Abstract
Choline is an essential nutrient that, via its metabolite betaine, serves as a donor of methyl groups used in fetal development to establish the epigenetic DNA and histone methylation patterns. Supplementation with choline during embryonic days (E) 11–17 in rats improves memory performance in adulthood and protects against age-related memory decline, whereas choline deficiency impairs certain cognitive functions. We previously reported that global and gene-specific DNA methylation increased in choline-deficient fetal brain and liver, and these changes in DNA methylation correlated with an apparently compensatory up-regulation of the expression of DNA methyltransferase Dnmt1. In the current study, pregnant rats were fed a diet containing varying amounts of choline (mmol/kg: 0 (deficient), 8 (control), or 36 (supplemented)) during E11–17, and indices of histone methylation were assessed in liver and frontal cortex on E17. The mRNA and protein expression of histone methyltransferases G9a and Suv39h1 were directly related to the availability of choline. DNA methylation of the G9a and Suv39h1 genes was up-regulated by choline deficiency, suggesting that the expression of these enzymes is under negative control by methylation of their genes. The levels of H3K9Me2 and H3K27Me3, tags of transcriptionally repressed chromatin, were up-regulated by choline supplementation, whereas the levels of H3K4Me2, associated with active promoters, were highest in choline-deficient rats. These data show that maternal choline supply during pregnancy modifies fetal histone and DNA methylation, suggesting that a concerted epigenomic mechanism contributes to the long term developmental effects of varied choline intake in utero.
Choline is an essential nutrient whose adequate supply in maternal diet during pregnancy is vital for normal development of the fetus (1), and studies in humans (2) and animals (3–6) indicate that its intake during gestation is especially important for the normal development and function of the central nervous system. For example, in a model that employs offspring of pregnant rats or mice consuming diets of varying choline content during only a 1-week period of the second half of gestation (E11–17),2 choline deficiency causes impairments in certain memory tasks (7), whereas choline supplementation improves memory and attention (7–14) and, remarkably, prevents age-related memory decline (3, 5, 14). In addition to behavioral changes, altered choline availability modifies fetal hippocampal cell proliferation (15, 16), apoptosis (17), and differentiation (18, 19). High E11–17 choline intake increases the size of cholinergic neurons in the adult basal forebrain (20) and enhances acetylcholine storage (14) and release (21), elevates brain concentrations of the neurotrophins nerve growth factor, brain-derived neurotrophic factor, and NT3 (22–24) and of IGF2 and its receptor IGF2R (25), lowers the stimulation threshold for induction of long term potentiation (26, 27), and enhances depolarization-induced mitogen-activated protein kinase (MAPK) activation and cAMP-response element-binding protein (CREB) phosphorylation (8) in postnatal hippocampus. Moreover, prenatally choline-supplemented animals are characterized by altered patterns of hippocampal and cerebral cortical gene expression during postnatal development and in adulthood (28) and have augmented adult hippocampal neurogenesis (23).
Choline serves as a metabolic precursor of certain phospholipids (e.g. phosphatidylcholine) and of the neurotransmitter, acetylcholine (3). However, its role as a methyl group donor may be key to understanding the means by which this nutrient causes the behavioral, neuroanatomical, and electrophysiological alterations observed in postnatal life following prenatal supplementation and deficiency. Choline becomes a source of methyl groups for enzymatic methylations after choline dehydrogenase-catalyzed oxidation to betaine (29). The latter compound can be used for the conversion of homocysteine to methionine and subsequently S-adenosylmethionine (AdoMet) (29) that, in turn, is the methyl group donor for most biological methylation reactions including the methylation of cytidines in CpG dinucleotide sequences of DNA and the methylation of lysine and arginine residues of histones (30).
We previously reported that AdoMet levels increased in liver and cerebral cortex of E17 fetuses whose mothers consumed a choline-supplemented diet since E11 (31). Surprisingly, however, global DNA methylation and methylation of a selected genomic locus known to regulate transcription, i.e. the DMR2 region of the insulin-like growth factor 2 (Igf2) gene, increased in choline-deficient animals. Remarkably, these changes in DNA methylation correlated with an apparently compensatory up-regulation of the expression of DNA methyltransferase 1 (Dnmt1) (31). Moreover, mRNA expression in brain and liver of another DNA methyltransferase, Dnmt3a, and of methyl CpG-binding domain 2 (Mbd2) protein as well as cerebral DNA methyltransferase, Dnmt3l, was inversely correlated to the intake of choline (31). Other investigators also reported that altered choline intake leads to changes in global and gene-specific DNA methylation via alterations in methyl group availability (32–34), and in mouse models, gestational consumption of diets highly enriched in compounds that serve as metabolic methyl group donors and cofactors (choline, betaine, methionine, folic acid, and vitamin B12) influences the phenotype of offspring that correlates with hypermethylation of the relevant genes (35–37).
The methylation of histone tails at specific lysine and arginine residues is essential for the epigenetic regulation of transcription, cell division, and the formation of heterochromatin (38–40). The addition of methyl groups to histones exerts different effects depending on which residue is methylated. Methylation of lysine 4 on histone 3 (H3K4) is associated with transcriptionally active genes (41–43), whereas methylation of lysine 9 and lysine 27 on histone 3 (H3K9 and H3K27, respectively) correlates with transcriptional repression (44, 45). Furthermore, the degree of methylation at certain residues results in distinct effects on chromatin state. The addition of two methyl groups to the ε-amino group of H3K9 is a hallmark of transcriptional repression in euchromatic regions during development (46), whereas trimethylation of the same residue is associated with pericentric heterochromatin (45, 47). The enzymes that catalyze these modifications also differ, with the histone methyltransferase G9a (KMT1C, EHMT2) responsible for dimethylation of H3K9 to H3K9Me2 and for trimethylation of H3K27 to H3K27Me3 (48), and histone methyltransferase Suv39h1 (KMT1A) responsible for trimethylation at H3K9 to H3K9Me3 (49).
Therefore, in addition to modulating DNA methylation, choline supply could affect the methylation of amino acid residues on histone tails (50, 51), leading to alterations in the expression of genes involved in growth and development. We examined several components of the histone 3 methylating machinery in E17 liver and cerebral cortex of rat fetuses derived from mothers consuming varying amounts of choline. The methylation of H3K9 and H3K27 and expression of G9a and Suv39h1 were directly related to the availability of choline. Consistent with our previous studies, DNA methylation of the G9a and Suv39h1 genes was dramatically up-regulated by choline deficiency. The latter finding points to the possibility that the expression of these histone methyltransferases is under negative control of methylation of their genes.
EXPERIMENTAL PROCEDURES
Dietary Intervention during Embryonic Days E11–E17—Several cohorts of pregnant Sprague-Dawley rats were divided into three groups of four animals per dietary group and fed a choline-supplemented, control, or choline-deficient diet during E11–17 of gestation (AIN-76A Rodent Purified Diet (Dyets Inc.)). Unlike commercial rat chows whose choline content is not controlled for, the AIN-76A diet was formulated to permit standardized studies using nutritionally adequate diet in rats and mice (52, 53). Choline was supplied in the food (as choline chloride) such that the choline-supplemented diet had 36 mmol/kg of choline, the control diet had 8 mmol/kg of choline, and the deficient diet had 0 mmol/kg of choline. The consumption of the choline-deficient diet during this period of pregnancy causes an over 50% reduction of the maternal choline and phosphocholine pools (54). The animals were housed individually with a 12-h light/dark cycle. The average litter size was not influenced by the diet (deficient, 12.4; control, 12.1; supplemented, 12.1 fetuses per dam). All animal procedures were performed in accordance with protocols approved by the Boston University School of Medicine Institutional Animal Care and Use Committee.
Dissection of the Embryonic Liver and Frontal Cortex—The pregnant dams were euthanized on E17 following anesthesia with CO2, and the uteri were dissected and placed in L15 medium (Invitrogen) on ice. The fronto-parietal cortex was dissected from the embryos, keeping the tissue on ice to minimize postmortem changes. Because at E17 the frontal cortex is not yet fully differentiated, the whole fronto-parietal cortex was taken, excluding the olfactory bulbs. The cortices from four embryos per mother were collected, pooled, and used in all subsequent experiments. For liver, a part of the right lobe of the embryonic liver was dissected. The livers of eight embryos per mother were collected, pooled, and used in all subsequent experiments. There was no effect of the diet on the average weight of the liver or brain (data not shown). Data are derived from material (DNA, RNA, and protein) obtained from four pregnant animals per dietary group.
Genomic DNA and Total RNA Isolation—For DNA, the tissue was frozen immediately on dry ice and stored at –70 °C until extraction. To obtain high yield pure genomic DNA from the liver, the Genomic tip 100/G kit (Qiagen) was used following the manufacturer's protocol. For frontal cortex, the DNeasy blood and tissue kit (Qiagen) was used following the manufacturer's protocol. The concentration of DNA was quantified via spectrophotometry at 260 nm.
For RNA, the tissue was homogenized immediately using a needle and syringe in ice-cold 4 m guanidine isothiocyanate solution, pH 7.0, containing 100 mm β-mercaptoethanol and 25 mm sodium citrate, placed on dry ice, and then stored at –70 °C. RNA was extracted using the phenol/chloroform method (55), precipitated with ethanol, and resuspended in nuclease-free water (Ambion) and quantified with the RiboGreen RNA quantitation reagent and kit (Molecular Probes) using a Victor3 multilabel plate reader (PerkinElmer Life Sciences).
Reverse Transcriptase-PCR—RNA was analyzed by reverse transcriptase PCR using Superscript One-Step RT-PCR with Platinum Taq (Invitrogen) according to the manufacturer's instructions. First-strand cDNA synthesis was performed with 10 ng of RNA, oligo(dT) primer and reverse transcriptase at 48 °C for 45 min. The following primer sets were used: G9a, forward, 5′-CAA GGA TGG TGA GGT CTA CTG C-3′; reverse, 5′-GCT CTT GAT ATC CCA GAA CCG-3′; Suv39h1, forward, 5′-ATC CCT GCA CAA GTT TGC C-3′; reverse, 5′-TTT GCG GAT CTT TTC CAG C-3′; β-actin, forward, 5′-CAC AGC TGA GAG GGA AAT C-3′; reverse, 5′-TCA GCA ATG CCT GGG TAC-3′. PCR was performed with a denaturing step for 2 min at 94 °C followed by 34 cycles of 1 min at 94 °C, 1 min at 58 °C, and 2 min at 70 °C and terminated by an elongation step at 72 °C for 7 min. The conditions of the reactions were determined to be in the linear range of the assay. The products were resolved on 10% Tris-buffered EDTA polyacrylamide gels and stained with ethidium bromide. The intensity of each band was quantified with a Kodak Image Station using Kodak ID software. The expression level was calculated as the percentage of the control after normalizing to β-actin.
Immunoblotting—Tissue extracts from E17 cortex and liver were prepared for Western blotting by adding lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 10% glycerol, 2 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride, 1 μg/ml leupeptin, 2 μg/ml aprotinin, 2 μg/ml pepstatin) to tissue followed by sonication and centrifugation. Protein concentration was determined by the BCA assay. Proteins (25–30 μg) were resolved on NuPAGE Novex 4–12% bis-Tris Midi gels in 1× NuPAGE MOPS-SDS running buffer (Invitrogen) and transferred to a nitrocellulose membrane using an iBLOT apparatus (Invitrogen). Membranes were blocked with 5% nonfat dry milk in 1× Tris-buffered saline containing 0.1% Tween 20 for 1 h and then probed with primary antibody overnight. The primary antibodies used were: polyclonal anti-H3 (AbCam) diluted 1:2000, anti-H3K4Me2 (AbCam) diluted 1:3000, and anti-G9a (AbCam) at 4 μg/ml, monoclonal anti-H3K9Me2 (AbCam) at 3 μg/ml, anti-H3K27Me3 (AbCam) at 3 μg/ml, anti-SUV39H1 (AbCam) at 4 μg/ml, and anti-β-actin (Sigma) diluted 1:2000. After treatment with primary antibody, the membranes were incubated in either anti-rabbit or anti-mouse horseradish peroxidase-conjugated secondary antibodies (Bio-Rad Laboratories) and detected after brief exposure to chemiluminescence reagents using a Kodak Image Station with Kodak ID software.
Methylation-specific PCR—One microgram of DNA from E17 liver and from frontal cortex was treated with sodium bisulfite using the EZ DNA methylation kit (Zymo Research) and analyzed by methylation-specific PCR to assay the methylation status of CpG islands located in the promoter regions of G9a and Suv39h1. Primers were designed using MethPrimer (56) to be specific for methylated or unmethylated CpGs in the template. For G9a, the forward primers spanned two CpGs, and the reverse primers contained three CpGs. The primer sequences were: methylated forward, 5′-GTT GTT GTT GGA GAA GGA GTT TC-3′; methylated reverse, 5′-GAC GAT AAC AAT AAC AAA AAC CGA-3′; unmethylated forward, 5′-TTG TTG GAG AAG GAG TTT TGA-3′; unmethylated reverse, 5′-AAC AAT AAC AAT AAC AAA AAC CAA A-3′. For Suv39h1, the forward primers spanned four CpGs within the promoter, and the reverse primers contained three CpGs. The primer sequences were: methylated forward, 5′-AGT AGC GAG TAT CGG CGT TC-3′; methylated reverse, 5′-GAA CCT AAA AAA CCT CGC GA-3′; unmethylated forward, 5′-TAA AAG TAG TGA GTA TTG GTG TTT GA-3′; unmethylated reverse, 5′-AAA CCT AAA AAA CCT CGC GA-3′. Each reaction was performed with 60 ng of template. The reaction conditions were: one cycle of 94 °C for 2 min, 1 min at annealing temperature, and 2 min at 70 °C, then 37 cycles of denaturing at 94 °C for 30 s, annealing for 40 s, and elongation at 72 °C for 1 min followed by one cycle of 70 °C for 10 min. The annealing temperatures were: G9a, methylated and unmethylated, 55.0 °C; Suv39h1, methylated, 59.0 °C, and unmethylated, 55.8 °C. The conditions of the reactions were determined to be in the linear range of the assay. The PCR products were resolved on 10% Tris-buffered EDTA polyacrylamide gels and stained with ethidium bromide. The intensity of each band was quantified with a Kodak Image Station using Kodak ID software. The methylation level was calculated using the ratio of the intensity of the methylated product divided by the unmethylated product.
Statistical Analysis—The data were analyzed by analysis of variance. The significant effects (p < 0.05) were further analyzed by the Tukey multiple comparison test using SYSTAT software (Systat Software Inc.). Data are presented as mean ± S.E.
RESULTS
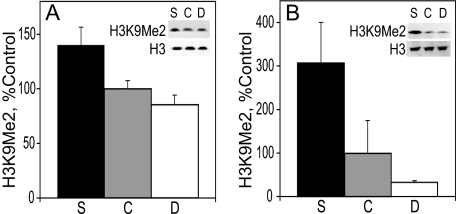
H3K9Me2 Levels in E17 Liver and Cortex—H3K9Me2 was analyzed by immunoblot, and its levels were normalized to those of total histone 3. The latter were not affected by the maternal diet (data not shown). H3K9Me2 is generally associated with transcriptional repression in euchromatic regions (48). In both liver and cortex, the level of H3K9Me2 increased with maternal choline intake. The level of H3K9Me2 in the liver of E17 embryos (Fig. 1A) from pregnant rats fed a choline-supplemented diet was 40% higher than in controls and 60% higher than in choline-deficient embryos (p < 0.04 and p < 0.02, respectively). The control and the choline-deficient animals had similar amounts of H3K9Me2. In E17 cortex (Fig. 1B), the amount of H3K9Me2 in choline-supplemented embryos was 9-fold higher than in choline-deficient embryos (p < 0.05). The levels of H3K9Me2 in the control group were in between the other two groups and were not statistically different from either one.
FIGURE 1.
H3K9Me2 in the liver (A) and cortex (B). H3K9Me2 protein levels on E17 were analyzed by immunoblot and normalized to total histone 3 (see example in blot insets). See “Results” for statistics. Bar and blot labels: S, choline-supplemented; C, controls; D, choline-deficient.
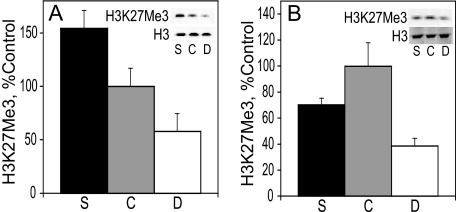
H3K27Me3 Levels in E17 Liver and Cortex—Like dimethylation of lysine 9, trimethylation of lysine 27 on histone 3 also facilitates transcriptional repression (44). In the liver of E17 embryos (Fig. 2A), H3K27Me3 was 2.7-fold higher in the choline-supplemented group than in the choline-deficient one (p < 0.008) with the control group exhibiting intermediate levels of H3K27Me3. In the frontal cortex, the levels of H3K27Me3 were similar in the control and the choline-supplemented groups, although the former tended to be higher. The levels of H3K27Me3 were significantly lower (by 60%) in the choline-deficient rats when compared with the controls (p < 0.01).
FIGURE 2.
H3K27Me3 in the liver (A) and cortex (B). H3K27Me3 protein levels were analyzed on E17 by immunoblot and normalized to total histone 3 (see example in blot insets). See “Results” for statistics. Bar and blot labels: S, choline-supplemented; C, controls; D, choline-deficient.
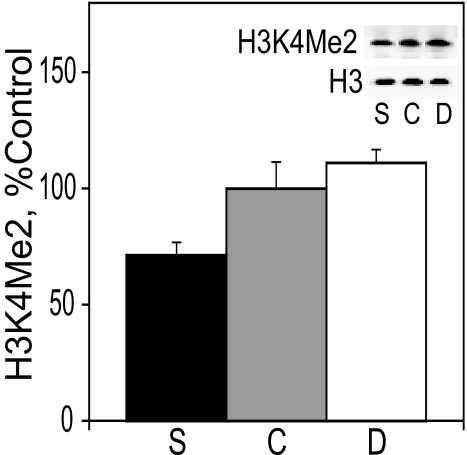
H3K4Me2 Levels in E17 Liver—Although H3K9Me2 and H3K27Me3 are hallmarks of transcriptionally repressed chromatin, histone 3 dimethylated on lysine 4 is present at the promoters of active genes (42). In contrast to the direct relationship between dietary choline intake and the modification of histone 3 associated with transcriptional repression (Figs. 1 and 2), the hepatic levels of H3K4Me2 were inversely related to the intake of choline. Specifically, the level of H3K4Me2 in the liver of E17 embryos (Fig. 3) from rats fed a choline-supplemented diet was 55% lower than in choline-deficient embryos (p < 0.05). We did not measure the levels of H3K4Me2 in cortex.
FIGURE 3.
H3K4Me2 in the liver. H3K4Me2 protein levels in E17 liver were analyzed by immunoblot and normalized to total histone 3 (see example in blot insets). See “Results” for statistics. Bar and blot labels: S, choline-supplemented; C, controls; D, choline-deficient.
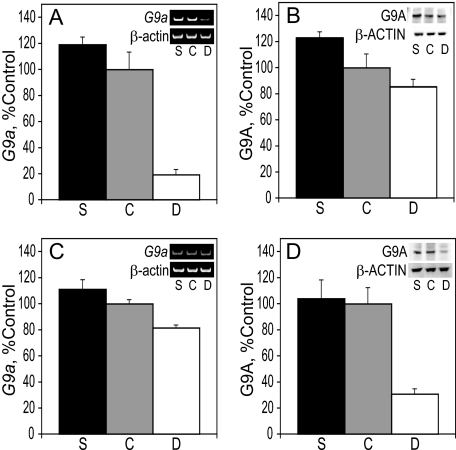
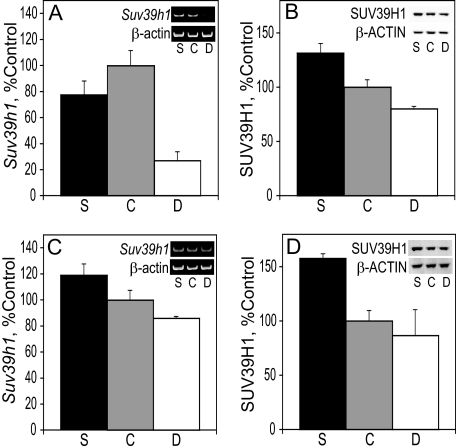
G9a Methyltransferase Levels in E17 Liver and Cortex—G9a (KMT1C) is a principal methyltransferase of H3K9 that catalyzes the addition of two methyl groups to this substrate in vivo (46). Reverse transcriptase-PCR and immunoblot were used to analyze the effects of maternal choline intake on the levels of G9a mRNA and protein, respectively, in the liver and cortex at E17. In both tissues, the levels of the G9a message and protein were directly related to the maternal choline intake. In the liver, G9a mRNA levels in choline-deficient rats was reduced to 18 and 15% of those found in choline-supplemented and control embryos, respectively (p < 0.0004 for both) (Fig. 4A). In the cortex, a 37% increase in G9a mRNA in choline-supplemented embryos was observed when compared with choline-deficient ones (p < 0.004) (Fig. 4C). The amount of G9A protein present in E17 liver and frontal cortex was consistent with the RT-PCR data. G9A in the liver of choline-supplemented embryos was 44% higher than in choline-deficient animals (p < 0.04) (Fig. 4B), and in the cortex, G9A protein levels in choline-deficient rats was reduced to ∼30% of those found in choline-supplemented and control embryos (p < 0.005 for both) (Fig. 4D).
FIGURE 4.
G9a in the liver and cortex. A, RNA was isolated from the liver of E17 embryos and used for RT-PCR of G9a and normalized to β-actin (see example in gel insets). B, G9A protein levels in E17 liver were analyzed by immunoblot and normalized to β-ACTIN (see example in blot insets). C, RNA was isolated from the cortex of E17 embryos and used for RT-PCR of G9a and normalized to β-actin (see example in gel insets). D, G9A protein levels in E17 cortex were analyzed by immunoblot and normalized to the levels ofβ-ACTIN (see example in blot insets). See “Results” for statistics. Bar labels: S, choline-supplemented; C, controls; D, choline-deficient.
Suv39h1 Methyltransferase Levels in E17 Liver and Cortex— The histone methyltransferase Suv39h1 (KMT1A) also catalyzes the methylation of H3K9 and thus contributes to transcriptional repression. Unlike G9a that tends to synthesize the H3K9Me2 product, Suv39h1 is reportedly responsible for the generation of H3K9Me3 (57). Reverse transcriptase-PCR was used to analyze the effects of maternal choline intake on the levels of Suv39h1 mRNA in the liver and frontal cortex at E17. In the liver of choline-deficient fetuses, Suv39h1 mRNA levels were one-fourth of those in the controls and one-third of those found in choline-supplemented rats (p < 0.001 and p < 0.01, respectively) (Fig. 5A). The latter two groups had similar levels of Suv39h1 mRNA (Fig. 5A). In the cortex, Suv39h1 mRNA expression was 40% higher in choline-supplemented fetuses than in choline-deficient animals (p < 0.015) (Fig. 5C). Consistent with the RT-PCR data, SUV39H1 protein levels were directly related to the intake of choline in both liver and cortex. In the liver, SUV39H1 protein levels were higher in choline-supplemented embryos by 31% when compared with controls and by 64% when compared with choline-deficient rats (p < 0.01 and p < 0.0004, respectively) (Fig. 5B). In the cortex, SUV39H1 protein was 1.8-fold higher in supplemented embryos relative to the deficient group (p < 0.0004) and 58% higher than in the controls (p < 0.001) (Fig. 5D).
FIGURE 5.
Suv39h1 in the liver and cortex. A, RNA was isolated from the liver of E17 embryos and used for RT-PCR of Suv39h1 and normalized to β-actin (see example in gel insets). B, SUV39H1 protein levels in E17 liver were analyzed by immunoblot and normalized to β-ACTIN (see example in blot insets). C, RNA was isolated from the cortex of E17 embryos and used for RT-PCR of Suv39h1 and normalized to β-actin (see example in gel insets). D, SUV39H1 protein levels in E17 cortex were analyzed by immunoblot and normalized to the levels of β-ACTIN (see example in blot insets). See “Results” for statistics. Bar labels: S, choline-supplemented; C, controls; D, choline-deficient.
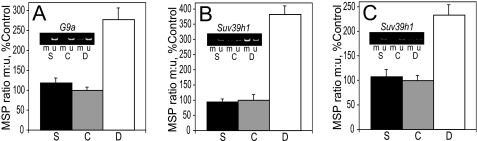
Methylation-specific PCR Analysis of G9a and Suv39h1 Promoter CpG Islands—Methylation-specific PCR was used to assay the methylation status of several CpGs within the promoter regions of G9a and Suv39h1 to examine the possibility that changes in the expression of these enzymes evoked by maternal choline might be mediated by altered DNA methylation. In E17 liver, the ratio of methylated to unmethylated template in the G9a promoter was 2.7-fold higher in choline-deficient fetuses when compared with controls (p < 0.0004) and 2.3-fold higher when compared with choline-supplemented rats (p < 0.0006) (Fig. 6A). There was no effect of the diet on the G9a promoter methylation in cortex (data not shown). In the liver (Fig. 6B), the ratio of methylated to unmethylated template in the Suv39h1 promoter was 3.8-fold higher in choline-deficient animals when compared with both control and supplemented rats (p < 0.0002). In the Suv39h1 promoter of E17 cortex (Fig. 6C), the ratio of methylated to unmethylated template was 2.3-fold higher in the choline-deficient embryos when compared with controls and 2.2-fold higher than choline-supplemented rats (p < 0.0011 and p < 0.0014, respectively).
FIGURE 6.
Methylation-specific PCR (MSP) analysis of G9a and Suv39h1 promoter CpG islands. A–C, genomic DNA isolated from E17 liver (A and B) or cortex (C) was treated with sodium bisulfite and analyzed by methylation-specific PCR. The inset shows examples of the PCR products obtained using the primers designed to amplify the methylated (m) and unmethylated (u) DNA templates. See “Results” for statistics. Bar and PCR product labels: S, choline-supplemented; C, controls; D, choline-deficient.
DISCUSSION
In mammals, one-carbon metabolism is dependent on the dietary supply of methyl group donors and cofactors: methionine, choline, folate, and vitamin B12 (58), and our previous studies showed that increased dietary choline consumption by pregnant rats elevates fetal hepatic and brain concentrations of AdoMet, the methyl-group-donating substrate of histone methyltransferases (31). Recent studies have shown that dietary methyl group deficiency in adult rats and mice leads to changes in the levels of methylated histones (51, 59–61). Here we found that gestational choline availability also affects histone methylation in the developing embryo. The levels of H3K9Me2 and H3K27Me3 in fetal liver and cerebral cortex were directly related to choline intake, and moreover, the changes in H3K9 methylation correlated with similar alterations in the expression of mRNA and protein of two histone methyltransferases, G9a and SUV39H1, that catalyze the addition of methyl groups to lysine 9 on histone 3. Thus, it appears that regulation of H3K9 methylation by choline supply may be mediated by two mechanisms that include parallel changes in the protein amounts of the methyltransferase enzymes and concentrations of their substrate, AdoMet. In contrast to the amounts of H3K9Me2 and H3K27Me3, the levels of hepatic H3K4Me2 varied inversely to choline intake. Dimethylation of H3K9 and trimethylation of H3K27 are associated with transcriptional repression (44, 48), whereas H3K4Me2 has been linked to transcriptional activation (42). Taken together, the observed pattern of histone methylation may suggest that increased prenatal choline availability leads to a generalized less transcriptionally permissive chromatin state than does choline deficiency. However, it is more likely that the chromatin changes are quite specific. Future studies utilizing chromatin immunoprecipitation techniques using antibodies specific to variously methylated histone 3 will help to resolve this question.
Our previous studies showed an apparently adaptive response of the fetus to the short E11–E17 period of altered dietary choline intake by the pregnant mother, which included changes in hepatic and cerebral cortical DNA methylation and modified expression of multiple genes, including those encoding DNA methyltransferases Dnmt1 and Dnmt3a as well as regulators of DNA methylation, Dnmt3l and Mbd2 (31). In those studies, we observed that in liver of E17 animals, choline deficiency caused a global- and gene-specific hypermethylation, likely caused by a compensatory induction of Dnmt1, and that the latter correlated with the hypomethylation of a CpG in its regulatory region (62) observed in the choline-deficient fetuses. In a recent study Pogribny et al. (61) found that a 36-week-period of methyl group deficiency (choline- and folate-free diet containing 0.18% methionine) in adult Fischer 344 rats caused epigenomic DNA and protein alterations in brain, including generalized DNA hypermethylation, hypomethylation at H3K9Me3 and H3K27Me3, and overexpression of DNMT3A. Moreover, these authors observed reduced levels of the SUV39H1 protein in brains of these animals and an increase in the levels of methyl-CpG-binding protein 2. Thus, based on the previous studies (31, 61) and the data presented here, it appears that there is a highly coordinated response to methyl group deprivation that is similar in the prenatal and adult animal. The current results confirm and extend our previous observations and show that the promoters of G9a and Suv39h1 were dramatically hypermethylated in choline-deficient embryos. At present, it is not known whether methylation of these promoter regions regulates the expression of G9a and Suv39h1. However, our data showing a relatively low expression of these genes in choline-deficient rats (that are characterized by DNA hypermethylation) are consistent with the notion that DNA methylation within regulatory regions of most genes reduces their expression (63–66). Thus, it is now possible to suggest a model whereby the supply of choline in utero causes multiple epigenomic adaptations. Specifically, when choline is in short supply, DNMT1 and DNMT3A expression is high, leading to hypermethylation of DNA and resulting in reduced expression of G9a and SUV39H1, causing low abundance of H3K9Me2 and H3K27Me3. (Note that although G9a can catalyze the methylation of H3K27 (48), the majority of H3K27Me3 is likely produced by KMT6/EZH2 (44).) In contrast, surfeit of choline causes lower expression of DNA methyltransferases, low DNA methylation, and a concomitant high expression of G9a and SUV39H1, resulting in high abundance of H3K9Me2 and H3K27Me3 further accelerated by high levels of AdoMet. Therefore, it is tempting to propose that low choline supply may down-regulate gene expression by DNA hypermethylation, whereas high choline intake may accomplish this by increased levels of repressive methylations on histone 3 (i.e. on H3K9 and H3K27).
There is growing evidence indicating that histone methylation, like DNA methylation, is a stable, heritable epigenomic chromatin modification that serves to regulate long term changes in gene expression. One mechanism for this stable inheritance involves the binding of the protein HP1 to methylated H3K9, which correlates with gene silencing in a variety of organisms (67–69). After binding, HP1 recruits SUV39H1, responsible for the initial methylation of H3K9. Thus, H3K9 methylation and HP1 is able to spread to successive nucleosomes in a self-propagating manner during mitosis and meiosis (70–72). Studies have shown that both DNA and histone methylation are critical for neural cell differentiation as well as high order cognitive functions such as learning and memory (73, 74). Interestingly, each modification appears to be involved in the regulation of the other. DNA methylation has been found to influence histone methylation, with the loss of methylated cytosines correlating to the loss of H3K9Me3 (75) and reductions in H3K4Me2 observed in regions of methylated CpGs (76). In turn, studies in Neurospora crassa (77) and Arabidopsis thaliana (78) have indicated that H3K9 methylation has the ability to direct DNA methylation. In mice, there is evidence that DNA methylation by DNMT3A and DNMT3B is dependent on SUV39H1-mediated histone methylation (79). It is becoming increasingly clear that DNA and histone methylation may have a mutually reinforcing relationship, with both modifications essential for long term regulation of the epigenome. Recently developed approaches that permit high throughput and high resolution analysis of DNA methylation concomitant with analysis of DNA-associated proteins, including specifically methylated histones, have revealed that the methylation patterns of histones and DNA correlate with one another and that DNA methylation of regulatory genomic domains changes dynamically as cells differentiate (80, 81). Our data suggest that the effects of the in utero supply of choline on the epigenome exhibit properties that can be considered as adaptations to the availability of the nutrient and may explain its long term actions. It will be interesting therefore to apply these novel techniques to explore in detail changes in the epigenomic landscape governed by the gestational intake of choline.
In addition to the well documented role of the adequate supply of choline in utero for the development and functioning of the adult rat brain (see above), we recently found that high E11–17 choline intake slows the growth of mammary tumors evoked by treatment with a carcinogen 7,12-dimethylbenz[α]anthracene in adulthood, and alters gene expression, and DNA methylation patterns within these tumors (82). These data pointed to the heretofore unanticipated role of adequate choline nutrition during pregnancy in prevention and/or amelioration of breast cancer in adult offspring. The current guidelines on choline intake advise women to consume increased amounts of choline during pregnancy and lactation (non-pregnant 425 mg/day, pregnant 450 mg/day, lactating 550 mg/day) (1). The individual requirement, however, may vary and will depend also on the genotype due to polymorphisms in genes encoding enzymes involved in choline and folate metabolism (6, 83). Significantly, the genes themselves may be subject to an epigenetic feedback regulation by choline.
This work was supported, in whole or in part, by National Institutes of Health Grant AG009525 from the NIA. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: E, embryonic day; AdoMet, S-adenosylmethionine; RT-PCR, reverse transcription-PCR; MOPS, 4-morpholinepropanesulfonic acid; bis-Tris, 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
References
- 1.Food and Nutrition Board (1998) Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Panthotenic Acid, Biotin, and Choline, National Academy Press, Washington, D.C. [PubMed]
- 2.Shaw, G. M., Carmichael, S. L., Yang, W., Selvin, S., and Schaffer, D. M. (2004) Am. J. Epidemiol. 160 102–109 [DOI] [PubMed] [Google Scholar]
- 3.Blusztajn, J. K. (1998) Science 281 794–795 [DOI] [PubMed] [Google Scholar]
- 4.Meck, W. H., and Williams, C. L. (2003) Neurosci. Biobehav. Rev. 27 385–399 [DOI] [PubMed] [Google Scholar]
- 5.McCann, J. C., Hudes, M., and Ames, B. N. (2006) Neurosci. Biobehav. Rev. 30 696–712 [DOI] [PubMed] [Google Scholar]
- 6.Zeisel, S. H. (2006) Annu. Rev. Nutr. 26 229–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meck, W. H., and Williams, C. L. (1997) Neuroreport 8 3045–3051 [DOI] [PubMed] [Google Scholar]
- 8.Mellott, T. J., Williams, C. L., Meck, W. H., and Blusztajn, J. K. (2004) FASEB J. 18 NIL412–NIL427 [DOI] [PubMed] [Google Scholar]
- 9.Meck, W. H., Smith, R. A., and Williams, C. L. (1988) Dev. Psychobiol. 21 339–353 [DOI] [PubMed] [Google Scholar]
- 10.Meck, W. H., Smith, R. A., and Williams, C. L. (1989) Behav. Neurosci. 103 1234–1241 [DOI] [PubMed] [Google Scholar]
- 11.Meck, W. H., and Williams, C. L. (1997) Neuroreport 8 2831–2835 [DOI] [PubMed] [Google Scholar]
- 12.Meck, W. H., and Williams, C. L. (1997) Neuroreport 8 3053–3059 [DOI] [PubMed] [Google Scholar]
- 13.Meck, W. H., and Williams, C. L. (1999) Dev. Brain Res. 118 51–59 [DOI] [PubMed] [Google Scholar]
- 14.Meck, W. H., Williams, C. L., Cermak, J. M., and Blusztajn, J. K. (2008) Front Integr. Neurosci. 1 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albright, C. D., Tsai, A. Y., Friedrich, C. B., Mar, M. H., and Zeisel, S. H. (1999) Dev. Brain Res. 113 13–20 [DOI] [PubMed] [Google Scholar]
- 16.Albright, C. D., Friedrich, C. B., Brown, E. C., Mar, M. H., and Zeisel, S. H. (1999) Dev. Brain Res. 115 123–129 [DOI] [PubMed] [Google Scholar]
- 17.Holmes-McNary, M. Q., Loy, R., Mar, M. H., Albright, C. D., and Zeisel, S. H. (1997) Dev. Brain Res. 101 9–16 [DOI] [PubMed] [Google Scholar]
- 18.Craciunescu, C. N., Albright, C. D., Mar, M. H., Song, J., and Zeisel, S. H. (2003) J. Nutr. 133 3614–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niculescu, M. D., Craciunescu, C. N., and Zeisel, S. H. (2005) Mol. Brain Res. 134 309–322 [DOI] [PubMed] [Google Scholar]
- 20.Williams, C. L., Meck, W. H., Heyer, D., and Loy, R. (1998) Brain Res. 794 225–238 [DOI] [PubMed] [Google Scholar]
- 21.Cermak, J. M., Holler, T., Jackson, D. A., and Blusztajn, J. K. (1998) FASEB J. 12 349–357 [DOI] [PubMed] [Google Scholar]
- 22.Sandstrom, N. J., Loy, R., and Williams, C. L. (2002) Brain Res. 947 9–16 [DOI] [PubMed] [Google Scholar]
- 23.Glenn, M. J., Gibson, E. M., Kirby, E. D., Mellott, T. J., Blusztajn, J. K., and Williams, C. L. (2007) Eur. J. Neurosci. 25 2473–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glenn, M. J., Kirby, E. D., Gibson, E. M., Wong-Goodrich, S., Mellott, T. J., Blusztajn, J. K., and Williams, C. L. (2008) Brain Res. 1237 110–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Napoli, I., Blusztajn, J. K., and Mellott, T. J. (2008) Brain Res. 1237 124–135 [DOI] [PubMed] [Google Scholar]
- 26.Jones, J. P., Meck, W. H., Williams, C. L., Wilson, W. A., and Swartzwelder, H. S. (1999) Dev. Brain Res. 118 159–167 [DOI] [PubMed] [Google Scholar]
- 27.Pyapali, G. K., Turner, D. A., Williams, C. L., Meck, W. H., and Swartzwelder, H. S. (1998) J. Neurophysiol. 79 1790–1796 [DOI] [PubMed] [Google Scholar]
- 28.Mellott, T. J., Follettie, M. T., Diesl, V., Hill, A. A., Lopez-Coviella, I., and Blusztajn, J. K. (2007) FASEB J. 21 1311–1323 [DOI] [PubMed] [Google Scholar]
- 29.Finkelstein, J. D., Martin, J. J., Harris, B. J., and Kyle, W. E. (1983) J. Nutr. 113 519–521 [DOI] [PubMed] [Google Scholar]
- 30.Chiang, P. K., Gordon, R. K., Tal, J., Zeng, G. C., Doctor, B. P., Pardhasaradhi, K., and McCann, P. P. (1996) FASEB J. 10 471–480 [PubMed] [Google Scholar]
- 31.Kovacheva, V. P., Mellott, T. J., Davison, J. M., Wagner, N., Lopez-Coviella, I., Schnitzler, A. C., and Blusztajn, J. K. (2007) J. Biol. Chem. 282 31777–31788 [DOI] [PubMed] [Google Scholar]
- 32.Wainfan, E., Dizik, M., Stender, M., and Christman, J. K. (1989) Cancer Res. 49 4094–4097 [PubMed] [Google Scholar]
- 33.Christman, J. K., Sheikhnejad, G., Dizik, M., Abileah, S., and Wainfan, E. (1993) Carcinogenesis 14 551–557 [DOI] [PubMed] [Google Scholar]
- 34.Niculescu, M. D., Craciunescu, C. N., and Zeisel, S. H. (2006) FASEB J. 20 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolff, G. L., Kodell, R. L., Moore, S. R., and Cooney, C. A. (1998) FASEB J. 12 949–957 [PubMed] [Google Scholar]
- 36.Waterland, R. A., and Jirtle, R. L. (2003) Mol. Cell. Biol. 23 5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterland, R. A., Dolinoy, D. C., Lin, J. R., Smith, C. A., Shi, X., and Tahiliani, K. G. (2006) Genesis 44 401–406 [DOI] [PubMed] [Google Scholar]
- 38.Jenuwein, T., and Allis, C. D. (2001) Science 293 1074–1080 [DOI] [PubMed] [Google Scholar]
- 39.Jenuwein, T. (2006) FEBS J. 273 3121–3135 [DOI] [PubMed] [Google Scholar]
- 40.Strahl, B. D., and Allis, C. D. (2000) Nature 403 41–45 [DOI] [PubMed] [Google Scholar]
- 41.Noma, K., Allis, C. D., and Grewal, S. I. (2001) Science 293 1150–1155 [DOI] [PubMed] [Google Scholar]
- 42.Schneider, R., Bannister, A. J., Myers, F. A., Thorne, A. W., Crane-Robinson, C., and Kouzarides, T. (2004) Nat. Cell Biol. 6 73–77 [DOI] [PubMed] [Google Scholar]
- 43.Strahl, B. D., Ohba, R., Cook, R. G., and Allis, C. D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14967–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao, R., Wang, L. J., Wang, H. B., Xia, L., Erdjument-Bromage, H., Tempst, P., Jones, R. S., and Zhang, Y. (2002) Science 298 1039–1043 [DOI] [PubMed] [Google Scholar]
- 45.Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C. P., Allis, C. D., and Jenuwein, T. (2000) Nature 406 593–599 [DOI] [PubMed] [Google Scholar]
- 46.Tachibana, M., Sugimoto, K., Fukushima, T., and Shinkai, Y. (2001) J. Biol. Chem. 276 25309–25317 [DOI] [PubMed] [Google Scholar]
- 47.Nakayama, T., Watanabe, M., Yamanaka, M., Hirokawa, Y., Suzuki, H., Ito, H., Yatani, R., and Shiraishi, T. (2001) Lab. Investig. 81 1049–1057 [DOI] [PubMed] [Google Scholar]
- 48.Tachibana, M., Sugimoto, K., Nozaki, M., Ueda, J., Ohta, T., Ohki, M., Fukuda, M., Takeda, N., Niida, H., Kato, H., and Shinkai, Y. (2002) Genes Dev. 16 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aagaard, L., Laible, G., Selenko, P., Schmid, M., Dorn, R., Schotta, G., Kuhfittig, S., Wolf, A., Lebersorger, A., Singh, P. B., Reuter, G., and Jenuwein, T. (1999) EMBO J. 18 1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, X. N., Mar, M. H., Song, J. N., and Zeisel, S. H. (2004) Dev. Brain Res. 149 121–129 [DOI] [PubMed] [Google Scholar]
- 51.Dobosy, J. R., Fu, V. X., Desotelle, J. A., Srinivasan, R., Kenowski, M. L., Almassi, N., Weindruch, R., Svaren, J., and Jarrard, D. F. (2008) Prostate 68 1187–1195 [DOI] [PubMed] [Google Scholar]
- 52.Bieri, J. G., Stoewsand, G. S., Briggs, G. M., Phillips, R. W., Woodard, J. C., and Kanapka, J. J. (1977) J. Nutr. 107 1340–1348874577 [Google Scholar]
- 53.Bieri, J. G. (1980) J. Nutr. 110 1726 [Google Scholar]
- 54.Zeisel, S. H., Mar, M. H., Zhou, Z. W., and Da Costa, K. A. (1995) J. Nutr. 125 3049–3054 [DOI] [PubMed] [Google Scholar]
- 55.Chomczynski, P., and Sacchi, N. (1987) Anal. Biochem. 162 156–159 [DOI] [PubMed] [Google Scholar]
- 56.Li, L. C., and Dahiya, R. (2002) Bioinformatics (Oxf.) 18 1427–1431 [DOI] [PubMed] [Google Scholar]
- 57.Rice, J. C., Briggs, S. D., Ueberheide, B., Barber, C. M., Shabanowitz, J., Hunt, D. F., Shinkai, Y., and Allis, C. D. (2003) Mol. Cell 12 1591–1598 [DOI] [PubMed] [Google Scholar]
- 58.Van den Veyver, I. B. (2002) Annu. Rev. Nutr. 22 255–282 [DOI] [PubMed] [Google Scholar]
- 59.Pogribny, I. P., Ross, S. A., Tryndyak, V. P., Pogribna, M., Poirier, L. A., and Karpinets, T. V. (2006) Carcinogenesis 27 1180–1186 [DOI] [PubMed] [Google Scholar]
- 60.Pogribny, I. P., Tryndyak, V. P., Muskhelishvili, L., Rusyn, I., and Ross, S. A. (2007) J. Nutr. 137 216S–222S [DOI] [PubMed] [Google Scholar]
- 61.Pogribny, I. P., Karpf, A. R., James, S. R., Melnyk, S., Han, T., and Tryndyak, V. P. (2008) Brain Res. 1237 25–34 [DOI] [PubMed] [Google Scholar]
- 62.Slack, A., Cervoni, N., Pinard, M., and Szyf, M. (1999) Eur. J. Biochem. 264 191–199 [DOI] [PubMed] [Google Scholar]
- 63.Nan, X., Ng, H. H., Johnson, C. A., Laherty, C. D., Turner, B. M., Eisenman, R. N., and Bird, A. (1998) Nature 393 386–389 [DOI] [PubMed] [Google Scholar]
- 64.Jones, P. L., Veenstra, G. J., Wade, P. A., Vermaak, D., Kass, S. U., Landsberger, N., Strouboulis, J., and Wolffe, A. P. (1998) Nat. Genet. 19 187–191 [DOI] [PubMed] [Google Scholar]
- 65.Eden, S., Hashimshony, T., Keshet, I., Cedar, H., and Thorne, A. W. (1998) Nature 394 842. [DOI] [PubMed] [Google Scholar]
- 66.Takizawa, T., Nakashima, K., Namihira, M., Ochiai, W., Uemura, A., Yanagisawa, M., Fujita, N., Nakao, M., and Taga, T. (2001) Dev. Cell 1 749–758 [DOI] [PubMed] [Google Scholar]
- 67.Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C., and Kouzarides, T. (2001) Nature 410 120–124 [DOI] [PubMed] [Google Scholar]
- 68.Lachner, M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. (2001) Nature 410 116–120 [DOI] [PubMed] [Google Scholar]
- 69.Snowden, A. W., Gregory, P. D., Case, C. C., and Pabo, C. O. (2002) Curr. Biol. 12 2159–2166 [DOI] [PubMed] [Google Scholar]
- 70.Cheutin, T., McNairn, A. J., Jenuwein, T., Gilbert, D. M., Singh, P. B., and Misteli, T. (2003) Science 299 721–725 [DOI] [PubMed] [Google Scholar]
- 71.Felsenfeld, G., and Groudine, M. (2003) Nature 421 448–453 [DOI] [PubMed] [Google Scholar]
- 72.Hall, I. M., Shankaranarayana, G. D., Noma, K., Ayoub, N., Cohen, A., and Grewal, S. I. (2002) Science 297 2232–2237 [DOI] [PubMed] [Google Scholar]
- 73.Lubin, F. D., Roth, T. L., and Sweatt, J. D. (2008) J. Neurosci. 28 10576–10586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ooi, L., and Wood, I. C. (2008) Biochem. J. 414 327–341 [DOI] [PubMed] [Google Scholar]
- 75.Reik, W., Dean, W., and Walter, J. (2001) Science 293 1089–1093 [DOI] [PubMed] [Google Scholar]
- 76.Higashimoto, K., Urano, T., Sugiura, K., Yatsuki, H., Joh, K., Zhao, W., Iwakawa, M., Ohashi, H., Oshimura, M., Niikawa, N., Mukai, T., and Soejima, H. (2003) Am. J. Hum. Genet. 73 948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tamaru, H., and Selker, E. U. (2001) Nature 414 277–283 [DOI] [PubMed] [Google Scholar]
- 78.Jackson, J. P., Lindroth, A. M., Cao, X., and Jacobsen, S. E. (2002) Nature 416 556–560 [DOI] [PubMed] [Google Scholar]
- 79.Lehnertz, B., Ueda, Y., Derijck, A. A. H. A., Braunschweig, U., Perez-Burgos, L., Kubicek, S., Chen, T. P., Li, E., Jenuwein, T., and Peters, A. H. F. M. (2003) Curr. Biol. 13 1192–1200 [DOI] [PubMed] [Google Scholar]
- 80.Meissner, A., Mikkelsen, T. S., Gu, H., Wernig, M., Hanna, J., Sivachenko, A., Zhang, X., Bernstein, B. E., Nusbaum, C., Jaffe, D. B., Gnirke, A., Jaenisch, R., and Lander, E. S. (2008) Nature 454 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gal-Yam, E. N., Egger, G., Iniguez, L., Holster, H., Einarsson, S., Zhang, X., Lin, J. C., Liang, G., Jones, P. A., and Tanay, A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 12979–12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kovacheva, V. P., Davison, J. M., Mellott, T. J., Rogers, A. E., Yang, S., O'Brien, M. O., and Blusztajn, J. K. FASEB J. (December 1, 2008) 10.1096/fj.08-122168 [DOI] [PMC free article] [PubMed]
- 83.Zeisel, S. H. (2008) Brain Res. 1237 5–11 [DOI] [PMC free article] [PubMed] [Google Scholar]