Abstract
PRC, a member of the PGC-1 coactivator family, is responsive to serum growth factors and up-regulated in proliferating cells. Here, we investigated its in vivo role by stably silencing PRC expression with two different short hairpin RNAs (shRNA1 and shRNA4) that were lentivirally introduced into U2OS cells. shRNA1 transductants exhibited nearly complete knockdown of PRC protein, whereas shRNA4 transductants expressed PRC protein at ∼15% of the control level. Complete PRC silencing by shRNA1 resulted in a severe inhibition of respiratory growth; reduced expression of respiratory protein subunits from complexes I, II, III, and IV; markedly lower complex I and IV respiratory enzyme levels; and diminished mitochondrial ATP production. Surprisingly, shRNA1 transductants exhibited a striking proliferation of abnormal mitochondria that were devoid of organized cristae and displayed severe membrane abnormalities. Although shRNA4 transductants had normal respiratory subunit expression and a moderately diminished respiratory growth rate, both transductants showed markedly reduced growth on glucose accompanied by inhibition of G1/S cell cycle progression. Microarray analysis revealed striking overlaps in the genes affected by PRC silencing in the two transductants, and the functional identities of these overlapping genes were consistent with the observed mitochondrial and cell growth phenotypes. The consistency between phenotype and PRC expression levels in the two independent transductant lines argues that the defects result from PRC silencing and not from off target effects. These results support a role for PRC in the integration of pathways directing mitochondrial respiratory function and cell growth.
Mitochondria are semiautonomous organelles that function as important sites of biological oxidation and ATP production. Central to this function is the electron transport chain and oxidative phosphorylation system, which is composed predominantly of five multisubunit protein complexes embedded in the mitochondrial inner membrane (1, 2). Mitochondria contain their own genetic system directed by a covalently closed circular DNA genome whose entire protein coding capacity is devoted to the production of 13 protein subunits of respiratory complexes I, III, IV, and V. Although essential, these subunits account for only a fraction of the protein composition with the majority of the respiratory subunits of nuclear origin (3, 4). Its bigenomic expression makes the respiratory apparatus unique among mitochondrial oxidative functions and poses an important biological problem in understanding the coordination of nuclear and mitochondrial gene expression.
A number of nucleus-encoded regulatory factors have been associated with the transcriptional control of both nuclear and mitochondrial genes that specify the respiratory apparatus in mammalian systems. Initial work identified nuclear respiratory factors, NRF-1 and NRF-2(GABP), as activators of nuclear cytochrome c and cytochrome oxidase genes. These factors have subsequently been associated with the expression of the majority of nuclear genes specifying mitochondrial respiratory function. These include genes encoding the respiratory subunits themselves but also those specifying protein import and assembly factors, heme biosynthetic enzymes, ribosomal proteins and tRNA synthetases of the mitochondrial translation system, and components of the mtDNA transcription and replication machinery (4, 5). The last group includes the nucleus-encoded mitochondrial transcription factors TFAM and the mtTFB isoforms, TFB1M and TFB2M, which act in conjunction with the mitochondrial RNA polymerase to maximize transcription from divergent heavy and light strand promoters (6, 7). Mice with a targeted disruption of Tfam (8) or Nrf-1 (9) have an embryonic lethal phenotype with loss of mtDNA, although lethality of the Nrf-1 embryos occurs at an earlier stage, suggesting that functions other than mitochondrial respiration are also disrupted (10). Recently, shRNA-mediated inhibition of both NRF-1 and NRF-2 has been linked to the down-regulation of all 10 nucleus-encoded cytochrome oxidase subunits, providing additional in vivo evidence for the importance of these factors in respiratory chain expression (11, 12).
Insight into how diverse transcription factors might be coordinated into a program of mitochondrial biogenesis came with the discovery of the PGC-1 (peroxisome proliferator-activated receptor γ coactivator-1) family of inducible coactivators. PGC-1α, the founding member of the family, displays a broad range of transcription factor interactions and biological functions (13, 14). It is induced during adaptive thermogenesis in brown fat and can interact directly with NRF-1 in enhancing mitochondrial respiration and biogenesis (15, 16). Ectopic overexpression of PGC-1α both in cultured cells and in mice results in dramatic increases in mitochondrial content and the expression of a number of genes important for mitochondrial respiratory function (16, 17). These properties are shared by PGC-1β, a close homologue of PGC-1α that also acts as a potent activator of NRF target genes (18). However, targeted disruption of either Pgc-1 coactivator in mice does not result in a dramatic mitochondrial biogenesis defect (19–22). In both cases, the homozygous knockouts were viable and fertile with no gross abnormalities in mitochondrial number or morphology. This may be explained by compensatory interactions among the PGC-1 family members or other adaptive changes in the transcriptional machinery. Interestingly, mice doubly deficient in Pgc-1α and Pgc-1β exhibit fetal arrest of mitochondrial biogenesis in both heart and brown fat associated with reduced cardiac output and postnatal mortality, suggesting that proper mitochondrial maturation requires both coactivators (23).
A third PGC-1 family coactivator, designated PRC (PGC-1-related coactivator), has limited overall sequence similarity with PGC-1α or -β but shares key structural features that define the family. These include a conserved amino-terminal transcriptional activation domain, a central proline-rich region, a functional binding site for host cell factor, and carboxyl-terminal R/S domain and RNA recognition motif (24). PRC binds NRF-1 and activates NRF-1 target genes but does not appear to be regulated significantly during adaptive thermogenesis. Rather, its expression is serum-induced in the absence of de novo protein synthesis and correlates with the cell proliferative cycle (25). Like the other PGC-1 coactivators, PRC binds HCF-1 (host cell factor-1), an abundant chromatin component that is required for cell cycle progression (26). HCF-1 is a NRF-2(GABP) coactivator (27) and exists in an in vivo complex with both PRC and NRF-2 (26). These in vivo interactions probably account for the trans-activation of NRF-2 target genes by PRC despite the absence of a direct interaction between PRC and NRF-2 subunits in vitro.
Here, the effects of shRNA-mediated PRC knockdown on respiratory function and mitochondrial biogenesis are investigated in stable lentiviral transductants of human U2OS cells. Loss of PRC results in a severe reduction in respiratory energy production associated with the reduced expression and assembly of respiratory complexes. Surprisingly, this respiratory defect is coincides with the proliferation of structurally defective mitochondria. In addition to the respiratory defect, PRC silencing was also implicated in the inhibition of cell cycle progression. Global changes in gene expression were consistent with both mitochondrial and growth phenotypes.
EXPERIMENTAL PROCEDURES
Cell Culture—U2OS cells and HEK-293 cells were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (Invitrogen). 293FT cells (Invitrogen) were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (HyClone), 1% penicillin/streptomycin (Invitrogen), and 1 mm Eagle's minimal essential medium nonessential amino acids (Mediatech, Inc.) with 500 μg/ml Geneticin (Invitrogen).
Generation of Lentiviral Transductants Expressing shRNA—Double-stranded oligonucleotides targeting the human PRC gene (PRCsh1S, CACCGCCATCAGGACATCACCATCACGAATGATGGTGATGTCCTGATGGC; PRCsh1AS, AAAAGCCATCAGGACATCACCATCATTCGTGATGGTGATGTCCTGATGGC; PRCsh4S, CACCGAGGCATTTGCAGCCATTGTTCAAGAGACAATGGCTGCAAATGCCTC; PRCsh4AS, AAAAGAGGCATTTGCAGCCATTGTCTCTTGAACAATGGCTGCAAATGCCTC) and a negative control sequence derived from the MISSION nontarget shRNA control vector (Sigma) (control shS, CACCCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTG; control shAS, AAAACAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTG) were ligated into the pENTR/U6 vector using the BLOCK-iT U6 RNAi2 entry vector kit (Invitrogen). The BLOCK-iT™ RNAi designer from the Invitrogen Web site was used to design shRNA sequences targeting PRC. The control hairpin contains four base pair mismatches to any known human or mouse gene (28). The resulting entry vectors were designated pENTR/PRCshRNA1, pENTR/PRCshRNA4, and pENTR/control shRNA. The lentiviral expression vectors pLenti/PRCshRNA1, pLenti/PRCshRNA4, pLenti/control shRNA, and pLenti-GW/U6-LaminshRNA were generated by transferring the U6-PRC, U6-control, and U6-Lamin RNA-mediated interference cassettes into the pLenti6/BLOCK-iT DEST vector using the LR recombination reaction. Lentiviral particles of these constructs were generated in 293FT cells using the BLOCK-iT Lentiviral RNAi expression system according to the manufacturer's protocol (Invitrogen). U2OS cells were transduced with each lentiviral construct at a multiplicity of infection of 10, and individual clones stably expressing each shRNA were selected with blasticidin.
Adenoviral Methods—The recombinant adenoviral plasmids Ad-GFP and Ad-PGC-1α were a kind gift from Dr. D. P. Kelly (Burnham Institute for Medical Research, Orlando, FL) (17). The plasmid Ad-NmycPRC was constructed using the AdEasy® basic kit from ATCC. An NH2-terminal c-Myc tag was incorporated into PRC full-length cDNA by PCR using FL-Bam-PRC/pBSII (24) as a template, resulting in NmycPRC/pBSII. NmycPRC/pAdTrack-CMV was generated by cloning the XhoI/NotI fragment of NmycPRC/pBSII into SalI/Not-digested pAdTrack-CMV. NmycPRC/pAdTrack-CMV was then used in a recombination reaction with pAdEasy-1 to produce Ad-NmycPRC. The adenoviral plasmids were linearized and transfected into the packaging cell line HEK-293 with Lipofectamine (Invitrogen), and the resulting viral stock was amplified to high titer. U2OS cells were infected, and the infection efficiency (95–100%) was determined by GFP expression 24 h after infection. Cells were harvested for preparation of RNA or protein extracts 72 h after infection.
Immunoblotting—Whole cell lysates were prepared in Non-idet P-40 lysis buffer as described previously (24). Extracts were subjected to denaturing gel electrophoresis, and the proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) with high molecular weight transfer buffer (50 mm Tris, 380 mm glycine, 0.1% SDS, and 20% methanol) in the Mini Trans-Blot Cell tank transfer system (Bio-Rad) for proteins of 70 kDa or larger or by using a Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad) with Towbin transfer buffer (29) for proteins under 70 kDa. The following primary antibodies were used: rabbit anti-PRC-(1047–1379) (25), rabbit anti-lamin A/C (a kind gift from Dr. R. D. Goldman, Northwestern University) (30), and rabbit anti-PGC-1α (a kind gift from Dr. D. P. Kelly, Burnham institute for Medical Research, Orlando, FL) (17). The relative levels of the five human OXPHOS complexes were determined by using the MitoProfile Human Total OXPHOS Complexes detection kit (MitoSciences), containing antibodies against MT-ND6 (NADH dehydrogenase subunit 6) of complex I, SDHB (succinate dehydrogenase iron-sulfur subunit) of complex II, UQCRC2 (ubiquinol-cytochrome c reductase core protein II) of complex III, MT-CO2 (cytochrome c oxidase subunit II) of complex IV, and ATP5A1 (subunit α of F1-ATPase) of complex V. The premixed mouse monoclonal antibody mixture was diluted to a working concentration of 7.2 μg/ml (MitoSciences). Human heart mitochondrial extract provided in the kit (0.5 μg/lane) was used as a positive control. All blots were visualized by SuperSignal West Pico chemiluminescent substrate (Pierce).
Flow Cytometry—For growth curves, 64000 U2OS cells were plated on day 0 in 6-cm dishes in Dulbecco's modified Eagle's medium (Invitrogen) containing either 5 mm glucose or 5 mm galactose (Sigma). Medium was changed daily, and on days 3, 4, and 5 cells were trypsinized, centrifuged at 4000 × g for 2 min, and resuspended in fresh medium. For absolute cell counting, 50 μl of AccuCount Blank particles, 5.0–5.9 μm (Spherotech), were mixed with 450 μl of the cell suspension, and samples were counted on a CyAn flow cytometer (Beckman Coulter). On day 5, cells were labeled in fresh medium containing 50 nm MitoTracker Green FM (Invitrogen) for 45 min at 37 °C immediately after trypsinization. After labeling, cells were centrifuged and resuspended in fresh medium, and AccuCount particles were added. The MitoTracker fluorescence of these cells was analyzed by flow cytometry using a CyAn flow cytometer (Beckman Coulter). Data were analyzed using Summit Software.
For DNA content analysis, exponentially grown lentiviral transductants were trypsinized, washed in phosphate-buffered saline, and fixed in 70% ice-cold ethanol. Cells were then washed with phosphate-buffered saline and stained with propidium iodide solution containing RNase A (50 μg/ml propidium iodide, 0.1% Triton X-100, 0.2 mg/ml RNase A). Cell cycle analysis was done on a Beckman Coulter Epics XL flow cytometer. Data were gated using pulse width and pulse area to exclude doublets, and the percentage of cells present in each phase of the cell cycle was calculated using Modfit software (Verity Software House).
Real Time Quantitative Reverse Transcription-PCR—Total RNA was purified using TRIzol reagent (Invitrogen). RNA samples were DNase-treated with the TURBO DNA-free kit (Ambion) and reverse transcribed into cDNA using the Taq-Man reverse transcription reagents (Applied Biosystems). Total genomic DNA was isolated using the GenElute Mammalian Genomic DNA miniprep kit (Sigma). Relative mRNA expression levels and relative mitochondrial DNA copy numbers were determined by Power SYBR Green (Applied Biosystems) quantitative real time PCR using the primer sets shown in Table 1. mRNA quantities were normalized to 18 S rRNA. The amount of mtDNA (as measured by amplification of the mtDNA-encoded COXI (cytochrome oxidase subunit I) gene and of a D-loop fragment) was normalized to 18 S rDNA. Assays for both the gene of interest and the 18 S control were performed in triplicate using an ABI PRISM 7900HT sequence detection system. Relative gene expression levels and relative mtDNA copy numbers were determined by the comparative Ct method using SDS version 2.1 software (Applied Biosystems).
TABLE 1.
List of primers used for quantitative real time PCR
| Gene name | Primer | Sequence (5′→3′) |
|---|---|---|
| PRC | hPRC sybr S | AGTGGTTGGGGAAGTCGAAG |
| hPRC sybr AS | CCTGCCGAGAGAGACTGAC | |
| CREB | hCREB sybr S | CCAGCAGAGTGGAGATGCAG |
| hCREB sybr AS | GTTACGGTGGGAGCAGATGAT | |
| Tfam | hTFAM sybr S | CCGAGGTGGTTTTCATCTGTC |
| hTFAM sybr AS | CAGGAAGTTCCCTCCAACGC | |
| TFB1M | hTFB1 M sybr S | CCTCCGTTGCCCACGATTC |
| hTFB1 M sybr AS | GCCCACTTCGTAAACATAAGCAT | |
| TFB2M | hTFB2 M sybr S | CGCCAAGGAAGGCGTCTAAG |
| hTFB2 M sybr AS | CTTTCGAGCGCAACCACTTTG | |
| Cytochrome c | hcytc sybr S2 | TGGGCCAAATCTCCATGGTCTCTT |
| hcytc sybr AS2 | TGCCTTTGTTCTTATTGGCGGCTG | |
| COXIV | hCOX4 sybr S | TTTAGCCTAGTTGGCAAGCGA |
| hCOX4 sybr AS | CCGATCCATATAAGCTGGGAGC | |
| SDHB | hSDHB sybr S | CCACAGCTCCCCGTATCAAG |
| hSDHB sybr AS | TCGGAAGGTCAAAGTAGAGTCAA | |
| UQCRC2 | UQCRC2 sybr 2S | TTCAGCAATTTAGGAACCACCC |
| UQCRC2 sybr 2AS | GTCACACTTAATTTGCCACCAAC | |
| ATP5A1 | ATP synthase sybr 2S | TACATGGGCTGAGGAATGTTCA |
| ATP synthase sybr 2AS | ACCAACTGGAACGTCCACAAT | |
| COXII | hCOX2 sybr S | ACAGATGCAATTCCCGGACGTCTA |
| hCOX2 sybr AS | GGCATGAAACTGTGGTTTGCTCCA | |
| MT-ND6 | hMT-ND6 sybr 2S | AGGATTGGTGCTGTGGGTGAAAGA |
| hMT-ND6 sybr 2AS | ATAGGATCCTCCCGAATCAACCCT | |
| Cytochrome b | hCYTB sybr S | AATTCTCCGATCCGTCCCTA |
| hCYTB sybr AS | GGAGGATGGGGATTATTGCT | |
| D-loop | hDloop-S | TTTCACGGAGGATGGTGGTCAA |
| hDloop-AS | ACCAACAAACCTACCCACCCTT | |
| COXI | hCOXI-S | AGGTTGAACAGTCTACCCTCCCTT |
| hCOXI-AS | GGCGTTTGGTATTGGGTTATGGCA | |
| 18 S rDNA | 18SrDNA-S | ACCAGAGCGAAAGCATTTGCCA |
| 18SrDNA-AS | TCGGCATCGTTTATGGTCGGAA |
Dipstick Immunoassay—U2OS transductants expressing the control shRNA or PRC shRNA1 were assayed for human complex I (NADH ubiquinone oxidoreductase) and complex IV (cytochrome c oxidase) using the respective MitoProfile dipstick assay kit (MitoSciences) according to the manufacturer's protocol. Briefly, cells were grown in Dulbecco's modified Eagle's medium containing 5 mm glucose for 3 days, washed with cold phosphate-buffered saline, and collected using a cell scraper. Total cell extract was prepared based on the manufacturer's protocol (MitoSciences). Levels of human complexes I and IV were determined using 5 μg and 2 μg of cell extract per dipstick, respectively. The signal intensity on each dipstick was measured using the ImageJ Image Processing and Analysis program (National Institutes of Health).
ATP Assay—Cells were plated at a density of 200,000 cells/6-cm dish. Two days after plating, half the cells were treated with oligomycin at a final concentration of 20 μg/μl (Sigma) for 3 h at 37 °C. Treated and untreated cells were trypsinized, counted, and diluted to 200,000 cells/ml. The cell suspension of 100 μl was used to measure steady-state ATP levels using an ATP bioluminescent somatic cell assay kit (Sigma) according to the manufacturer's instructions in duplicate. An ATP calibration curve was prepared with the provided ATP standard stock. The amount of light emitted was measured with a Monolight 2010 luminometer (Analytical Luminescence Laboratory).
Transmission Electron Microscopy—U2OS cells were pelleted, embedded in 2% agar, and fixed in 2.5% glutaraldehyde in 0.1 m sodium cacodylate (pH 7.4) overnight at room temperature. Cells were rinsed in 0.1 m sodium cacodylate (pH 7.4), postfixed in 2% osmium in 0.1 m sodium cacodylate (pH 7.4) for 1 h, rinsed in distilled water, and prestained with uranyl acetate for 30 min. Cells were then washed in distilled water and dehydrated in ascending grades of ethanol. After three changes of propylene oxide, cells were infiltrated in a mixture of propylene oxide and Epon/Araldite (1:1 ratio) for 1 h, embedded in Epon/Araldite resin, and placed in a 60 °C oven overnight to polymerize. Thick sections (1 μm) were cut and stained with toluidine blue O for examination and selection of specific regions for further analysis. Thin sections (90 nm) were stained with uranyl acetate and lead citrate and examined in a JEOL JEM 1220 transmission electron microscope. Gatan DigitalMicrograph software was used for digital imaging. For morphometric analysis, a grid was placed over the digital image of 10 different cells/sample, and the mitochondrial content as a percentage of the cytoplasm was estimated (31).
Microarray Analysis—Total RNA was isolated from PRC shRNA1, PRC shRNA4, and control shRNA transductants using TRIzol (Invitrogen) and DNase-treated with the TURBO DNA-free kit (Ambion). RNA samples were further purified using the RNeasy MinElute Cleanup kit (Qiagen). Integrity of the RNA was evaluated using the Agilent 2100 BioAnalyzer. 500 ng of RNA was used to perform in vitro transcription in the presence of biotin UTP with the Illumina TotalPrep RNA amplification kit (Ambion). The amplified, labeled RNA (1.5 μg) was then hybridized in triplicate to an Illumina Whole-Genome Sentrix Human-6 v2 Expression BeadChip and detected according to the Illumina user manual. Data normalization was performed using the statistical modeling language R through the BioConductor Lumi package. Quality control of the hybridization was performed using intensity box plots and sample clustering. To identify differentially expressed genes, routines implemented in the Limma package were applied to fit linear models to the normalized expression values. The variance used in the t score calculation was corrected by an empirical Bayesian method for better estimation under small sample size (32). To control the effects of multiple testing, the false discovery rate was limited to 5% (false discovery rate-adjusted p value < 0.05) to identify probe sets that are statistically significant between control and PRC shRNA1 or -4 samples. Gene functional categories were assigned using the GO data base (available on the World Wide Web) and the NCBI Entrez Gene data base (available on the World Wide Web).
RESULTS
Isolation of Lentiviral Transductants of U2OS Cells Exhibiting Partial or Complete PRC Silencing—The in vivo function of PRC was investigated by constructing lentiviruses designed to express two different shRNAs, shRNA1 and shRNA4, targeting different regions of the PRC mRNA. Human U2OS cells were infected with these viruses, and a number of stable transductants were selected in the presence of blasticidin. U2OS cells were chosen because they exhibit proper serum-regulated expression of PRC mRNA and protein, which are both down-regulated in serum-starved cells and rapidly up-regulated upon serum stimulation of quiescent cells (25). Approximately 20 individual shRNA1 and shRNA4 transductants were screened for PRC protein levels by immunoblotting. Two of these were chosen for further study. As shown in Fig. 1A (lane 4), one of the shRNA1 transductants had complete or nearly complete knockdown of PRC protein. Although none of the shRNA4 transductants showed complete PRC knockdown, one expressed ∼15% of control levels (Fig. 1A, lane 5), as estimated by densitometry. The inhibition of PRC expression was specific in that lamin A/C protein levels were not altered in either transductant (lanes 4 and 5). In addition, a negative control shRNA that lacks identity with any known mouse or human sequence failed to affect the expression of PRC or the lamin A/C control (lane 2), and a lentiviral transductant expressing a shRNA specific to lamin exhibited complete knockdown of lamin expression without affecting PRC (lane 3). In addition, as shown in Fig. 1B, PGC-1α is undetectable by immunoblotting in wild type U2OS cells (lane 3) and in the lentiviral transductants (lanes 4–6) under conditions where PGC-1α protein expressed from Ad-PGC-1α is easily detected (lane 2). Note that the intense antibody reactive band below PGC-1α is probably a nonspecific cross-reacting species, because it is expressed in the Ad-GFP control (lane 1). Thus, the effects of PRC in these cells can be studied in the absence of detectable PGC-1α protein expression.
FIGURE 1.
Silencing of PRC expression in U2OS cells by lentivirally delivered short hairpin RNAs. A, total cell extracts were prepared from wild type U2OS cells (wt) and from stable lentiviral transductants expressing the control shRNA, lamin shRNA, PRC shRNA1, or PRC shRNA4. PRC and lamin A/C proteins were detected by immunoblotting following denaturing gel electrophoresis using rabbit anti-PRC-(1047–1379) and rabbit anti-lamin A/C antibodies, respectively, following division of the membrane at the indicated position (dashed line). Molecular mass standards in kilodaltons are indicated at the left. B, total cell extract was prepared from wild type U2OS cells, adenovirus-infected U2OS cells expressing either GFP (Ad-GFP) or PGC-1α (Ad-PGC-1α), and U2OS cell lentiviral transductants expressing control shRNA, PRC shRNA1, or PRC shRNA4. PGC-1α protein was detected by immunoblotting following denaturing gel electrophoresis using rabbit anti-PGC-1α serum. The arrows indicate the positions of PRC, PGC-1α, and lamin A/C.
Additional confirmation that the shRNAs target PRC expression specifically was obtained from Ad-NmycPRC infected cells. As shown in Fig. 2A, PRC is abundantly overexpressed in Ad-NmycPRC-infected control transductants (lane 2) compared with those infected with Ad-GFP (lane 1). By contrast, PRC protein is undetectable in shRNA1 transductants infected with Ad-NmycPRC (lane 4) or the Ad-GFP control (lane 3). This result is not explained by a general defect in adenovirus expression in the shRNA1 transductant, because PGC-1α can be overexpressed specifically from Ad-PGC-1α in the PRC shRNA1 transductant (lanes 5 and 6). Finally, a side-by-side comparison of Ad-NmycPRC-infected transductants in Fig. 2B shows overexpression of PRC in the control (lane 1), complete silencing of PRC in the shRNA1 transductant (lane 2), and partial silencing of PRC expression in the shRNA4 transductant (lane 3). This parallels the relative level of PRC silencing observed in the uninfected transductant lines (Fig. 1A). The results establish that the PRC shRNAs can silence PRC protein levels specifically, even when PRC is overexpressed from a potent nonchromosomal transcriptional unit.
FIGURE 2.
Silencing of adenovirus-expressed PRC. A, immunoblots of total cell extracts from the control shRNA and PRC shRNA1 transductants infected with either adenovirus Ad-NmycPRC, Ad-PGC-1α, or the control Ad-GFP. PGC-1α protein was detected using rabbit anti-PGC-1α serum. B, comparison of PRC expression in Ad-NmycPRC-infected control, shRNA1, and shRNA4 transductants. In both panels, PRC protein was detected using rabbit anti-PRC-(1047–1379) serum.
Growth Inhibition Associated with PRC Silencing—Both the shRNA1 and -4 transductants showed a markedly reduced growth rate on glucose as a carbon source compared with that of wild type cells or the control transductant (Fig. 3A). Glucose growth does not require mitochondrial respiratory function, and the growth inhibition by even partial loss of PRC suggests that PRC may affect cell growth by mechanisms that are independent of its effects on mitochondrial respiration.
FIGURE 3.
Growth of PRC shRNA transductants and controls on either glucose or galactose as the primary carbon source. A, growth of U2OS wild type (wt; open squares) and lentiviral transductants expressing either the control shRNA (control; closed squares), PRC shRNA1(sh1; closed circles), or PRC shRNA4 (sh4; open circles) on glucose media. B, flow cytometric DNA content analysis of control shRNA, PRC shRNA1, and PRC shRNA4 U2OS transductants stained with propidium iodide. Representative DNA histograms are shown. Quantitative assessment of the cell cycle distributions are shown in each panel. Values are the averages ± S.E. for three independent determinations. C, growth of U2OS wild type (open squares) and lentiviral transductants expressing either the control shRNA (closed squares), PRC shRNA1 (closed circles), or PRC shRNA4 (open circles) on galactose media. Growth curves in A and B represent the averages ± S.E. for three separate determinations.
Flow cytometry was performed to determine whether the glucose growth defect is associated with a specific stage of the cell cycle. As shown in Fig. 3B, both the shRNA1 and -4 transductants exhibit a significantly higher proportion of G1 phase cells and a lower proportion of S phase cells compared with the control shRNA transductant. This is consistent with a PRC-dependent defect in the G1 to S transition. There was no significant effect on the percentage of cells in G2/M phase, and no sub-G1 population of apoptotic cells is present in any of the histograms. Hence, the decrease in cell proliferation in cells where PRC is silenced cannot be explained by induction of apoptosis. It is notable that although both PRC shRNA transductants exhibit significant cell cycle inhibition in the G1 to S transition, the degree to which G1 cells accumulate is related to the degree of PRC silencing in the two transductant lines. A higher accumulation of G1 stage cells occurs in the shRNA1 transductant, which exhibits complete PRC silencing than in the shRNA4 transductant, where PRC silencing is incomplete. This proportional response to PRC silencing argues that the PRC deficiency is responsible for the growth phenotype in these independent transductant lines.
In contrast to growth on glucose, the shRNA1 and -4 transductants are markedly different in their rate of respiratory growth on galactose, which mainly depends on mitochondrial ATP production (33). The growth rate of the shRNA4 transductant on galactose relative to the controls is diminished significantly (Fig. 3C). However, the shRNA1 transductant displays a much more severe respiratory growth inhibition on galactose (Fig. 3C), suggesting that complete PRC silencing results in a severe defect in mitochondrial respiratory function. The negative control transductant had a growth rate similar to that of wild type on both glucose and galactose, demonstrating that the growth defects are unrelated to lentiviral transduction or to the selection. The apparent PRC dose-dependent galactose growth inhibition displayed by the shRNA4 and shRNA1 transductants supports the conclusion that PRC is responsible for the inhibition of respiratory growth in these cell lines.
Mitochondrial Respiratory Chain Deficiency Associated with Loss of PRC Expression—PRC is a transcriptional coactivator that operates, at least in part, through the nuclear respiratory factors, NRF-1 and NRF-2, to activate the expression of NRF target genes. Therefore, it is likely that the respiratory growth defect in the shRNA1 transductant is associated with reduced expression of mRNAs encoding NRF target genes required for mitochondrial respiratory function. As shown in Fig. 4A, complete silencing of PRC protein expression in the shRNA1 transductant is accompanied by only a partial reduction of PRC mRNA under conditions where the cAMP-response element-binding protein control is unchanged, suggesting that shRNA1 reduces PRC protein expression through both transcriptional and translational inhibition. The diminished PRC mRNA expression is associated with a reduction in the mRNAs encoding the mitochondrial transcription factors (TFAM, TFB1M, and TFB2M). The biggest decrease is in TFB2M, whose expression has recently been correlated with the transcription and replication of mtDNA in both human (34) and Drosophila (35) cells. Here, we observe the down-regulation of three mitochondrial transcripts (Fig. 4A; COXII, ND6, and cytochrome b (Cytb)) but no significant change in mtDNA copy number normalized to 18 S rDNA (Fig. 4B). The maintenance of mtDNA levels despite reductions in TFB2M and mitochondrial transcripts is surprising in light of the coupling of mtDNA transcription and replication (7). Nevertheless, the results suggest that PRC control over respiratory gene expression contributes to the respiratory growth defect.
FIGURE 4.
Steady-state mRNA expression of regulatory and structural genes involved in mitochondrial respiratory function and estimation of mtDNA content. A, gene expression was monitored by quantitative real time PCR using total RNA prepared from control shRNA and PRC shRNA1 transductants. The battery of genes examined represent nuclear regulatory factors (PRC and cAMP-response element-binding protein), mitochondrial transcription factors (TFAM, TFB1M, and TFB2M), and nucleus-encoded (cytochrome c, COXIV, SDHB, UQCRC2, and ATP5A1) and mitochondrion-encoded (COXII, ND6, and cytochrome b) respiratory subunits. Relative steady-state mRNA levels were normalized to 18 S rRNA as an internal control, and values are expressed relative to the shRNA control, which was assigned a value of 1. Values are the averages ± S.E. for at least three separate determinations. B, content of mtDNA was estimated by quantitative real time PCR using total DNA prepared from control shRNA and PRC shRNA1 transductants. Relative mtDNA content was determined using probes specific for the mitochondrial D-loop or the COXI transcriptional unit. Values (gray bars) were normalized to 18 S rDNA as an internal control and expressed relative to those obtained from the shRNA control, which was assigned a value of 1, as the averages ± S.E. for at least three separate determinations. Values were also normalized to the MitoTracker Green FM fluorescence per cell (black bars) derived from the values in Table 2.
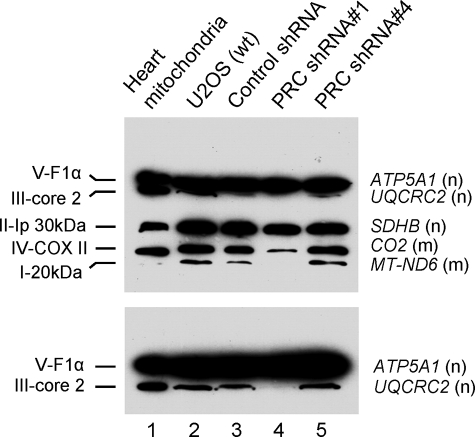
Given the respiratory growth defect, one might expect a deficiency in the expression, assembly, or function of the respiratory chain in the PRC shRNA1 transductant. This was investigated by immunoblotting using a battery of antibodies directed against key subunits of all five respiratory complexes. The subunits were chosen because they are labile unless they are assembled into their respective complexes, and thus their steady-state levels are thought to reflect those of the complexes in which they function. As shown in Fig. 5, similar levels of the five subunits are detected in cell extracts from the wild-type U2OS cells (lane 2), control shRNA (lane 3) and PRC shRNA4 transductants (lane 5). This suggests that there are no major differences in the expression or assembly of the five respiratory complexes in these cells. However, the PRC shRNA1 transductant exhibits markedly diminished expression of several subunits (lane 4). These include major reductions in COXII of complex IV, ND6 of complex I, and core protein 2 of complex III as well as a less pronounced but reproducible reduction in SDHB of complex II (Fig. 5). These changes do not result from a generalized reduction in respiratory gene expression, because the α-subunit of F1-ATPase (ATP5A1) of complex V is expressed normally. It is interesting to note that both nuclear (SDHB, core 2) and mitochondrial (COXII, ND6) gene products exhibit diminished steady-state levels, suggesting that PRC affects the expression and/or the assembly of the products of both genomes. The reduced levels of COXII and ND6 correlate with reduced transcripts levels (Fig. 4A), although the changes in protein expression appear quantitatively larger. Surprisingly, core protein 2 (Fig. 5) but not its mRNA (Fig. 4A) is dramatically down-regulated, whereas both SDHB protein (Fig. 5) and mRNA (Fig. 4A) are reduced in the shRNA1 transductant. The reasons for the discrepancy between mRNA and protein for these nuclear gene products are unclear but may reflect gene-specific differences in transcription and post-transcriptional controls. PRC may control steady-state expression of respiratory subunits through multiple means. For example, it may affect the expression of import or assembly factors or the mitochondrial or cytosolic translation machinery.
FIGURE 5.
Mitochondrial respiratory subunit expression in lentiviral transductants and controls. Total cell extract was prepared from wild type U2OS cells (wt) and lentiviral transductants expressing a control shRNA, PRC shRNA1, or PRC shRNA4. The indicated subunits from each of the five respiratory complexes were detected by immunoblotting following denaturing gel electrophoresis using a mixture of mouse monoclonal antibodies directed against each subunit. Heart mitochondrial extract was run as a control. Subunit designations for the respective complexes (I–V) are indicated at the left, and gene names along with their nuclear (n) or mitochondrial (m) assignment are indicated at the right. The lower panel shows an immunoblot from an extended run of an electrophoresis gel used to resolve V-F1α and III-core2.
The diminished expression of respiratory chain subunits in the PRC shRNA1 transductants was accompanied by significantly decreased respiratory enzyme levels. As shown in Fig. 6A, the levels of both NADH ubiquinone oxidoreductase (complex I) and cytochrome oxidase (complex IV) were reduced by ∼60% in the PRC shRNA1 transductant compared with the control. This result is consistent with the diminished levels of both ND6 (complex I) and COXII (complex IV) in the shRNA1 transductant. In addition, the steady-state level of oligomycin-sensitive ATP, which represents the mitochondrially produced ATP fraction, was reduced by ∼50% in the PRC shRNA1 transductant compared with control shRNA or PRC shRNA4 transductants (Fig. 6B), in keeping with similar reductions in respiratory enzyme levels (Fig. 6A). These results are consistent with the defect in respiratory growth and establish that loss of PRC is associated with a severe defect in the expression and function of the respiratory apparatus.
FIGURE 6.
Respiratory enzyme and ATP levels in lentiviral transductants. A, levels of respiratory complexes I and IV in total cell extracts were quantitated by dipstick assay (MitoSciences). The top band represents the positive control signal; the bottom band represents the specific signal from the respective complexes. Membranes were scanned for densitometric analysis, and the percentage of enzyme per unit of protein in the PRC shRNA1 transductant relative to the control shRNA is expressed as the average ± S.E. for three separate determinations. B, steady-state ATP levels were quantitated in total cell extracts using a bioluminescent assay. Values for oligomycin-sensitive ATP levels for control shRNA, PRC shRNA1, and PRC shRNA4 transductants are shown as the average ± S.E. for three separate determinations.
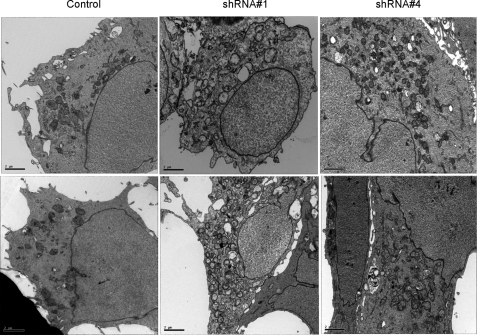
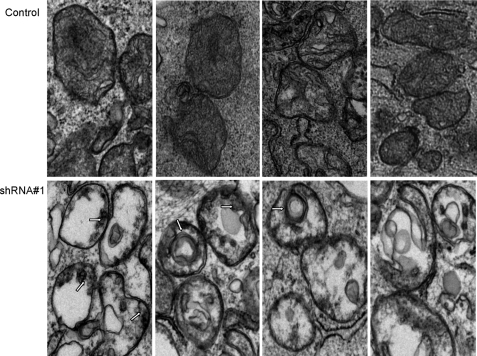
Changes in Mitochondrial Number and Morphology Associated with PRC Silencing—The PGC-1 family coactivators are thought to function as positive regulators of respiratory gene expression and mitochondrial biogenesis (4). Thus, the diminished expression and function of the respiratory chain in the PRC shRNA1 transductant may reflect a generalized down-regulation of mitochondrial biogenesis in these cells. Mitochondrial number and morphology were examined by transmission electron microscopy. As shown in Fig. 7, the cells are generally asymmetric, with the majority of cytoplasm occupying one side of the cell. The cytoplasm of the control transductants contains a number of electron-dense mitochondria that exhibit the classical double membrane structure with cristae visualized as invaginations of the inner membrane that extend into the matrix (Figs. 7 and 8). In contrast, the shRNA1 transductants contain an increased number of double membrane organelles that bear little resemblance to the control mitochondria (Fig. 7). In addition to being more abundant, these organelles exhibit much reduced electron density and, instead of recognizable cristae, contain a granular internal structure. Electron micrographs of PRC shRNA4 transductants show mitochondrial profiles that are intermediate between control and shRNA1 cells with a modestly elevated mitochondrial content and a modestly reduced electron density relative to the control transductants, confirming that PRC is mediating the changes in mitochondrial number and morphology. Closer examination of control and shRNA1 mitochondria reveals striking structural differences (Fig. 8). Instead of well formed cristae extending into the mitochondrial matrix, as observed in the controls, the shRNA1 mitochondria show blebbing of the inner mitochondrial membrane as if formation of proper cristae were truncated. In many cases, the matrix space is completely devoid of any internal structure. In others, circular multimembranous structures are found in the matrix spaces of a subset of shRNA1 mitochondria. This may reflect an abnormality in organelle biogenesis in these cells.
FIGURE 7.
Evaluation of mitochondrial number and morphology by electron microscopy. U2OS cells stably infected with recombinant lentiviruses expressing a control shRNA, PRC shRNA1, or PRC shRNA4 were analyzed by transmission electron microscopy. Bars, 2 μm.
FIGURE 8.
Mitochondrial ultrastructure in lentiviral transductants. The panels show representative electron micrographs of mitochondria from lentiviral U2OS transductants. The upper panels are from the control shRNA transductant, whereas the lower panels are from the PRC shRNA1 transductant. The arrows indicate the blebbing of the inner mitochondrial membrane and the circular multimembranous structures seen in some mitochondria of the PRC shRNA1 transductant.
The mitochondrial abundance was compared quantitatively by flow cytometric analysis using MitoTracker Green FM and by morphometric assessment of mitochondrial profiles. As shown in Table 2, the shRNA1 transductant has twice the Mito-Tracker Green FM fluorescence on a per cell basis compared with the control. The MitoTracker Green FM fluorescence of the shRNA4 transductant is intermediate between that of the control and shRNA1. Interestingly, an independent shRNA1 isolate had a PRC expression level equivalent to that in shRNA4 and displayed similar growth retardation and Mito-Tracker staining (not shown). Thus, the level of PRC expression coincides with the severity of the phenotype.
TABLE 2.
Mitochondrial content in lentiviral transductants
| Lentiviral transductant | MitoTracker Green FM fluorescencea(mean ± S.E.) | Mitochondrial contentb(% cytoplasmic area ± S.E.) |
|---|---|---|
| % | ||
| Control shRNA | 647 ± 125 (1.0)c | 16 ± 1 (1.0) |
| PRC shRNA1 | 1360 ± 84 (2.1) | 34 ± 1 (2.1) |
| PRC shRNA4 | 855 ± 58 (1.3) | 19 ± 2 (1.3) |
Mean fluorescence intensity of MitoTracker Green FM as quantifies by flow cytometry is the average of at least three separate determinations ± S.E.
Mitochondrial content expressed as a percentage of cytoplasmic area ± S.E. is derived from morphometrical analysis of 10 digital electron micrographs for each transductant, such as those shown in Fig. 8
The numbers in parentheses indicate the -fold differences in mitochondrial content relative to the control
The mitochondrial accumulation of MitoTracker Green FM is independent of membrane potential and thus can be used to measure organelle content independent of function (36, 37). The mitochondrial content as a percentage of the cytoplasm was also estimated morphometrically for 10 different cells/transductant line (Table 2). The results also show a 2-fold greater abundance of mitochondrial content in the shRNA1 transductants and a slight increase in the shRNA4 transductants compared with the controls, confirming the increase in mitochondrial abundance estimated by flow cytometry. Thus, the loss of PRC is associated with a proliferation of structurally abnormal mitochondria.
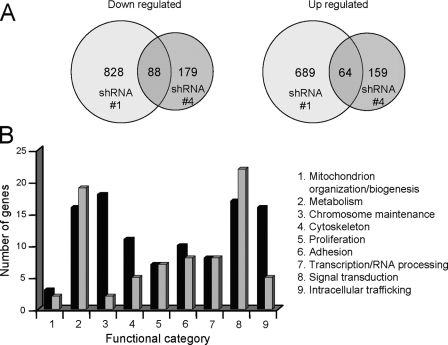
Changes in Gene Expression Profiles Associated with Complete or Partial PRC Silencing—Global changes in gene expression associated with PRC silencing were determined by microarray analysis. As summarized in Fig. 9A, complete PRC silencing in the shRNA1 transductant is associated with significant changes (-fold ≥1.5 or ≤1.5, p < 0.01, false discovery rate p < 0.05) in the expression of 1517 genes. As might be expected, the majority of these were down regulated. By contrast, incomplete PRC silencing in the shRNA4 transductant results in the altered expression of 338 genes with about the same proportion down-regulated (Fig. 9A). Although far fewer genes exhibit altered expression in the shRNA4 transductant, 49% of the down-regulated genes and 40% of the up-regulated genes in the shRNA4 transductant are shared with the shRNA1 transductant (for a complete list, see Table S1). This striking overlap argues that similar PRC-dependent functions are affected in both of these independent transductants.
FIGURE 9.
Summary of gene expression patterns associated with PRC silencing as assessed by microarray analysis. A, Venn diagrams representing the number of significantly down-regulated (left) and up-regulated (right) mRNAs expressed in PRC shRNA1 and -4 transductants compared with the shRNA control. The number of genes common to shRNA1 and -4 transductants is shown in the region of overlap in each diagram. These are listed in Table S1. B, bar graph depicts the assignment of genes that are down-regulated (black bars) or up-regulated (gray bars) in both transductant lines to nine functional categories.
All but about 10% of the overlapping genes, which could not be assigned a function, were grouped into nine functional categories (Fig. 9B). Only a few of the overlapping genes between shRNA1 and -4 transductants were related to mitochondrial function. As shown in Table S2, 37 genes related to mitochondrial respiratory function, including respiratory proteins, import and assembly factors, and components of the mitochondrial translational machinery, were down-regulated significantly (p < 0.01, false discovery rate p < 0.05) in the shRNA1 transductant. Of these, only eight were also down-regulated in the shRNA4 transductant. This difference is consistent with the relative severity of the respiratory chain and mitochondrial morphology defects in the shRNA1 transductant.
Also notable is the relatively large number of down-regulated genes among those shared between the two tranductants in the chromosome maintenance category (Fig. 9B). Many of these are genes of the histone clusters. As shown in Table S3, some of the most highly down-regulated genes in the shRNA1 transductant are histone cluster genes, and the vast majority of these are also down-regulated in the shRNA4 transductant. The -fold differences reflect the degree of PRC silencing in the two transductant lines, supporting the conclusion that the reduced histone expression is a PRC-dependent phenotype. In addition, histone mRNA expression is known to be tightly coupled to cell cycle progression and DNA synthesis (38). The striking overlap between shRNA1 and -4 transductants in the down-regulation of histone cluster genes (Table S3) is consistent with similarities between the two lines in the inhibition of G1/S progression. Thus, the global changes in gene expression revealed by microarray are consistent with the cell growth and mitochondrial phenotypes.
DISCUSSION
Transcriptional expression of the respiratory chain in mammalian systems relies upon the interplay of nuclear transcription factors and the PGC-1 family of regulated coactivators (4, 5, 39). Ubiquitous transcription factors, exemplified by NRF-1, NRF-2(GABP), and ERRα (estrogen-related receptor), target many nuclear genes required for the direct expression of the respiratory complexes as well as many others that play indirect roles in assembly, protein import, heme biosynthesis, and mtDNA maintenance. The PGC-1 coactivators are thought to impose integrative regulatory control on the pathway through their induction by environmental signals and their trans-activation of respiratory genes through the NRFs and other transcription factors. In this manner, they function as positive regulators of respiratory chain expression and mitochondrial biogenesis.
PRC is a growth-regulated member of the PGC-1 family (24, 25). It has the characteristics of an immediate early gene in that it is rapidly induced in response to serum stimulation of quiescent fibroblasts in the absence of de novo protein synthesis and down-regulated upon serum withdrawal or contact inhibition. PRC is indistinguishable from the other PGC-1 family members in its ability to trans-activate NRF target genes (40) and to interact with HCF-1 (26). This, along with its immediate early expression, suggests that PRC may be involved in the expression of the respiratory apparatus in response to growth-regulatory signals. This possibility is supported by the observation that a PRC subfragment containing the NRF-1/cAMP-response element-binding protein binding site inhibits respiratory growth when expressed in trans (25). Here, we show that complete PRC silencing in a PRC shRNA-expressing lentiviral transductant is associated with a severe respiratory deficiency marked by slow growth on galactose, defective respiratory subunit expression, and reduced respiratory enzyme levels and ATP production. These changes coincide with reductions in the expression of mRNAs encoding mitochondrial transcription factors (TFAM, TFB2M) as well as nucleus- and mitochondrion-encoded respiratory subunits. Moreover, mitochondria in the PRC-deficient cells were more abundant than in the control cells and displayed severe structural abnormalities.
RNA interference-based methodology is sometimes associated with nonspecific silencing effects and activation of the interferon response. In this case, we find no evidence for non-specific silencing. Complete PRC silencing is observed in shRNA1 transductants infected with Ad-NmycPRC, which massively overproduces PRC in the control shRNA transductant. Interestingly, PRC expressed from Ad-NmycPRC is still observed in shRNA4 cells but is much reduced compared with control level. Thus, shRNA1 eliminates PRC expression whether it occurs from the endogenous gene or from a potent extrachromosomal transcriptional unit, whereas shRNA4 partially reduces PRC expression in both scenarios. This silencing is PRC-specific because PGC-1α is abundantly expressed in the shRNA1 and shRNA4 transductants upon infection with Ad-PGC-1α.
Additional evidence supports the conclusion that the phenotypes of transductants expressing either shRNA1 or shRNA4, targeting different regions of the PRC mRNA, are specific to PRC silencing. Both the PRC shRNA1 transductant, showing complete loss of PRC protein, and the shRNA4 transductant, expressing residual levels of PRC, display similar glucose growth deficiencies. This growth inhibition coincides with the increased accumulation of G1 stage cells and reduced S phase cells indicative of inhibition of G1/S progression. The degree of G1 accumulation was related to PRC expression, being lowest in the control transductant and highest in the shRNA1 transductant. In addition, the shRNA1 and -4 transductants exhibit striking overlap in the down-regulation of histone cluster genes, whose expression is highly sensitive to diminished progression through S phase (38). Again, the histone mRNA levels in the two transductants reflect their relative levels of PRC expression. Interestingly, PRC exists in a complex with HCF-1 (26), a major chromatin-associated coactivator that is required for progression through G1 (41). PRC is also expressed as an immediate early gene in response to serum growth factors (25). These properties along with the growth phenotype observed here are consistent with the suggestion that PRC is a regulated component of a transcription complex that mediates cell cycle progression (26). Currently, the mechanism by which PRC silencing disrupts cell cycle dynamics is not well understood. It may affect cell cycle regulators that control the progression from G1 to S phase or interfere directly with transcription of the histone genes.
Although the shRNA4 transductant had only modestly reduced growth on galactose and no detectable loss of respiratory subunit expression or ATP production, it did have changes in mitochondrial content and morphology that were intermediate between the control and shRNA1 transductants. Thus, although reduction of PRC to ∼15% of the control level can impair growth and affect mitochondrial profiles, it is sufficient to support respiratory function. This is consistent with the microarray data showing reduced expression in the shRNA4 transductant of only a small subset of genes that contribute to mitochondrial respiratory function (Table S2). In contrast, the severe respiratory chain deficiency in the shRNA1 transductant coincides with the reduced expression of many genes required for oxidative activities, protein import and assembly, and mitochondrial translation revealed by the microarray (Table S2). Once again, the phenotype is consistent with the gene expression profiles and reflects the relative levels of PRC expression in the two transductant lines. The results argue that the phenotypic changes are mediated by specific PRC silencing. Finally, the respiratory phenotype does not result from activation of the interferon response. There is no global up-regulation of interferon-stimulated genes (42, 43), such as OAS1, OAS2, OAS3, STAT1, or IRF3, in U2OS cells stably expressing PRC shRNA1 or -4 (not shown). Therefore, the observed changes in mitochondrial function and morphology and in cell growth result from reduced PRC expression and not from nonspecific silencing or activation of the interferon response.
In several instances, the reductions in mRNA expression seen in the shRNA1 cells differ from those of their respective proteins. Notably, PRC protein is undetectable in the shRNA1 transductant, whereas PRC mRNA is reduced by half. This suggests that PRC shRNA1 is inhibitory at both the transcriptional and post-transcriptional level, most probably through translational inhibition. Translational repression is most commonly observed with short interfering RNAs having imperfect complementarity with their target sequences (44, 45). However, a small interfering RNA with perfect base complementarity to its target in the geminin gene also had a greater inhibitory effect on geminin protein expression relative to its mRNA, suggesting that gene silencing in this case occurs predominantly at the translational level (46). This is similar to what we see here with PRC shRNA1, which has perfect sequence complementarity to a single target site in the PRC coding region. It is also noteworthy that PRC mRNA has a relatively rapid decay rate with a half-life of approximately 2 h (25). Messenger RNAs that turn over rapidly may be more susceptible to shRNA-mediated translational repression (47).
The reductions in mitochondrial respiratory subunit expression also appear more pronounced at the protein level, especially for UQCRC2 encoding core protein 2 of complex III, where the dramatic down-regulation of the protein occurs in the absence of any mRNA reduction. These discrepancies suggest that PRC may affect the expression of these subunits through mechanisms not restricted to transcriptional inhibition. For example, PRC may control NRF target genes involved in mitochondrial translation or the import or assembly of respiratory subunits (Table S2). It remains to be determined whether the respiratory defect results from relatively modest reductions in the expression of many genes or from a large reduction in a few genes or even a single gene.
The PGC-1 coactivators are thought to function as positive regulators of mitochondrial biogenesis. This conclusion comes largely from gain of function experiments where PGC-1α or -β is ectopically expressed in cultured cells or transgenic mice. Under these conditions, dramatic increases in respiratory gene expression, energy production, mitochondrial volume, and cristae density have been observed (16, 17, 48). The respiratory chain deficiencies observed here upon loss of PRC protein are consistent with a role for PRC as a positive regulator of respiratory chain expression. The severity of the defect contrasts with the relatively subtle changes in respiratory function observed in mice deficient in either PGC-1α (19, 20) or PGC-1β (21, 22). This is probably explained by the fact that U2OS cells have no detectable PGC-1α and thus are possibly more dependent on PRC to maintain basal respiratory function. Moreover, most of the available evidence points to PGC-1α and -β as mediators of differentiation-induced mitochondrial content. For example, shRNA-mediated silencing of PGC-1β in a preadipocyte cell line derived from PGC-1α null mice led to a failure to establish and maintain differentiated levels of mitochondrial density in mature brown adipocytes (49). In the absence of both coactivators, mitochondrial content was maintained at the preadipocyte level. Loss of either coactivator alone had a relatively modest effect, but together they provide complementary functions in maintaining the differentiated level. Mice with ablations of both Pgc-1α and Pgc-1β exhibit normal early fetal formation of mitochondria but show an arrest in perinatal mitochondrial biogenesis in the heart and brown adipose tissue and die shortly after birth as a result of heart failure (23). PRC may be required for the early mitochondrial maturation and maintenance in these mice. In PGC-1β-deficient mice, there is compensation for the loss of PGC-1β by PGC-1α (23). Here, we find no evidence of an increase in PGC-1α mRNA or protein levels in cells where PRC is silenced, suggesting that PGC-1α cannot compensate for the loss of PRC in this system. Thus, synergies among members of the coactivator family may set the upper and lower limits for mitochondrial content in a given physiological context.
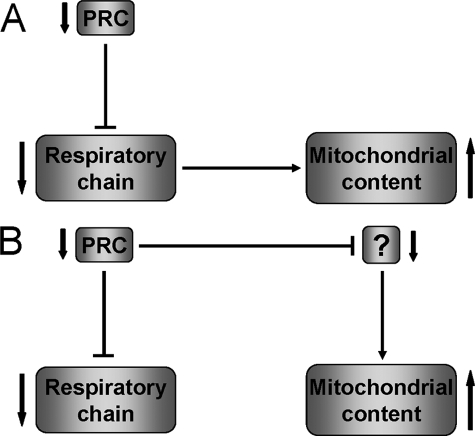
Gain of function experiments performed with PGC-1α and -β suggest a tight coordination between expression of the respiratory chain and the biogenesis of the organelle. Overexpression of either coactivator increases the expression of respiratory subunits concomitant with organelle biogenesis (16, 17, 48). Thus, it is surprising that shRNA-mediated PRC knockdown results in an increase in mitochondrial content as measured by uptake of MitoTracker Green FM and by morphometric analysis. This indicates that organelle biogenesis is not positively linked to the PRC-dependent expression of the respiratory chain in this system. It is notable that the elevation in mitochondrial density is associated with a severe morphological defect consisting of markedly disrupted cristae structure, blebbing of the inner mitochondrial membrane, and reduced electron density within the matrix. This increase in structurally defective mitochondria may be a compensatory response to the respiratory chain deficiency mediated by the loss of PRC. One possibility, depicted in Fig. 10A, is that organelle biogenesis increases in response to the respiratory defect mediated by the loss of PRC by a pathway that is PRC-independent. In this scenario, anything that interferes with mitochondrial respiratory function might be expected to lead to increased organelle biogenesis. Although the expression of some respiratory chain genes is altered in response to various respiratory inhibitors, there is little evidence for a retrograde pathway mediating the up-regulation of mitochondrial biogenesis in cultured cells (50, 51). Alternatively, as shown in Fig. 10B, PRC may regulate both the respiratory chain and an unidentified regulator(s) that modulates organelle biogenesis. Loss of PRC may lead to the down-regulation of this putative factor and the consequent increase in mitochondrial content. In this case, both the respiratory chain and organelle biogenesis are under the control of PRC. The absence of PGC-1α in these cells may also be a contributing factor. PGC-1α has recently been implicated in the ability of cells to recover from reduced ATP levels that were depleted because of treatment with a chemical uncoupler (52). Interestingly, increases in abnormal mitochondria are also observed in the muscle fibers of patients with certain mitochondrial myopathies leading to ragged red fibers as a diagnostic indicator. These are thought to arise as a compensatory response to respiratory chain deficiencies caused by mutation of mitochondrial DNA (53). In these patients, extremely high levels of mutant mtDNA accumulate in the muscle fiber. An accumulation of abnormal mitochondria associated with ragged red fibers and cytochrome oxidase deficiency is also seen in mice with skeletal muscle-specific disruption of Tfam expression (54). This is associated with progressively reduced levels of mtDNA in skeletal muscle. A similar increase in abnormal mitochondria with normal mtDNA copy number but increased mtDNA transcripts is seen in heart-specific mTERF3 knock-out mice (55). These cases contrast with the current findings, where mtDNA levels normalized to 18 S rDNA remain the same in the PRC knock-down and the control, despite a reduction in mitochondrial transcript levels and respiratory subunits. It is possible that the mtDNA replication machinery cannot respond to the respiratory defect because of the absence of PRC or PGC-1α.
FIGURE 10.
Alternative models depicting the association of respiratory chain expression with mitochondrial biogenesis. A, silencing of PRC results in the down-regulation of respiratory chain expression and function. This leads to an increase in organelle content as a PRC-independent compensatory response to the loss of respiratory function. In this case, PRC regulates mitochondrial biogenesis indirectly through its effects on the respiratory apparatus. B, silencing of PRC results in the down-regulation of both the respiratory chain and unidentified target(s) that control organelle content. Thus, the response to the loss of respiratory function occurs through a PRC-dependent pathway. In this case, PRC exerts direct control over respiratory chain expression and the factors that regulate organelle biogenesis.
In conclusion, these results show that PRC functions as an important regulator of respiratory chain expression and mitochondrial biogenesis in cells where PGC-1α is absent. Therefore, PRC resembles the other PGC-1 family coactivators in controlling mitochondrial respiratory function through its induction by environmental signals and its trans-activation of respiratory genes through the NRFs and other transcription factors. In addition to their importance to mitochondrial function, these coactivators have a broad specificity and are involved in the regulation of many other cellular activities (4, 18). Here, we show that PRC may control cell growth by mechanisms that are independent of its effects on mitochondrial function. It will be of interest to further understand the biological role of PRC and to examine the functional redundancy between the different members of the PGC-1 coactivator family.
Supplementary Material
Acknowledgments
We thank Robert D. Goldman (Northwestern Medical School) for the lamin A/C antibody and Daniel P. Kelly (Burnham institute for Medical Research) for the PGC-1α adenoviral plasmid and antibody. We are grateful to the Flow Cytometry Core Facility for help with the fluorescence-activated cell sorting analysis. Lennell Reynolds from the Cell Imaging Facility provided valuable assistance with the electron microscopy. We thank the Center for Genetic Medicine for processing the Illumina Beadchip and the BioInformatics Core for statistical analysis of the data generated from the microarray study.
This work was supported, in whole or in part, by National Institutes of Health Grant GM32525-26. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3.
Footnotes
The abbreviations used are: RNAi, RNA interference; shRNA, short hairpin RNA; mtDNA, mitochondrial DNA; TFB, mitochondrial transcription factor B; GFP, green fluorescent protein; Ad, adenovirus.
References
- 1.Hatefi, Y. (1985) Annu. Rev. Biochem. 54 1015-1069 [DOI] [PubMed] [Google Scholar]
- 2.Balaban, R. S. (1990) Am. J. Physiol. 258 C377-C389 [DOI] [PubMed] [Google Scholar]
- 3.Wallace, D. C. (2005) Annu. Rev. Genet. 39 359-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scarpulla, R. C. (2008) Physiol. Rev. 88 611-638 [DOI] [PubMed] [Google Scholar]
- 5.Kelly, D. P., and Scarpulla, R. C. (2004) Genes Dev. 18 357-368 [DOI] [PubMed] [Google Scholar]
- 6.Gaspari, M., Larsson, N. G., and Gustafsson, C. M. (2004) Biochim. Biophys. Acta 1659 148-152 [DOI] [PubMed] [Google Scholar]
- 7.Bonawitz, N. D., Clayton, D. A., and Shadel, G. S. (2006) Mol. Cell 24 813-825 [DOI] [PubMed] [Google Scholar]
- 8.Larsson, N. G., Wang, J. M., Wilhelmsson, H., Oldfors, A., Rustin, P., Lewandoski, M., Barsh, G. S., and Clayton, D. A. (1998) Nat. Genet. 18 231-236 [DOI] [PubMed] [Google Scholar]
- 9.Huo, L., and Scarpulla, R. C. (2001) Mol. Cell. Biol. 21 644-654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cam, H., Balciunaite, E., Blias, A., Spektor, A., Scarpulla, R. C., Young, R., Kluger, Y., and Dynlacht, B. D. (2004) Mol. Cell 16 399-411 [DOI] [PubMed] [Google Scholar]
- 11.Ongwijitwat, S., Liang, H. L., Graboyes, E. M., and Wong-Riley, M. T. (2006) Gene (Amst.) 374 39-49 [DOI] [PubMed] [Google Scholar]
- 12.Dhar, S. S., Ongwijitwat, S., and Wong-Riley, M. T. (2008) J. Biol. Chem. 283 3120-3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puigserver, P., and Spiegelman, B. M. (2003) Endocr. Rev. 24 78-90 [DOI] [PubMed] [Google Scholar]
- 14.Handschin, C., and Spiegelman, B. M. (2006) Endocr. Rev. 27 728-735 [DOI] [PubMed] [Google Scholar]
- 15.Puigserver, P., Wu, Z., Park, C. W., Graves, R., Wright, M., and Spiegelman, B. M. (1998) Cell 92 829-839 [DOI] [PubMed] [Google Scholar]
- 16.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C., and Spiegelman, B. M. (1999) Cell 98 115-124 [DOI] [PubMed] [Google Scholar]
- 17.Lehman, J. J., Barger, P. M., Kovacs, A., Saffitz, J. E., Medeiros, D. M., and Kelly, D. P. (2000) J. Clin. Invest. 106 847-856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, J., Handschin, C., and Spiegelman, B. M. (2005) Cell Metab. 1 361-370 [DOI] [PubMed] [Google Scholar]
- 19.Lin, J., Wu, P., Tarr, P. T., Lindenberg, K. S., St-Pierre, J., Zhang, C., Mootha, V. K., Jäger, S., Vianna, C. R., Reznick, R. M., Cui, L., Manieri, M., Donovan, M. X., Wu, Z., Cooper, M. P., Fan, M. L., Rohas, L. M., Zavacki, A. M., Cinti, S., Shulman, G. I., Lowell, B. B., Krainc, D., and Spiegelman, B. M. (2004) Cell 119 121-135 [DOI] [PubMed] [Google Scholar]
- 20.Leone, T. C., Lehman, J. J., Finck, B. N., Schaeffer, P. J., Wende, A. R., Boudina, S., Courtois, M., Wozniak, D. F., Sambandam, N., Bernal-Mizrachi, C., Chen, Z., Holloszy, J. O., Medeiros, D. M., Schmidt, R. E., Saffitz, J. E., Abel, E. D., Semenkovich, C. F., and Kelly, D. P. (2005) PLoS Biol. 3 672-687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lelliott, C. J., Medina-Gomez, G., Petrovic, N., Kis, A., Feldmann, H. M., Bjursell, M., Parker, N., Curtis, K., Campbell, M., Hu, P., Zhang, D., Litwin, S. E., Zaha, V. G., Fountain, K. T., Boudina, S., Jimenez-Linan, M., Blount, M., Lopez, M., Meirhaeghe, A., Bohlooly, Y., Storlien, L., Stromstedt, M., Snaith, M., Oresic, M., Abel, E. D., Cannon, B., and Vidal-Puig, A. (2006) PLoS Biol. 4 2042-2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonoda, J., Mehl, I. R., Chong, L. W., Nofsinger, R. R., and Evans, R. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5223-5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai, L., Leone, T. C., Zechner, C., Schaeffer, P. J., Kelly, S. M., Flanagan, D. P., Medeiros, D. M., Kovacs, A., and Kelly, D. P. (2008) Genes Dev. 22 1948-1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersson, U., and Scarpulla, R. C. (2001) Mol. Cell. Biol. 21 3738-3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vercauteren, K., Pasko, R. A., Gleyzer, N., Marino, V. M., and Scarpulla, R. C. (2006) Mol. Cell. Biol. 26 7409-7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vercauteren, K., Gleyzer, N., and Scarpulla, R. C. (2008) J. Biol. Chem. 283 12102-12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel, J. L., and Kristie, T. M. (2000) EMBO J. 19 683-690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moffat, J., Grueneberg, D. A., Yang, X., Kim, S. Y., Kloepfer, A. M., Hinkle, G., Piqani, B., Eisenhaure, T. M., Luo, B., Grenier, J. K., Carpenter, A. E., Foo, S. Y., Stewart, S. A., Stockwell, B. R., Hacohen, N., Hahn, W. C., Lander, E. S., Sabatini, D. M., and Root, D. E. (2006) Cell 124 1283-1298 [DOI] [PubMed] [Google Scholar]
- 29.Towbin, H., Staehelin, T., and Gordon, J. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 4350-4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moir, R. D., Montag-Lowy, M., and Goldman, R. D. (1994) J. Cell Biol. 125 1201-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weibel, E. R. (1969) Int. Rev. Cytol. 26 235-302 [DOI] [PubMed] [Google Scholar]
- 32.Smyth, G. K. (2004) Stat. Appl. Genet. Mol. Biol. 3 Article 3 [DOI] [PubMed]
- 33.Robinson, B. H. (1996) Methods Enzymol. 264 454-464 [DOI] [PubMed] [Google Scholar]
- 34.Cotney, J., Wang, Z., and Shadel, G. S. (2007) Nucleic Acids Res. 35 4042-4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adan, C., Matsushima, Y., Hernandez-Sierra, R., Marco-Ferreres, R., Fernandez-Moreno, M. A., Gonzalez-Vioque, E., Calleja, M., Aragon, J. J., Kaguni, L. S., and Garesse, R. (2008) J. Biol. Chem. 283 12333-12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Septinus, M., Berthold, T., Naujok, A., and Zimmermann, H. W. (1985) Histochemistry 82 51-66 [DOI] [PubMed] [Google Scholar]
- 37.Metivier, D., Dallaporta, B., Zamzami, N., Larochette, N., Susin, S. A., Marzo, I., and Kroemer, G. (1998) Immunol. Lett. 61 157-163 [DOI] [PubMed] [Google Scholar]
- 38.Marzluff, W. F., and Duronio, R. J. (2002) Curr. Opin. Cell Biol. 14 692-699 [DOI] [PubMed] [Google Scholar]
- 39.Puigserver, P. (2005) Int. J. Obesity 29 55-59 [DOI] [PubMed] [Google Scholar]
- 40.Gleyzer, N., Vercauteren, K., and Scarpulla, R. C. (2005) Mol. Cell. Biol. 25 1354-1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wysocka, J., Reilly, P. T., and Herr, W. (2001) Mol. Cell. Biol. 21 3820-3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bridge, A. J., Pebernard, S., Ducraux, A., Nicoulaz, A. L., and Iggo, R. (2003) Nat. Genet. 34 263-264 [DOI] [PubMed] [Google Scholar]
- 43.Sledz, C. A., and Williams, B. R. (2004) Biochem. Soc. Trans. 32 952-956 [DOI] [PubMed] [Google Scholar]
- 44.Zeng, Y., Yi, R., and Cullen, B. R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9779-9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doench, J. G., Petersen, C. P., and Sharp, P. A. (2008) Genes Dev. 17 438-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saxena, S., Jonsson, Z. O., and Dutta, A. (2003) J. Biol. Chem. 278 44312-44319 [DOI] [PubMed] [Google Scholar]
- 47.Valencia-Sanchez, M. A., Liu, J., Hannon, G. J., and Parker, R. (2006) Genes Dev. 20 515-524 [DOI] [PubMed] [Google Scholar]
- 48.St. Pierre, J., Lin, J., Krauss, S., Tarr, P. T., Yang, R. J., Newgard, C. B., and Spiegelman, B. M. (2003) J. Biol. Chem. 278 26597-26603 [DOI] [PubMed] [Google Scholar]
- 49.Uldry, M., Yang, W., St-Pierre, J., Lin, J., Seale, P., and Spiegelman, B. M. (2006) Cell Metab. 3 333-341 [DOI] [PubMed] [Google Scholar]
- 50.Butow, R. A., and Avadhani, N. G. (2004) Mol. Cell 14 1-15 [DOI] [PubMed] [Google Scholar]
- 51.Biswas, G., Guha, M., and Avadhani, N. G. (2005) Gene (Amst.) 354 132-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohas, L. M., St-Pierre, J., Uldry, M., Jager, S., Handschin, C., and Spiegelman, B. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7933-7938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallace, D. C. (1992) Annu. Rev. Biochem. 61 1175-1212 [DOI] [PubMed] [Google Scholar]
- 54.Wredenberg, A., Wibom, R., Wilhelmsson, H., Graff, C., Wiener, H. H., Burden, S. J., Oldfors, A., Westerblad, H., and Larsson, N. G. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15066-15071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park, C. B., sin-Cayuela, J., Camara, Y., Shi, Y., Pellegrini, M., Gaspari, M., Wibom, R., Hultenby, K., Erdjument-Bromage, H., Tempst, P., Falkenberg, M., Gustafsson, C. M., and Larsson, N. G. (2007) Cell 130 273-285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.