdk5 participates in the regulation of nociceptive signaling, and the expression of Cdk5 and its activator, p35, are up-regulated in nociceptive neurons during peripheral inflammation. The aim of our current study was to identify the proinflammatory molecules that regulate Cdk5/p35 activity in response to inflammation. We constructed a vector that contains the mouse p35 promoter driving luciferase expression. We transiently transfected this vector in PC12 cells to test the effect of several cytokines on p35 transcriptional activity and Cdk5 activity. Our results indicate that tumor necrosis factor-α (TNF-α) activates p35 promoter activity in a dose- and time-dependent manner and concomitantly up-regulates Cdk5 activity. Because TNF-α is known to activate ERK1/2, p38 MAPK, JNK, and NF-κB signaling pathways, we examined their involvement in the activation of p35 promoter activity. MEK inhibitor, which inhibits ERK activation, decreased p35 promoter activity, whereas the inhibitors of p38 MAPK, JNK, and NF-κB increased p35 promoter activity, indicating that these pathways regulate p35 expression differently. The mRNA and protein levels of Egr-1, a transcription factor, were increased by TNF-α treatment, and this increase was dependent on ERK signaling. In a mouse model of inflammation-induced pain in which carrageenan injection into the hind paw causes hypersensitivity to heat stimuli, TNF-α mRNA was increased at the site of injection. These findings suggest that TNF-α-mediated regulation of Cdk5 activity plays an important role in inflammation-induced pain signaling.
Cyclin-dependent kinase 5 (Cdk5)2 is a proline-directed serine/threonine kinase that belongs to the family of cyclin-dependent protein kinases (1). Cdk5 kinase activity is predominantly found in postmitotic neurons (2, 3), where its activators, p35 and p39, are predominantly expressed (2–4). We reported earlier that cellular p35 level is the main limiting factor for the Cdk5 kinase activity (5). Cdk5 phosphorylates a spectrum of proteins, most of them associated with cell morphology, synaptic activity, neuronal survival, and apoptosis (1, 6). A majority of known substrates of Cdk5 are cytoskeletal elements, signaling molecules, or regulatory proteins (6). Cdk5 is most abundant in the nervous system, and it appears to be indispensable for neural development and function (7). The deregulation of Cdk5 activity has been implicated in neurodegenerative diseases of the mammalian nervous system (1, 6, 8, 9). We have previously reported that Cdk5 activity participates in the regulation of nociceptive signaling (10). The expressions of Cdk5 and p35 as well as Cdk5 kinase activity are increased in nociceptive primary afferent neurons during peripheral inflammation. In p35 knock-out mice (p35-/-) responses to a nociceptive heat stimulus are attenuated, whereas in p35-overexpressing transgenic mice, these responses are potentiated, indicating an important role of Cdk5 in nociceptive process (10–13). Recently, we have demonstrated that Cdk5-mediated phosphorylation of transient receptor potential vanilloid 1 (TRPV1) at threonine 407 can modulate agonist-induced calcium influx (14). TRPV1, a ligand-gated cation channel highly expressed in small diameter sensory neurons, is activated by heat, protons, and capsaicin. The phosphorylation of TRPV1 provides a versatile regulation of intracellular calcium levels, critical for its function in responding to noxious stimuli (14).
Several cytokines are known to mediate chronic pain caused by inflammation or sensory nerve damage (15). In animal models of inflammatory or neuropathic pain, the levels of interleukin-6 (IL-6), interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α) are elevated in the spinal cord and at the site of injury (16–19). It has also been reported that the mRNA levels of IL-6, IL-1β, and TNF-α were elevated in inflamed hind paw of rats injected with intraplantar carrageenan (20). TNF-α is a pro-inflammatory cytokine produced by astrocytes, microglia, and neurons (21). TNF-α exerts its biological functions through the action of two main receptors, TNF receptors 1 and 2, and both are expressed in dorsal root ganglia (DRG) (20). Through these receptors, TNF-α can induce activation of the NF-κB pathway (22) and MAPK pathways such as the extracellular signal-regulated kinase 1/2 (ERK1/2), p38 MAPK, and c-Jun N-terminal kinase (JNK) (23). Because activation of the ERK1/2 signaling pathway has been implicated in the regulation of Cdk5 activity through induction of p35 protein expression (24–26), we investigated the role of TNF-α in the regulation of Cdk5/p35 expression. In the present study, we demonstrate that p35 expression is increased through ERK1/2 and Egr-1 pathways. These findings suggest that during peripheral inflammation, TNF-α induces Cdk5 kinase activity by increasing the levels of p35, which can sensitize TRPV1-positive sensory neurons.
EXPERIMENTAL PROCEDURES
Animals—Mice were maintained in a C57BL/6 × 129/SvJ hybrid background and were housed with a 12 h light/dark cycle and given water and food ad libitum. All of the animal procedures were conducted in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals. Inflammation was induced by injecting carrageenan Lambda type IV (Sigma). Carrageenan (4% in phosphate-buffered saline) was suspended by sonication in saline and injected subcutaneously in a volume of 25 μl into the left plantar hind paw by using an insulin syringe with a 28-gauge needle. Contralateral right hind paws served as the controls. The mice were euthanized by carbon dioxide inhalation at 24 h after carrageenan injection. The mid-central plantar portion of the hind paw, which included both skin and underlying muscle, was removed and processed for RNA extraction. DRG from lumbar region L4 and L5 were removed for RNA extraction.
Materials—Mouse recombinant TNF-α, leukemia inhibitory factor (LIF), oncostatin M (OSM), IL-1β, interferon γ (IFN-γ), histone H1, SP600125, and α-tubulin antibody were obtained from Sigma. Human recombinant IL-6, PD98059, SB203580, and NF-κB inhibitor were obtained from Calbiochem, and rat recombinant β nerve growth factor was obtained from R & D Systems (Minneapolis, MN). Protein quantification reagents were obtained from Bio-Rad, and enhanced chemiluminescence reagents for Western blot analysis were purchased from Thermo Scientific (Rockford, IL).
Antibodies—Antibodies to Cdk5, p35, and Egr-1 and secondary antibodies (HP-conjugated goat anti-mouse, anti-rabbit antibodies) were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Antibodies to ERK1/2, phospho-ERK1/2, p38 MAPK, phospho-p38 MAPK, JNK, phospho-JNK, NF-κB p65, and phospho-NF-κB p65 were obtained from Cell Signaling Technology (Beverly, MA). Antibody to β-actin was obtained from Chemicon (Temecula, CA).
Cell Culture—The PC12 cell line (derived from pheochromocytoma of rat adrenal medulla) and Neuro-2a cell line (derived from mouse neuroblastoma) were obtained from American Type Culture Collection (Rockville, MD). PC12 and Neuro-2a cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum plus 5% horse serum (Hyclone Laboratories, Logan, UT).
Preparation of the p35 Promoter-Luciferase Reporter Plasmid—We constructed a p35 promoter-LUC vector by inserting a 1,219-bp mouse p35 promoter into the pGL3 enhancer luciferase vector from Promega (Madison, WI). Briefly, pBluescript II SK(-) p35 promoter vector (27) was digested with XbaI and XhoI, and a 1,219-bp fragment containing the p35 promoter was cloned between the NheI and XhoI sites of pGL3 enhanced vector.
Transient Transfection and Reporter Activity Assays—One hour before transfection, medium with serum was replaced by Dulbecco's modified Eagle's medium plus 0.1% bovine serum albumin. Transfection of the reporter plasmid into PC12 cells was done using FuGENE 6 transfection reagent (Roche Applied Science). Five hundred nanograms (ng) of p35 promoter-LUC and 250 ng of control vector Renilla luciferase expressed under the constitutive promoter of thymidine kinase (RL-TK; Promega, Madison, WI) were cotransfected into ∼4 × 104 cells. After the transfection, the cells were treated with either IL-6 (50 ng/ml), LIF (50 ng/ml), OSM (100 ng/ml), IL-1β (10 ng/ml), INF-γ (100 ng/ml), or TNF-α (50 ng/ml) for 24 h. The proteins were extracted from the treated cells, and the luciferase activity was measured with a dual luciferase reporter assay system (Promega, Madison, WI), and the results were presented as the relative p35 promoter activity, which was calculated by dividing the mean value of p35 promoter luciferase activity by the mean value of Renilla luciferase activity. In the second set of experiments, we treated PC12 cells with inhibitors of MAPKs (PD98059, SB203580, and SP600125) or NF-κB inhibitor prior to TNF-α treatment and measured luciferase activity 24 h after.
RNA Isolation and Real Time PCR—PC12 cells were grown in 60-mm culture dishes and were incubated for 0, 0.25, 0.5, 1, 2, 3, 6, 9, 15, or 24 h in serum-free medium with TNF-α (25 ng/ml). After discarding growth medium, total RNA was isolated from the cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Following TURBO DNA-free (Ambion, Austin, TX) digestion of total RNA sample, to remove contaminated genomic DNA, oligo(dT) primed synthesis of cDNA from 1 μg of total RNA was made using Super-Script™ III reverse transcriptase (Invitrogen). For detection of TNF-α mRNA, PCR was performed using the following primers: TNF-α sense (S): 5′-GAT CTC AAA GAC AAC CCA ACT AGT-3′ and TNF-α antisense (AS) 5′-CTC CAG CTG GAA GAC TCC TCC CAG-3′. The PCR consisted of 35 cycles of 30 s each at 94 °C, 60 °C, and 72 °C. For detection of Egr-1, p35, and Cdk5 mRNA we used real time PCR, and the following reaction mixture was used for these PCR samples: 1xIQ™ Sybr®Green Super Mix (Bio-Rad), 100–200 nm of each primer and 1 μl of cDNA. cDNA was amplified and analyzed in triplicate using Opticon Monitor Chromo 4 (Bio-Rad). The following primers were used to amplify and measure the amount of mouse mRNA by real time reverse transcription-PCR: Egr-1 S: 5′-CCC TTC CAG GGT CTG GAG AAC CGT-3′, Egr-1 AS: 5′-GGG GTA CTT GCG CAT GCG GCT GGG-3′, p35 S: 5′-GCC CTT CCT GGT AGA GAG CTG-3′, p35 AS: 5′-GTG TGA AAT AGT GTG GGT CGG C-3′, Cdk5 S: 5′-GGC TAA AAA CCG GGA AAC TC-3′ and Cdk5 AS: 5′-CCA TTG CAG CTG TCG AAA TA-3′ (28). mRNA levels were standardized by using the following primers to GAPDH: GAPDH S: 5′-AAT GTG TCC GTC GTG GAT CTG A-3′ and GAPDH AS: 5′-GAT GCC TGC TTC ACC ACC TTC T-3′.
RNA Interference Analysis—A chemically synthesized siGENOME set of four duplex of small interference RNA (siRNA) for rat Egr-1 was obtained from Dharmacon, Inc. (Chicago, IL). The pool contained four RNA duplexes, and their sequences are as follows: 5′-UAA AGG CUC UUA AUA ACA C-3′;5′-CGA CAG CAG UCC CAU UUA C-3′;5′-UGA CAU CGC UCU GAA UAA C-3′; and 5′-CGA AUC UGC AUG CGU AAU U-3′. Both negative and positive GAPDH siRNA duplexes were also included in the assay (Dharmacon, Inc., Chicago, IL). Delivery of siRNAs into PC12 cells was accomplished using Dharmafect1 reagent (Dharmacon, Inc). Using a siRNA-negative control coupled to fluorescein isothiocyanate, we determined by fluorescence a transfection efficiency of 65 ± 8% (means ± S.D.). As a positive control we used GAPDH siRNA. This protocol knocked down GAPDH expression at both mRNA and protein levels. At 20 h after transfection with Egr1 siRNA duplexes at concentration of 100 nm each siRNA, the cultures were treated with either 100 ng/ml TNF-α or distilled water for 1, 3, and 6 h. The cells were then used for extraction of total RNA, and analysis of mRNA expression for Egr1 and p35 by real time PCR were conducted as described above.
Immunoblot Analysis—Tissue homogenates were lysed in T-PER buffer (Pierce) with protease inhibitor mixture tablets and phosphatase inhibitor mixture tablets PhosSTOP (Roche Applied Science). Protein concentration of the supernatant was determined by using Bradford Protein Assay (Bio-Rad). The proteins were separated by 4–12% SDS-PAGE gels and transferred to nitrocellulose membranes (Invitrogen). The membranes were soaked in a blocking buffer (5% nonfat dry milk in phosphate-buffered saline with 0.05% Tween 20 (PBST) for 1 h at room temperature and then incubated overnight at 4 °C with the appropriate primary antibody diluted in the blocking buffer. The membranes were washed in PBST and incubated for 1 h at room temperature with the secondary antibodies diluted in blocking buffer. Immunoreactivity was detected by SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). The membranes were stripped for 15 min at room temperature with Re-blot Plus Strong Solution (Chemicon) and retested with α-tubulin or β-actin antibodies to normalize for protein loading. The optical densities of the bands were quantified by using an image analysis system with Scion Image Alpha 4.0.3.2 software (Scion Corporation, Frederick, MD).
Cdk5 Kinase Activity Assay—Cdk5 kinase activity was measured as described (10). In brief, 150 μg of protein from PC12 or neuro-2a cells was dissolved in T-PER buffer and immunoprecipitated with anti-Cdk5 antibody (5 μg). Immunoprecipitated proteins were washed in kinase buffer (20 mm Tris·HCl, pH 7.4, 10 mm MgCl2, 1 mm EDTA, 10 μm NaF, and 1 μm Na2VO3) and mixed with kinase assay mixture (100 mm Tris·HCl, pH 7.4, 50 mm MgCl2, 5 mm EDTA, 50 μm NaF, 5 μm Na2VO3, and 5 mm dithiothreitol), and Histone H1 (1 μg/μl) as a substrate, and the kinase activity was quantified as described (10).
Hargreaves Test—Radiant heat from a focused projector lamp was used to measure basal sensitivity to noxious thermal stimulation. Each mouse was placed in an individual glass chamber (12.5 × 12.5 × 12.5 cm) with transparent outer walls to allow for experimental observation and a 3/16-inch-thick (1 inch = 2.54 cm) glass floor. The mice were acclimated for at least 1 h before testing. The stimulus was a high intensity beam (750 mA) from a projector lamp bulb aimed at the plantar surface of the mid-hind paw of a mouse after habituation. Paw withdrawal latency was tested to the nearest 0.1 s, and each mouse was tested until the 15-s cut-off was reached.
Statistical Analysis—All of the experiments were performed a minimum of three times. Statistical evaluation was done with GraphPad Prism software, version 4.0 (GraphPad, San Diego, CA). Significant differences between experiments were assessed by univariate ANOVA (more than two groups) or unpaired t test (two groups). ANOVA was followed by a t test using a Bonferroni α-correction for multiple comparisons, where α was set to 0.05.
RESULTS
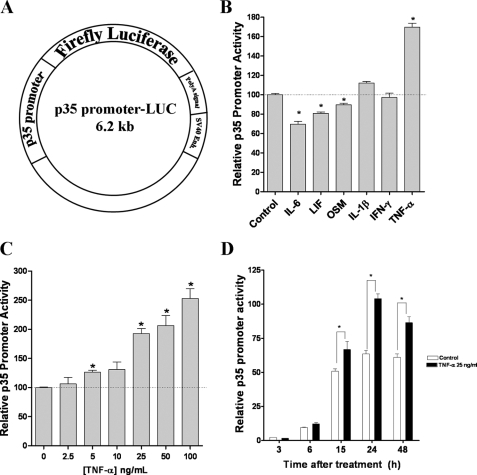
TNF-α Regulates p35 Promoter Activity—Because the level of p35 expression is the main limiting factor for Cdk5 activity (5), we developed a cell-based assay to screen proinflammatory molecules for their effects on p35 promoter activity. In brief, we constructed a p35 promoter-LUC vector by cloning a 1,219-bp fragment of mouse p35 promoter (27) into luciferase pGL3 enhancer vector (Fig. 1A). We used the rat pheochromocytoma PC12 cell line for this study because several groups have reported that it expresses receptors for a variety of cytokines (29–32). PC12 cells, transiently cotransfected with p35 promoter-LUC vector and RL-TK vector (control for transfection efficiency), were treated with indicated cytokine for 24 h, and then luciferase activity was measured. We found that of the cytokines tested, TNF-α significantly increased p35 promoter activity (Fig. 1B). TNF-α (50 ng/ml) treatment resulted in an 80% increase in the luciferase activity, whereas other cytokines did not enhance the activity. IL-6, LIF, and OSM treatment resulted in a slight inhibition of p35 promoter activity (Fig. 1B). Treatment with IFN-γ or IL-1β did not have any obvious effect on p35 promoter activity (Fig. 1B). Subsequently, we analyzed the effect of different concentrations of TNF-α over p35 promoter activity and found that TNF-α increased p35 promoter activity in a dose-dependent manner (Fig. 1C). TNF-α treatment increased p35 promoter activity at a concentration of 5 ng/ml followed by a linear increase with higher concentrations up to 100 ng/ml. At a concentration of 100 ng/ml, p35 promoter activity increased by 150% as compared with the control. Next, we analyzed the time course of the TNF-α-mediated increase of p35 promoter activity. Transiently transfected PC12 cells treated with TNF-α (25 ng/ml) showed a time-dependent with increase in p35 promoter activity (Fig. 1D). TNF-α treatment significantly increased p35 promoter activity beginning at 6 h and reached a peak at 24 h. Together, these results indicated that TNF-α treatment strongly induced p35 promoter activity in PC12 cells in a dose- and time-dependent manner.
FIGURE 1.
TNF-α regulates p35 promoter activity. A, schematic representation of the p35 promoter-LUC vector (6.2 kb). It consists of 1,219-bp of mouse p35 promoter driving luciferase firefly expression. B, PC12 cells were transiently transfected with 0.5 and 0.25 μg of p35 promoter-LUC and Renilla luciferase, respectively. After transfection, the cells were incubated with either 50 ng/ml of IL-6, 50 ng/ml of LIF, 100 ng/ml of OSM, 10 ng/ml of IL-1β, 100 ng/ml of IFN-γ, 50 ng/ml of TNF-α, or medium alone (control) for 24 h, and relative p35 promoter activity was calculated by dividing the mean value of p35 promoter luciferase activity by the mean value of Renilla luciferase activity. C, TNF-α treatment at different concentrations increased p35 promoter activity in a dose-dependent manner at 24 h. D, TNF-α treatment (25 ng/ml) increased p35 promoter activity in a time-dependent manner. All of the data are presented as the means and S.E. (n = 3–7). *, p < 0.01 (Bonferroni's test after ANOVA).
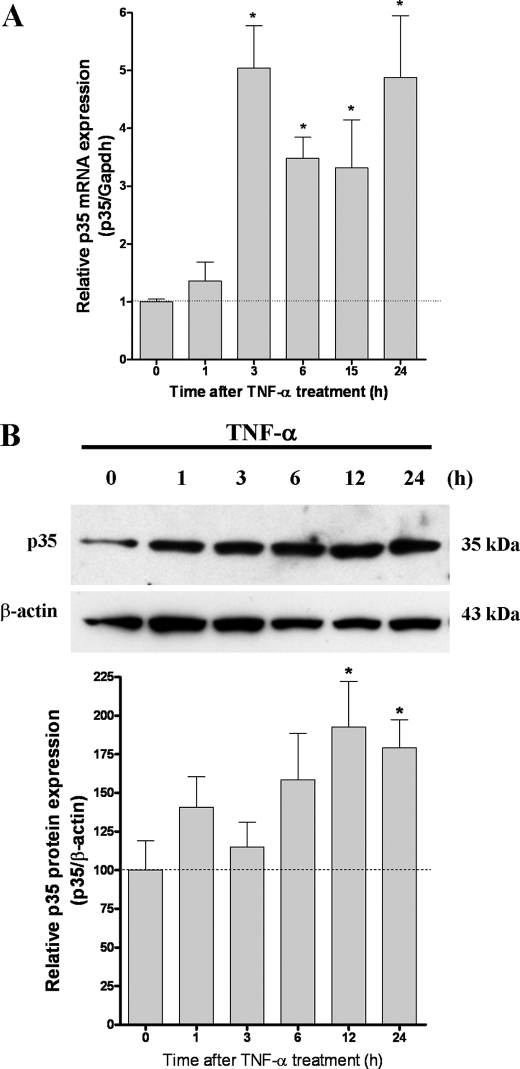
TNF-α Treatment Significantly Increases p35 Expression in PC12 Cells—To further confirm that TNF-α activates p35 promoter activity, we examined endogenous levels of p35 mRNA and protein at different time points during treatment with TNF-α. The level of p35 mRNA increased significantly within 3 h after TNF-α treatment and remained elevated until 24 h (Fig. 2A). Subsequently, p35 protein levels increased significantly at 12 and 24 h following TNF-α treatment (Fig. 2B). On the other hand, Cdk5 mRNA and protein levels did not change significantly following TNF-α treatment (data not shown).
FIGURE 2.
TNF-α treatment results in a significant increase in p35 expression. A, real time PCR analysis of p35 RNA levels normalized against GAPDH. Total RNA was obtained from PC12 cells treated with TNF-α (25 ng/ml) for the time indicated. After reverse transcription we carried out real time PCR with specific primers for p35 and GAPDH. The bars are the means and S.E. of seven independent experiments measured in triplicate. *, p < 0.01 (Bonferroni's test after ANOVA). B, Western blot analysis for p35 protein levels. PC12 cells were treated with TNF-α (50 ng/ml) for the time indicated. Western blot analysis indicated increase in p35 protein levels after treatment with TNF-α. β-actin was used as a loading control. The results are quantified in the bar graphs shown below B. The bars are the means and S.E. of three independent experiments. *, p < 0.05 by using t test.
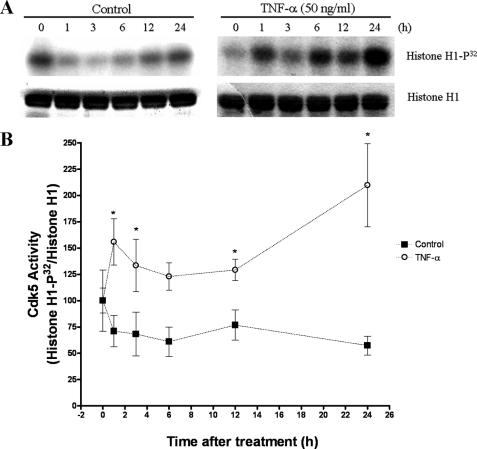
TNF-α Treatment Increases Cdk5 Activity in PC12 Cells—Because p35 protein levels regulate Cdk5 kinase activity (5), we analyzed whether the TNF-α-mediated increase in p35 expression results in increased Cdk5 activity. We immunoprecipitated Cdk5 protein from control or TNF-α-treated cells and then assayed kinase activity by using histone H1 as a substrate (10). In control cells, Cdk5 kinase activity decreased slightly after serum deprivation (Fig. 3A, left panel). In contrast, TNF-α-treated cells showed an increased Cdk5 kinase activity (Fig. 3A, right panel). Cdk5 activity increased as early as 1 h after TNF-α treatment and peaked at 24 h (Fig. 3B). We also found that Cdk5 activity increased in the Neuro-2a cell line after TNF-α treatment (data not shown). Together, these results indicated that TNF-α-induced expression of p35 resulted in increased Cdk5 kinase activity.
FIGURE 3.
TNF-α treatment increases Cdk5 activity. A, PC12 cells were treated with TNF-α (50 ng/ml) for the times indicated. The untreated PC12 cells served as controls. We immunoprecipitated Cdk5 with anti-Cdk5 antibody and assayed its kinase activity, using histone H1 as a substrate. B, quantification of the increase in Cdk5 activity after TNF-α treatment in PC12 cells. The values are shown as the means and S.E. of four to seven independent experiments. *, p < 0.05 by using t test.
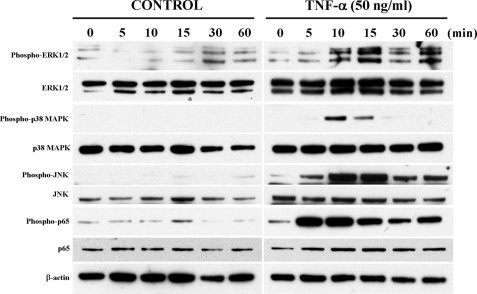
TNF-α-mediated Activation of ERK1/2, p38 MAPK, JNK, and NF-κB Signaling Pathways Differentially Regulates p35 Promoter Activity—TNF-α is known to activate MAPK pathways, such as ERK1/2, p38 MAPK, JNK, and NF-κB pathways (22, 23, 33). We examined the influence of these pathways on TNF-α-mediated activation of p35 promoter activity. By Western blot analysis, we observed a rapid activation of ERK1/2, p38 MAPK, JNK, and NF-κB p65 pathways within 5–10 min following TNF-α treatment of PC12 cells (Fig. 4). The activation of ERK1/2 was sustained, whereas the activation of p38 MAPK, JNK, and NF-κB p65 was transient. As reported earlier (24) we also found that nerve growth factor activated these pathways in PC12 cells (data not shown). To confirm this further, prior to TNF-α treatment we tested inhibitors of these pathways in the PC12 cells. We tested MEK inhibitor PD98059, an upstream regulator of ERK1/2 (34); p38 MAPK inhibitor SB203580 (35); JNK inhibitor SP600125 (36); and an NF-κB inhibitor (37). We found that all of these inhibitors clearly attenuated the activation of these pathways by TNF-α (data not shown). Next, we assayed the effects of different concentrations of MAPK inhibitors and the NF-κB inhibitor on p35 promoter activity in the presence or absence of TNF-α in PC12 cells transiently transfected with p35-LUC and RL-TK vectors. MEK inhibitor PD98059 decreased basal p35 promoter activity significantly at 30 and 60 μm (Fig. 5A, left panel) in the absence of TNF-α. In the presence of PD98059 (30 μm) plus TNF-α (50 ng/ml), we observed an 85% reduction in p35 promoter activity as compared with TNF-α treatment alone (Fig. 5A, right panel). This indicated that ERK1/2 positively regulated p35 promoter activity. On the other hand, p38 MAPK inhibitor SB203580 (1, 10, and 20 μm), JNK inhibitor SP600125 (10, 20, and 40 μm), and the NF-κB inhibitor (0.02, 0.2, and 1 μm) increased basal p35 promoter activity (Fig. 5, B–D), and this activity increased several fold in the presence of TNF-α (50 ng/ml) plus SB203580 (20 μm), or SP600125 (20 μm), or NF-κB inhibitor (0.2 μm) as compared with TNF-α treatment alone (Fig. 5, B–D, right panels), indicating that p38 MAPK, JNK, and NF-κB pathways negatively regulate p35 promoter activity.
FIGURE 4.
TNF-α induced activation of ERK1/2, p38 MAPK, JNK, and NF-κB signaling pathways. Western blot analysis for protein levels of phospho-ERK1/2, total ERK1/2, phospho-p38 MAPK, total p38 MAPK, phospho-JNK, total JNK, phospho-p65, and total p65 in PC12 cells treated with TNF-α (100 ng/ml) for the times indicated. As seen, TNF-α treatment induced a rapid activation of all these pathways.
FIGURE 5.
TNF-α-dependent activation of ERK1/2, p38 MAPK, JNK, and NF-κB pathways differently regulates p35 promoter activity. A, relative p35 promoter activity was measured in PC12 cells incubated with different concentrations of MEK inhibitor PD98059 (panel left) or with PD98059 (30 μm) plus TNF-α (50 ng/ml) (panel right). B, relative p35 promoter activity was measured in PC12 cells treated with different concentrations of p38 MAPK inhibitor SB203580 (left panel) or with SB203580 (20 μm) plus TNF-α (50 ng/ml) (right panel). C, relative p35 promoter activity was measured in PC12 cells treated with different concentrations of JNK inhibitor SP600125 (left panel) or SP6000125 (10 μm) plus TNF-α (50 ng/ml) (right panel). D, relative p35 promoter activity was measured in PC12 cells treated with different concentrations of NF-κB inhibitor (left panel) or NF-κB inhibitor (0.2 μm) plus TNF-α (50 ng/ml) (right panel). All of the data are presented as the means and S.E. (n = 3–7). *, p < 0.05 by using t test.
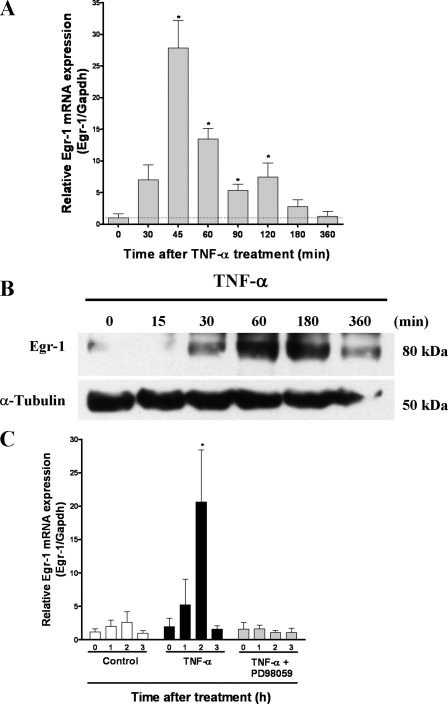
TNF-α Treatment Increases Egr-1 mRNA and Protein Levels—Because the p35 promoter region contains several putative sequence elements, including the binding site for transcription factor Egr-1 (also known as NGFI-A, Zif268, and Krox24, (27)), we investigated the time course of Egr-1 expression after TNF-α treatment. First, we measured the Egr-1 mRNA levels by real time PCR after TNF-α treatment. Egr-1 mRNA levels increased within 30 min, reached a plateau at 45 min, and decreased to a basal level at 6 h (Fig. 6A). In addition, the Egr-1 protein levels increased after 30 min following TNF-α treatment and remained high until 6 h (Fig. 6B). Because it was reported earlier that the Egr-1 promoter region contains a serum response element, and expression of Egr-1 is regulated by the ERK1/2 pathway (38), we examined the effects of PD98059 (30 μm) on Egr-1 mRNA levels in PC12 cells treated with TNF-α (25 ng/ml). Preincubation of transfected PC12 cells with PD98059 1 h before TNF-α stimulation blocked the increase in Egr-1 mRNA levels (Fig. 6C), indicating that TNF-α induces Egr-1 expression through the ERK1/2 pathway.
FIGURE 6.
TNF-α increases Egr-1 mRNA and protein levels. A, Real time PCR analysis of Egr-1 RNA levels normalized against GAPDH. Total RNA was obtained from PC12 cells treated with TNF-α (25 ng/ml) for the indicated time. After reverse transcription, we carried out real time PCR with specific primers for Egr-1 and GAPDH. The bars are the means and S.E. of three independent experiments measured in triplicate. *, p < 0.05 by using t test. B, Western blot analysis for Egr-1. PC12 cells were treated with TNF-α (50 ng/ml) for the time indicated. Western blot analysis showed Egr-1 protein levels were increased after treatment with TNF-α. α-tubulin was used as a loading control. C, PC12 cells were treated with 50 ng/ml TNF-α or with 30 μm PD98059 for 1 h before treatment with TNF-α (50 ng/ml) for 0, 1, 2, and 3 h. Total RNA was isolated and subjected to reverse transcription and real time PCR for Egr-1 and GAPDH. The bars are the means and S.E. of three independent experiments measured in triplicate. *, p < 0.05 (Bonferroni's test after ANOVA).
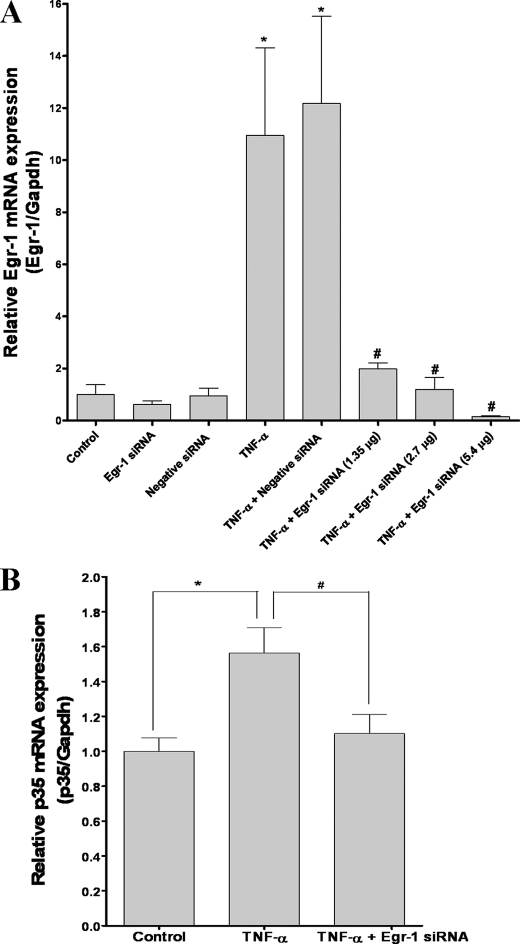
Depletion of Endogenous Egr-1 by RNA Interference Decreases p35 Expression—We have provided evidence that TNF-α-induced Egr-1 activity is associated with increased p35 promoter activity, induction of p35 transcripts, and increased Cdk5 activity. To confirm whether Egr-1 is essential for TNF-α-induced p35 expression, we employed siRNA-mediated gene silencing to knockdown Egr-1 levels in PC12 cells. We used a chemically synthesized siGENOME set of four duplexes of siRNA for rat Egr-1. The siRNA pool included four siRNA duplexes to maximize their silencing efficiency. Using Dharmafect 1 reagent we successfully delivered the siRNA duplexes into PC12 cells with a transfection efficiency of 65 ± 8% (mean ± S.D.) as determined using a fluorescein isothiocyanate-labeled negative siRNA duplex. This protocol also decreased the expression of GAPDH at both the mRNA (84%) and protein levels (55%) (data not shown) in cell transfected with GAPDH siRNA duplex. The Egr-1 mRNA levels increased about 10-fold after 1 h of TNF-α treatment (p < 0.01), but this effect was reduced in cells transfected with 1.35, 2.7, and 5.4 μg of all four Egr-1 siRNA prior to TNF-α treatment (Fig. 7A). This effect was specific for Egr-1 siRNA because nonspecific siRNA (negative siRNA) did not inhibit increased in Egr-1 mRNA levels following TNF-α treatment (Fig. 7A). The reduction of Egr-1 expression was accompanied by a decrease in endogenous p35 mRNA levels. TNF-α treatment (3 h) increased p35 mRNA levels (p < 0.01), whereas their levels were significantly decreased in PC12 cells transfected with Egr-1 siRNA and treated with TNF-α (Fig. 7B). Similarly, p35 mRNA levels increased after 6 h of TNF-α treatment, and this effect was blocked by Egr-1 siRNA (data not shown). These results indicate that Egr-1 activation is specifically essential for induction of p35 expression in response to TNF-α.
FIGURE 7.
siRNA-mediated knockdown of Egr-1 levels reduces TNF-α-induced p35 mRNA expression. A, real time PCR analysis of Egr-1 RNA levels normalized against GAPDH. Total RNA was obtained from PC12 cells transfected with a pool of four Egr-1 siRNAs at different concentrations (1.35, 2.7, and 5.4 μg) or with negative control siRNA. The cells were treated after 24 h with TNF-α (100 ng/ml) for 1 h. After reverse transcription, we carried out real time PCR with specific primers for Egr-1 and GAPDH. The bars are the means and S.E. of three to eight independent experiments measured in triplicate. *, mean control compared with all bars (p < 0.01); #, mean TNF-α compared with TNF-α plus EGR-1 siRNA (p < 0.05) (Bonferroni's test after ANOVA). B, real time PCR analysis of p35 transcription levels normalized against GAPDH. Total RNA was obtained from PC12 cells transfected for 24 h with a pool of four Egr-1 siRNAs and treated for 3 h with TNF-α (100 ng/ml), and after reverse transcription, real time PCR with specific primers for p35 and GAPDH were conducted. The bars are the means and S.E. of seven independent experiments measured in triplicate. *, statistically significant difference between control and TNF-α-treated group (p < 0.01); #, statistically significant difference between TNF-α-treated group compared with TNF-α plus EGR-1 siRNA-treated group (p < 0.05) (Bonferroni's test after ANOVA).
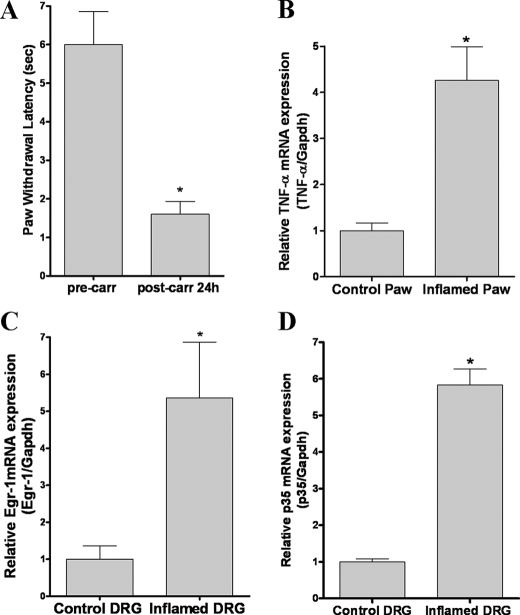
In Vivo Carrageenan-induced Inflammation Increases TNF-α, Egr-1, and p35 mRNA Expression—To extend our in vitro experiments to an animal model, we inflamed mouse hind paws unilaterally using intraplantar carrageenan injection and then tested paw withdrawal latency from a noxious heat stimulus prior to and 24 h post-injection. As expected, we observed reduced paw withdrawal latencies (i.e. faster withdrawal, hyperalgesia) in inflamed paws compared with the same paws before injection (Fig. 8A). Next we harvested plantar skin and muscle from the inflamed and uninjected (control) paws and measured TNF-α mRNA expression levels. We found that TNF-α mRNA levels were four times higher in inflamed paws as compared with control paws (Fig. 8B). Then we separately collected L4-L5 DRGs from the inflamed and control sides and measured Egr-1 and p35 mRNA expression levels. We found that Egr-1 mRNA levels (Fig. 8C) and p35 mRNA levels (Fig. 8D) were elevated in DRGs from inflamed side more than five times compared with DRG from the control side. Together with our previous report, in which carrageenan-induced inflammation increased Cdk5 activity in DRGs (10), these results suggest that TNF-α regulates an increase in Cdk5 activity during inflammation-induced pain through induction of Egr-1 and p35 expression.
FIGURE 8.
Carrageenan-induced inflammation in mice increases TNF-α, Egr-1, and p35 mRNA levels. A, inflammation was induced by injecting carrageenan into the left hind paw of wild-type mice and the latency of paw withdrawal was measured before and 24 h after-injection with carrageenan. Carrageenan-induced inflammation increased mRNA levels of TNF-α in the inflamed paw (B), and Egr-1 (C), and p35 (D) mRNA levels in DRGs from the left side (inflamed) as compared with those from the control right side. All of the data are presented as the means and S.E. *, p < 0.05 by using t test.
DISCUSSION
Cdk5 was recently shown to be a key regulator in inflammation-induced pain signaling (10–14). We identified its direct involvement in phosphorylation of the TRPV1 receptor, which possibly regulates influx of calcium ions in heat-sensitive neurons during nociceptive signaling (14). Because of these findings and other recent reports, Cdk5 is now considered as a potential target for developing new analgesics (10–14, 39, 40). However, it is still unclear how Cdk5 activity is regulated during inflammation-induced pain signaling. It is not clear which modulators that are triggered by inflammation affect Cdk5 activity. To address these questions in the present study, we focused on the regulation of p35 expression, which is a limiting factor for Cdk5 kinase activity (5). We analyzed a group of cytokines for their effects on p35 promoter activity and found that TNF-α is a major regulator of p35 expression and subsequently of Cdk5 activity. Furthermore, we also investigated the mechanism underlying the stimulation of the p35 promoter by TNF-α. We demonstrate that through activation of the ERK1/2 pathway, TNF-α induces sustained and robust expression of p35 in PC12 cells, thereby increasing Cdk5 kinase activity. The activation of ERK1/2 by TNF-α leads to an increase of Egr-1 expression and subsequent elevation of p35 expression.
During inflammation or neuropathic pain, proinflammatory cytokines are released by activated immune cells (15, 41). In damaged peripheral nerves, as in other tissues, macrophages are recruited by chemotactic molecules and release many proinflammatory cytokines, including IL-1β, IL-6, and TNF-α among others (15–19, 41). Both mRNA and protein levels of these cytokines were elevated in the inflamed hind paw of rats injected with carrageenan (20), and we found that TNF-α mRNA levels were also elevated in the hind paws of mice injected with carrageenan. Interestingly, we also noted increased levels of Cdk5 and p35 proteins, and enhanced Cdk5 activity in the spinal cord, DRG, and trigeminal ganglia from rats injected with carrageenan in hind paws (10). To examine which cytokine regulates Cdk5 activity, we developed a cell-based assay and identified that TNF-α activates p35 promoter activity in a dose- and time-dependent manner. We also found that treatment of transfected PC12 cells with IL-6, LIF, or OSM resulted in slight inhibition of p35 promoter activity. We did not observe any significant effect on p35 promoter activity following treatment of PC12 cells with IFN-γ or IL-1β. Our findings on the effects of these cytokines on transcriptional activation of p35 differ from previous reports in which IL-6 was found to affect p35 protein levels in hippocampal neurons, and IFN-γ increased p35 protein levels in neuroblastoma Paju cells (25, 26). These disparities could be due to differences in the cell type and assay system used in these studies.
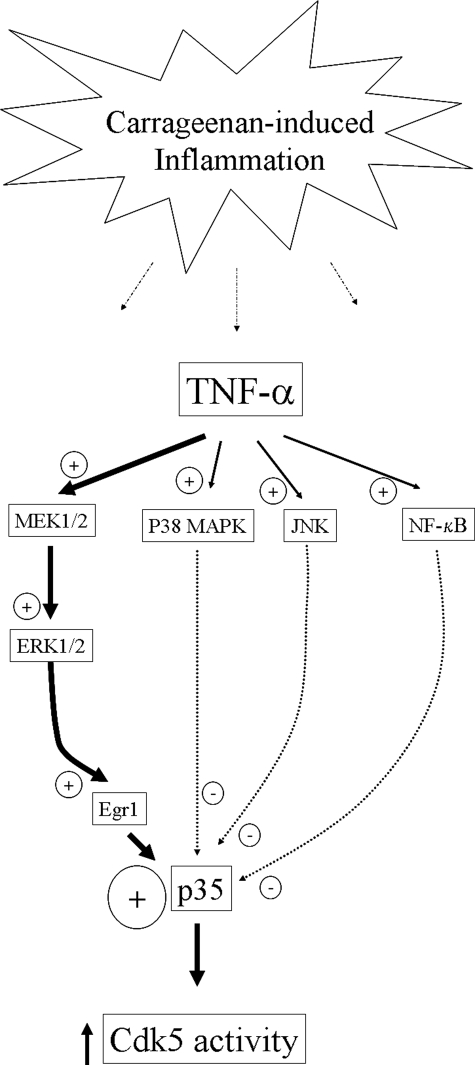
TNF-α regulates MAPKs and NF-κB signaling pathways in cell lines and primary neuronal cultures (23, 31, 33, 42–44). In this report we show that TNF-α induces activation of ERK1/2, p38 MAPK, JNK, and NF-κB signaling pathways in PC12 cells. ERK1/2 activation by TNF-α resulted in the induction of Egr-1, a transcription factor that binds to the promoter region of p35 and induces p35 expression. In addition, the increase in p35 expression in response to Egr-1 induction is consistent with previous findings that suggest involvement of ERK1/2 in regulation of Cdk5 pathways (22–24). It is also reported that p35/p25 protein levels and Cdk5 activity are reduced in Egr-1 knock-out mouse brains (45). These observations coincide well with our results that TNF-α induces an increase in p35 and Egr-1, and this increase is inhibited by PD98059. Most importantly, when we knocked down Egr-1 mRNA levels with a pool of 4 Egr-1-specific siRNA, TNF-α treatment did not result in increased levels of Egr-1 and p35 mRNA. Additionally, we showed that TNF-α treatment induced activation of p38 MAPK, JNK, and NF-κB signaling pathways in PC12 cells. In contrast to the ERK1/2 pathway, treatment with an inhibitor of the p38 MAPK pathway (SB203580), an inhibitor of JNK (SP600125), or an NF-κB inhibitor increased p35 promoter activity, suggesting a suppressive role for these pathways over p35 promoter activity. We speculate that the activation of these pathways might induce an unknown repressor for the p35 promoter or suppress the expression or activity of Egr-1. Thus, although TNF-α is able to modulate several signaling pathways, resulting in either positive or negative regulation, the cumulative result is a significant increase in p35 promoter activity with a subsequent increase in Cdk5 activity (Fig. 9). Most importantly, the MEK-ERK1/2-Egr-1 pathway seems to be the predominant pathway by which TNF-α activates the p35 promoter.
FIGURE 9.
Proposed TNF-α-mediated regulation of p35 expression and Cdk5 activity. During inflammation, TNF-α is released from immune cells recruited at the site of the injury. TNF-α binds to its receptor and activates MAPKs (ERK1/2, p38 MAPK, and JNK) and NF-κB signaling pathways. TNF-α-mediated ERK1/2 activation increases both Egr-1 mRNA and protein levels and, subsequently, p35 mRNA and protein levels. On the other hand, the activation of p38 MAPK, JNK, and NF-κB pathways inhibits p35 promoter activity. Thus, although TNF-α is able to modulate several signaling pathways, resulting in either positive or negative regulation, the end result is a significant increase in p35 promoter activity with a concomitant increase in Cdk5 activity.
TNF-α is known to be released after induction of inflammation (19, 20). Carrageenan-induced inflammation in the hind paws of mice was accompanied by an early increase in TNF-α and subsequent increase in paw volume because of edema, resulting in mechanical allodynia (17, 43) and thermal hyperalgesia. Paw volume and associated mechanical allodynia were significantly decreased following carrageenan injections to TNF receptor 1 knock-out mice or to the wild-type mice pretreated with either TNF-α antibody or with the inhibitor of TNF-α synthesis (46). Furthermore, mechanical allodynia was significantly enhanced in TNF-α-overexpressing transgenic mice compared with that of the wild-type mice, suggesting an important role for TNF-α in the pathogenesis of neuropathic pain (47). All of these observations point toward a key role for TNF-α in pain signaling during inflammation. In conclusion, we have shown that TNF-α can regulate Cdk5 activity, and this effect is mediated by MAPKs through subsequent activation of Egr-1 and p35 expression. Because TNF-α released from inflamed tissue could regulate Cdk5 activity, inhibiting TNF-α activity or its effect on p35 expression may serve as a novel therapeutic strategy for the prevention and treatment of neuropathic pain. High throughput screening of potential transcriptional regulators of p35 expression using our cell-based assay will accelerate identifications of new class of analgesics for treatment of neuropathic pain.
Acknowledgments
We thank Drs. Toshio Ohshima and Tej K. Pareek for helpful discussions.
This work was supported, in whole or in part, by the National Institutes of Health, Division of Intramural Research of NIDCR and NINDS. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Cdk, cyclin-dependent kinase; TRPV, transient receptor potential vanilloid; IL, interleukin; TNF, tumor necrosis factor; ERK, extracellular-signal regulated kinase; Egr, early gene response; LIF, leukemia inhibitory factor; OSM, oncostatin M; IFN, interferon; MAPK, mitogen-activated protein kinase; MEK, mitogen-activated protein kinase/ERK kinase; JNK, c-Jun N-terminal kinase; DRG, dorsal root ganglia; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siRNA, small interference RNA; ANOVA, analysis of variance.
References
- 1.Dhavan, R., and Tsai, L. H. (2001) Nat. Rev. Mol. Cell. Biol. 2 749-759 [DOI] [PubMed] [Google Scholar]
- 2.Tsai, L. H., Delalle, I., Caviness, V. S., Jr., Chae, T., and Harlow, E. (1994) Nature 371 419-423 [DOI] [PubMed] [Google Scholar]
- 3.Lew, J., Huang, Q. Q., Qi, Z., Winkfein, R. J., Aebersold, R., Hunt, T., and Wang, J. H. (1994) Nature 371 423-426 [DOI] [PubMed] [Google Scholar]
- 4.Humbert, S., Lanier, L. M., and Tsai, L. H. (2000) Neuroreport 11 2213-2216 [DOI] [PubMed] [Google Scholar]
- 5.Takahashi, S., Ohshima, T., Cho, A., Sreenath, T., Iadarola, M. J., Pant, H. C., Kim, Y., Nairn, A. C., Brady, R. O., Greengard, P., and Kulkarni, A. B. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 1737-1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhariwala, F. A., and Rajadhyaksha, M. S. (2008) Cell. Mol. Neurobiol. 28 351-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohshima, T., Ward, J. M., Huh, C. G., Longenecker, G., Veeranna, Pant, H. C., Brady, R. O., Martin, L. J., and Kulkarni, A. B. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 11173-11178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maccioni, R. B., Otth, C., Concha, II, and Munoz, J. P. (2001) Eur. J. Biochem. 268 1518-1527 [DOI] [PubMed] [Google Scholar]
- 9.Cruz, J. C., and Tsai, L. H. (2004) Trends. Mol. Med. 10 452-458 [DOI] [PubMed] [Google Scholar]
- 10.Pareek, T. K., Keller, J., Kesavapany, S., Pant, H. C., Iadarola, M. J., Brady, R. O., and Kulkarni, A. B. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 791-796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pareek, T. K., and Kulkarni, A. B. (2006) Cell Cycle 5 585-588 [DOI] [PubMed] [Google Scholar]
- 12.Saikkonen, B., Pareek, T. K., Agarwal, N., Molinolo, A., Kriete, M., and Kulkarni, A. B. (2007) Cell Cycle 7 750-753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pareek, T. K., and Kulkarni, A. B. (2008) in Cyclin Dependent Kinase 5 (Cdk5) (Tsai, L. H., and Ip, N., eds) pp. 211-226, Springer, New York
- 14.Pareek, T. K., Keller, J., Kesavapany, S., Agarwal, N., Kuner, R., Pant, H. C., Iadarola, M. J., Brady, R. O., and Kulkarni, A. B. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 660-665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers, R. R., Campana, W. M., and Shubayev, V. I. (2006) Drug Discov. Today 11 8-20 [DOI] [PubMed] [Google Scholar]
- 16.Schafers, M., Sorkin, L. S., Geis, C., and Shubayev, V. I. (2003) Neurosci. Lett. 347 179-182 [DOI] [PubMed] [Google Scholar]
- 17.Arruda, J. L., Colburn, R. W., Rickman, A. J., Rutkowski, M. D., and DeLeo, J. A. (1998) Brain Res. Mol. Brain Res. 62 228-235 [DOI] [PubMed] [Google Scholar]
- 18.Safieh-Garabedian, B., Poole, S., Allchorne, A., Winter, J., and Woolf, C. J. (1995) Br. J. Pharmacol. 115 1265-1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loram, L. C., Fuller, A., Fick, L. G., Cartmell, T., Poole, S., and Mitchell, D. (2007) J. Pain 8 127-136 [DOI] [PubMed] [Google Scholar]
- 20.Yang, H. Y., Mitchell, K., Keller, J. M., and Iadarola, M. J. (2007) J. Neurochem. 103 1628-1643 [DOI] [PubMed] [Google Scholar]
- 21.Cheng, B., Chistakos, S., and Mattson, M. P. (1994) Neuron 12 139-153 [DOI] [PubMed] [Google Scholar]
- 22.Baud, V., and Karin, M. (2001) Trends Cell Biol. 11 372-377 [DOI] [PubMed] [Google Scholar]
- 23.Barbin, G., Roisin, M. P., and Zalc, B. (2001) Neurochem. Res. 26 107-112 [DOI] [PubMed] [Google Scholar]
- 24.Harada, T., Morooka, T., Ogawa, S., and Nishida, E. (2001) Nat. Cell Biol. 3 453-459 [DOI] [PubMed] [Google Scholar]
- 25.Quintanilla, R. A., Orellana, D. I., Gonzalez-Billault, C., and Maccioni, R. B. (2004) Exp. Cell Res. 295 245-257 [DOI] [PubMed] [Google Scholar]
- 26.Song, J. H., Wang, C. X., Song, D. K., Wang, P., Shuaib, A., and Hao, C. (2005) J. Biol. Chem. 280 12896-12901 [DOI] [PubMed] [Google Scholar]
- 27.Ohshima, T., Kozak, C. A., Nagle, J. W., Pant, H. C., Brady, R. O., and Kulkarni, A. B. (1996) Genomics 35 372-375 [DOI] [PubMed] [Google Scholar]
- 28.Hawasli, A. H., Benavides, D. R., Nguyen, C., Kansy, J. W., Hayashi, K., Chambon, P., Greengard, P., Powell, C. M., Cooper, D. C., and Bibb, J. A. (2007) Nat. Neurosci. 10 880-886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marz, P., Gadient, R. A., and Otten, U. (1996) Brain Res. 706 71-79 [DOI] [PubMed] [Google Scholar]
- 30.Cheshire, J. L., and Baldwin, A. S., Jr. (1997) Mol. Cell. Biol. 17 6746-6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mielke, K., and Herdegen, T. (2002) Mol. Cell. Neurosci. 20 211-224 [DOI] [PubMed] [Google Scholar]
- 32.Shu, H. F., Wang, B. R., Bi, H., Pei, J. M., Wang, X., Fan, J., and Ju, G. (2007) Respir. Physiol. Neurobiol. 157 187-195 [DOI] [PubMed] [Google Scholar]
- 33.Takahashi, N., Kikuchi, S., Shubayev, V. I., Campana, W. M., and Myers, R. R. (2006) Spine 31 523-529 [DOI] [PubMed] [Google Scholar]
- 34.Pang, L., Sawada, T., Decker, S. J., and Saltiel, A. R. (1995) J. Biol. Chem. 270 13585-13588 [DOI] [PubMed] [Google Scholar]
- 35.Cuenda, A., Rouse, J., Doza, Y. N., Meier, R., Cohen, P., Gallagher, T. F., Young, P. R., and Lee, J. C. (1995) FEBS Lett. 364 229-233 [DOI] [PubMed] [Google Scholar]
- 36.Bennett, B. L., Sasaki, D. T., Murray, B. W., O'leary, E. C., Sakata, S. T., Xu, W., Leisten, J. C., Motiwala, A., Pierce, S. A., Satoh, Y., Bhagwat, S. S., Manning, A. M., and Anderson, D. W. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13681-13686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tobe, M., Isobe, Y., Tomizawa, H., Nagasaki, T., Takahashi, H., Fukazawa, T., and Hayashi, H. (2003) Bioorg. Med. Chem. 11 383-391 [DOI] [PubMed] [Google Scholar]
- 38.Hodge, C., Liao, J., Stofega, M., Guan, K., Carter-Su, C., and Schwartz, J. (1998) J. Biol. Chem. 273 31327-31336 [DOI] [PubMed] [Google Scholar]
- 39.Glicksman, M. A., Cuny, G. D., Liu, M., Dobson, B., Auerbach, K., Stein, R. L., and Kosik, K. S. (2007) Curr. Alzheimer Res. 4 547-549 [DOI] [PubMed] [Google Scholar]
- 40.Yang, Y. R., He, Y., Zhang, Y., Li, Y., Li, Y., Han, Y., Zhu, H., and Wang, Y. (2007) Pain 127 109-120 [DOI] [PubMed] [Google Scholar]
- 41.Zhang, J. H., and Huang, Y. G. (2006) Chin. Med. J. (Engl.) 119 930-938 [PubMed] [Google Scholar]
- 42.Lee, H. J., Cho, J. W., Kim, S. C., Kang, K. H., Lee, S. K., Pi, S. H., Lee, S. K., and Kim, E. C. (2006) Cytokine 35 67-76 [DOI] [PubMed] [Google Scholar]
- 43.Ulfhammer, E., Larsson, P., Karlsson, L., Hafnkelsdóttir, T., Bokarewa, M., Tarkowski, A., and Jern, S. (2006) J. Thomb. Haemostasis 4 1781-1789 [DOI] [PubMed] [Google Scholar]
- 44.Friedrichsen, S., Harper, C. V., Semprini, S., Wilding, M., Adamson, A. D., Spiller, D. G., Nelson, G., Mullins, J. J., White, M. R., and Davis, J. R. (2006) Endocrinology 147 773-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, T., Chalifour, L. E., and Paudel, H. K. (2007) J. Biol. Chem. 282 6619-6628 [DOI] [PubMed] [Google Scholar]
- 46.Rocha, A. C., Fernandes, E. S., Quintao, N. L., Campos, M. M., and Calixto, J. B. (2006) Br. J. Pharmacol. 148 688-695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeLeo, J. A., Rutkowski, M. D., Stalder, A. K., and Campbell, I. L. (2000) Neuroreport 11 599-602 [DOI] [PubMed] [Google Scholar]