Abstract
Protein kinase D1 (PKD1) is a physiologically important signaling enzyme that is activated via protein kinase C-dependent trans-phosphorylation of the activation loop at Ser744 and Ser748 followed by PKD1 autophosphorylation at Ser916. Although PKD-Ser916 autophosphorylation is widely used to track cellular PKD activity, this study exposes conditions leading to increased PKD-Ser(P)916 immunoreactivity without an associated increase in PKD activity in cardiomyocytes that heterologously overexpress catalytically inactive PKD1 and in cardiomyocytes treated with Gö6976 (a PKD inhibitor that competes with ATP). In each case, PKD1 is detected as a Ser916-phosphorylated enzyme that lacks kinase activity. In vitro kinase assays reconcile these seemingly discrepant findings by demonstrating that PKD1-Ser916 autophosphorylation can proceed via either an intermolecular reaction or an intramolecular autophosphorylation that requires only very low ATP concentrations that do not support target substrate phosphorylation. Additional studies show that Ser744 and Ser748 are targets for a protein kinase C-independent autocatalytic phosphorylation and that the PKD1-S744A/S748A mutant is a Ser916-phosphorylated enzyme that is not active toward heterologous substrates. In contrast, PKD1-S916A is an active kinase that autophosphorylates at Ser744. However, the S916A substitution leads to a Ser748 phosphorylation defect and a prolonged cellular PKD1 signaling response. Collectively, these results implicate PKD1-Ser744 phosphorylation in the phorbol 12-myristate 13-acetate-dependent mechanism that increases PKD1 activity toward physiologically relevant substrates. We show that PKD1-Ser916 autophosphorylation does not necessarily correlate with PKD1 activity. Rather, autophosphorylation at Ser916 is required for subsequent autophosphorylation at Ser748. Finally, this study exposes a novel role for Ser916 and/or Ser748 autophosphorylation to terminate the cellular PKD1 signaling response.
Protein kinase D1 (PKD1)2 is the founding member of a family of three related serine/threonine kinases that share a similar structural architecture and control a large number of biological processes that influence cell growth, differentiation, migration, and apoptosis (1, 2). PKDs have an N-terminal regulatory domain containing tandem cysteine-rich C1A/C1B domains that bind diacylglycerol-/phorbol ester-enriched membranes with high affinity and a pleckstrin homology (PH) domain that participates in protein-protein interactions. PH domain-dependent autoinhibitory intramolecular interactions maintain the enzyme in an inactive state, with low basal activity, in resting cells. PKD isoforms are activated by agonists that promote diacylglycerol accumulation and activate novel PKC (nPKC) isoforms at membranes. nPKCs activate PKD isoforms by phosphorylating a pair of highly conserved serine residues (Ser744/Ser748 in PKD1, nomenclature based upon rodent sequence) in the kinase domain activation loop. This post-translational modification relieves autoinhibition and stabilizes the activation loop in a conformation that is optimized for catalysis. PKD1 then undergoes a series of autophosphorylation reactions at a cluster of serine residues at Ser205/Ser208 and Ser219/Ser223 in the regulatory C1A/C1B interdomain region and at Ser916 at the extreme C terminus. These autophosphorylation reactions create docking sites for PKD1 binding partners and influence PKD1 localization within the cell (3, 4). A recent study also identified PKD1 autophosphorylation at the activation loop (primarily at Ser748) during the chronic phase of PKD1 activation in bombesin-treated COS-7 cells (5); the relative roles of autocatalytic versus PKC-dependent activation loop phosphorylation in other cellular contexts has never been considered.
PKD has emerged as a physiologically important signaling enzyme in many cell types. However, the list of known PKD substrates remains relatively short. We and others recently implicated PKD as a CREB-Ser133 kinase that regulates Cre-dependent transcriptional responses (6, 7). PKD also functions as a physiologically relevant HDAC5 kinase (8). HDAC5 is a signal-responsive repressor of pathological cardiac remodeling (9, 10). PKD neutralizes the antihypertrophic actions of HDAC5, leading to the activation of a pathologic gene program that culminates in cardiac hypertrophy and ventricular remodeling. PKD also phosphorylates cardiac troponin I (cTnI), the “inhibitory” subunit of the troponin complex that “fine-tunes” myofilament function to hemodynamic load. cTnI contains three functionally distinct phosphorylation clusters at Ser23/Ser24, Ser43/Ser45, and Thr144. PKD-dependent cTnI phosphorylation has been mapped to Ser23/Ser24, a modification that desensitizes the myofilament to Ca2+ and leads to functionally important changes in contractile performance (2, 11, 12). Finally, PKD phosphorylates heat shock protein 27 (HSP27); a PKD-HSP27 phosphorylation pathway has been implicated in the vascular endothelial growth factor-dependent angiogenic response (13, 14).
PKD1 phosphorylation at Ser916 is viewed as an obligatory autocatalytic reaction. Immunoblotting with a phosphorylation site-specific antibody (PSSA) that recognizes PKD-Ser916 phosphorylation is widely used as a convenient surrogate method to track PKD activity (in place of more cumbersome direct enzyme activity measurements). This approach is based upon early studies showing that phorbol ester-dependent PKD activation is accompanied by an increase in PKD-Ser916 phosphorylation, that constitutively active forms of PKD1 (such as the PH domain-deleted or S744E/S748E-substituted mutants) are constitutively Ser916-phosphorylated under resting conditions in several cell types, and that catalytically inactive PKD1-D733A and K618M mutants display a Ser916 phosphorylation defect in some experimental models (15). The notion that PKD-Ser916 phosphorylation faithfully reports PKD activity in cells has generally remained unchallenged, although the literature is littered with isolated reports hinting that this conclusion may not necessarily be valid under all experimental conditions (16–18). This study exposes differences in the control of PKD1-Ser916 phosphorylation and PKD1 activity in cardiomyocytes, further undermining the assumption that PKD-Ser916 immunoreactivity faithfully reports PKD activation in all cellular contexts. We also identify a PKD1 activation loop Ser744 and Ser748 autophosphorylation mechanism that might assume functional importance when nPKC isoforms are down-regulated. Finally, we identify a novel role for PKD1-Ser916 autophosphorylation to prime PKD1 for a subsequent autophosphorylation reaction at Ser748. Although autophosphorylation reactions at Ser748 and/or Ser916 are not required for PMA-dependent PKD1 activation, this study exposes a novel role for Ser916 and/or Ser748 autophosphorylation to limit the duration of the cellular PKD1 signaling response.
EXPERIMENTAL PROCEDURES
Materials—Antibodies were from the following sources: PKD1-Ser(P)744/Ser(P)748, PKD1-Ser(P)916, PKD1, CREB-Ser(P)133, ERK, cardiac troponin I-Ser(P)23/Ser(P)24, and HSP27-Ser(P)82 from Cell Signaling Technology; PKD1-Ser(P)742 (numbering based upon human sequence, corresponding to rodent PKD1-Ser748) from Abcam; CREB-MBP fusion protein from Invitrogen; and syntide-2 from Sigma. Recombinant His6-tagged full-length PKD1 expressed in Sf21 cells and purified using Ni2+/nitrilotriacetic acid-agarose enzyme was purchased from Upstate Biotechnology, Inc.; the purity of this preparation (assessed by SDS-PAGE and Coomassie Blue staining) is 73%. PMA and PDBu were from Sigma. Other chemicals were reagent grade.
Cardiomyocyte Culture—Cardiomyocytes were isolated from hearts of 2-day-old Wistar rats by a trypsin dispersion procedure that uses a differential attachment procedure followed by irradiation to enrich for cardiomyocytes (19). Cells were plated on protamine sulfate-coated culture dishes at a density of 5 × 106 cells/100-mm dish and grown in minimum Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum for 4 days and then serum-deprived for 24 h prior to experiments.
Adenoviral Infections—Cardiomyocytes were infected with adenoviral constructs that drive expression of HA-tagged kinase-dead PKD1 (KD-PKD1, generously provided by Drs. Terry Rogers and William Randall, University of Maryland) or β-galactosidase as a control. Infections were performed on cultures grown for 5 days in minimum Eagle's medium supplemented with 10% fetal calf serum (with protein extracts prepared 48 h following infections) according to methods described previously (6).
PKD1 Mutants—Plasmids that drive expression of HA-tagged WT-PKD1, PKD1-SS/EE (harboring phosphomimetic substitutions at Ser744/Ser748 in the activation loop), PKD1-SS/AA (harboring nonphosphorylatable alanines at Ser744/Ser748 in the activation loop), PKD1-ΔPH (PH domain deletion), PKD1-ΔC1 (an N-terminal deletion mutant that lacks the C1 domain), and PKD1-S916A generated by the Toker laboratory were obtained from Addgene. A plasmid that drives expression of GFP-tagged full-length PKD1 was generously provided by Drs. David E. Clapham and Elena Oancea and has been described previously (20). PKD1 expression plasmids were introduced into COS-7 or HEK293 cells by Effectene transfection reagent (Qiagen) according to the instruction manual. Cells were grown for 24 h and lysed in RIPA buffer containing 1 mm sodium orthovanadate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml benzamidine, 0.5 mm phenylmethylsulfonyl fluoride, 5 μm pepstatin A, and 0.1 μm calyculin. Cell extracts were subjected to immunoprecipitation with anti-HA tag-agarose (Roche Applied Science).
In Vitro Kinase Assays—In vitro kinase assays were performed with 0.1 μg of recombinant human PKD1 or with PKD1 immunoprecipitated from 150 μg of starting cell extract. Incubations were performed in 110 μl of a reaction buffer containing 30 mm Tris-Cl, pH 7.5, 5.45 mm MgCl2, 0.65 mm EDTA, 0.65 mm EGTA, 0.1 mm dithiothreitol, 1.09 mm sodium orthovanadate, 0.1 μm calyculin, 0.55 μm protein kinase inhibitor, 217 mm NaCl, 3.6% glycerol, and [γ-32P]ATP (10 μCi, 66 μm, unless indicated otherwise). The reaction buffer used for in vitro kinase assays depicted in Figs. 3, 4, 5, 6, 7 and Fig. 9 contained 89 μg/ml phosphatidylserine (PS) and 175 nm PMA. Assay buffer used in Fig. 8 contained 89 μg/ml PS plus 175 nm PMA, 89 μg/ml PS plus 200 nm PDBu, or 30 μg/ml dextran sulfate as indicated. Assays contained either 4 μg of troponin complex (consisting of equimolar concentrations of cardiac troponin I, cardiac troponin T, and cardiac troponin C, generously provided by Drs. John Solaro and Marius Sumandea) or recombinant human CREB-maltose binding protein fusion construct (CREB-MBP, 1 μg per assay, BIOSOURCE). Incubations were for 30 min at 30 °C. PKD1 autophosphorylation was tracked with the Cell Signaling Technology anti-PKD1-Ser(P)744/Ser(P)748 (which is reported to track primarily Ser744 phosphorylation), the Cell Signaling Technology anti-PKD1-Ser(P)916 PSSA, and the Abcam anti-PKD1-Ser(P)742 antibody (which was recently characterized as relatively selective for Ser742 in human PKD1, corresponding to Ser748 in rodent PKD1 (5)).
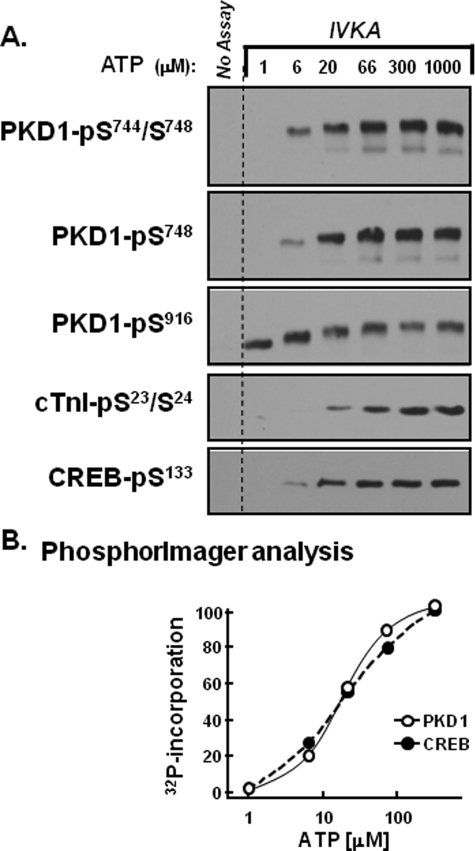
FIGURE 3.
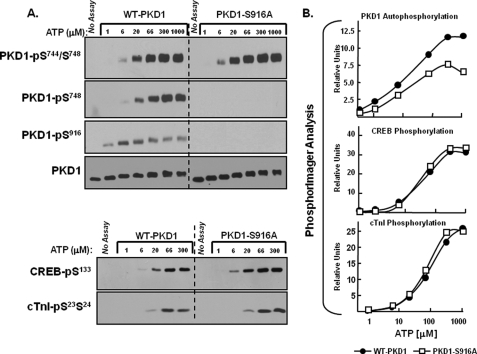
Distinct ATP requirements for PKD1 autophosphorylation at Ser916 and Ser744/Ser748. 0.1 μg of recombinant human PKD1 (Upstate Biotechnology, Inc.) was subjected to in vitro kinase assays (IVKA) according to “Experimental Procedures.” Assays were performed in parallel with either a recombinant human CREB-maltose-binding protein fusion construct (CREB-MBP, 1 μg per assay, BIOSOURCE) or 4 μg of troponin complex (consisting of equimolar cTnI, cTnC, and cTnT) as substrate. A, depicts Western blots for PKD1-Ser744/Ser748, PKD1-Ser748, PKD1-Ser916, CREB-Ser133, and cTnI-Ser23/Ser24 phosphorylation. PKD1 activity also was tracked by PhosphorImager analysis as 32P incorporation into PKD1 or CREB and is expressed as a percent of maximal activity at 300 μm ATP in B. Results for PKD1 autophosphorylation were identical in assays without and with CREB or cTnI. Results are from a single experiment and are representative of data obtained in five separate kinase assays.
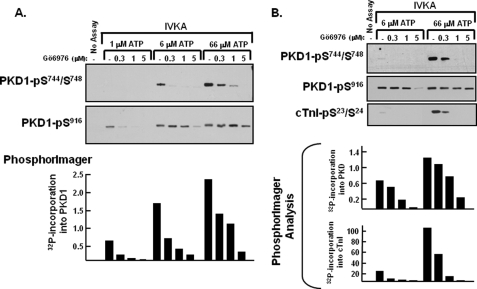
FIGURE 4.
Gö6976 inhibits PKD1-Ser744/Ser748 autophosphorylation and PKD1 phosphorylation of cTnI, without inhibiting PKD1-Ser916 autophosphorylation. In vitro kinase assays (IVKAs) were performed with the indicated concentrations of ATP and Gö6976. All assays contained tracer amounts of [γ-32P]ATP to track 32P incorporation into PKD1 and cTnI, which is quantified in the bar graphs. The results were replicated in three separate experiments.
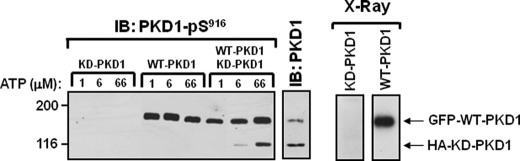
FIGURE 5.
Distinct ATP requirements for WT-PKD1-Ser916 autophosphorylation and KD-PKD1-Ser916 phosphorylation in trans by WT-PKD1. GFP-tagged WT-PKD1 and HA-tagged KD-PKD1 were heterologously overexpressed in COS-7 cells, immunoprecipitated, and subjected to in vitro kinase assays with varying amounts of ATP. All assays contained tracer amounts of [γ-32P]ATP to track 32P incorporation into WT-PKD1 or KD-PKD1 and to screen for possible co-precipitating contaminating proteins. Proteins were resolved by SDS-PAGE, subjected to autoradiography, and probed for PKD1 and PKD1-Ser(P)916 immunoreactivity according to “Experimental Procedures.” Similar results were obtained in three separate experiments. IB, immunoblot.
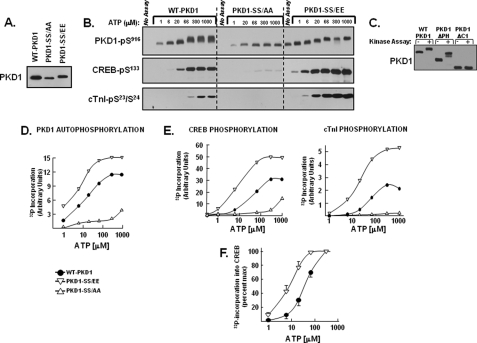
FIGURE 6.
PKD1 activity is regulated by activation loop phosphorylation. WT-PKD1, PKD1-S744E/S748E (PKD1-SS/EE), PKD1-S744A/S748A (PKD1-SS/AA), PKD1-ΔPH, and PKD1-ΔC1 enzymes heterologously overexpressed in HEK293 cells were immunoprecipitated for kinase assays with buffers containing the indicated ATP concentrations (and tracer amounts of [γ-32P]ATP) and either CREB or troponin complex as substrate according to “Experimental Procedures.” A, immunoblotting for levels of WT-PKD1, PKD1-SS/EE, and PKD1-SS/AA transgene expression. B, immunoblotting to track WT-PKD1, PKD1-SS/AA, PKD1-SS/EE autophosphorylation at Ser916 as well as PKD1 phosphorylation of either CREB or cTnI (which were examined in separate kinase assays performed in parallel with either CREB or troponin complex as substrate). PKD1 autophosphorylation was identical without or with either substrate. C, immunoblotting to track mobility shifts resulting from in vitro phosphorylation of WT-PKD1, PKD1-ΔPH, or PKD1-ΔC1; kinase assays were performed with buffers containing 66 μm ATP. D and E, quantification by PhosphorImager of 32P incorporation into WT-PKD1 or PKD1 mutants harboring activation loop substitutions (D) or 32P incorporation into CREB or cTnI (E). The results are from the experiment depicted in B, with data replicated in three separate kinase assays. F, dose-response curves for 32P incorporation into CREB by WT-PKD1 and PKD1-S744E/S748E. Results represent averaged data from three separate experiments and are normalized to correct for differences in maximal activity. The figure shows that S744E/S748E substitutions induce a significant left shift in the ATP requirement for CREB phosphorylation (p < 0.05, by analysis of variance).
FIGURE 7.
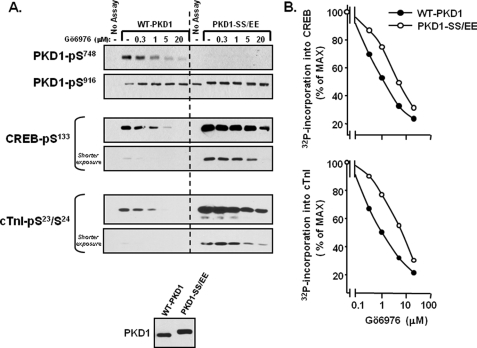
PKD1-S744E/S748E is refractory to inhibition by Gö6976, compared with WT-PKD1. WT-PKD1 and PKD1-S744E/S748E (PKD1-SS/EE) were heterologously overexpressed in HEK293 cells and then immunoprecipitated. Equal amounts of WT-PKD1 and PKD1-SS/EE enzyme were recovered (A, bottom) and used in kinase assays containing 50 μm ATP (and tracer amounts of [γ-32P]ATP) without and with the indicated concentrations of Gö6976 and either CREB or troponin complex as substrate. Immunoblotting for PKD1, CREB, and cTnI phosphorylation is depicted in A. Because CREB-Ser133 and cTnI-Ser23/Ser24 phosphorylation is considerably higher in assays with PKD1-S744E/S748E than in assays with WT-PKD1, shorter exposures are included to show Gö6976 inhibition of PKD1-S744E/S748E-dependent CREB-Ser133 and cTnI-Ser23/Ser24 phosphorylation. B, PhosphorImager analysis was performed to quantify 32P incorporation into CREB and cTnI. Results are normalized to correct for differences in maximal 32P incorporation into CREB and cTnI in assays with WT-PKD1 and PKD1-SS/EE. Results are from a single experiment and were replicated in two other experiments.
FIGURE 9.
The PKD1-S916A mutant is an active enzyme that autophosphorylates at Ser744, but not Ser748. WT-PKD1 and PKD1-S916A were heterologously overexpressed in HEK293 cells and then immunoprecipitated for kinase assays in buffers containing the indicated concentrations of ATP (and tracer amounts of [γ-32P]ATP) according to “Experimental Procedures.” Immunoblot analysis of PKD1 protein expression and PKD1, CREB-Ser133, and cTnI-pSer23/Ser24 phosphorylation are depicted in A, with the results of this experiment quantified in B. The result was replicated in two separate experiments.
FIGURE 8.
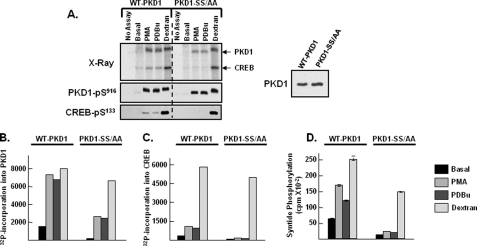
PKD1-S744A/S748A enzyme autophosphorylates at Ser916, but it does not effectively phosphorylate target substrates. WT-PKD1 and PKD1-S744A/S748A (PKD1-SS/AA) were heterologously overexpressed in HEK293 cells and then immunoprecipitated for kinase assays in buffers containing 50 μm ATP, without or with PMA, PDBu, or dextran sulfate according to “Experimental Procedures.” Kinase assays were performed in parallel with CREB or syntide-2 as substrate. A, left, autoradiography showing WT-PKD1 and PKD1-SS/AA autophosphorylation and phosphorylation of CREB (top) as well as immunoblotting for PKD1-Ser916 and CREB-Ser133 phosphorylation (bottom). A, right, immunoblotting to show that the recovery of WT-PKD1 and PKD1-SS/AA enzymes was similar. B and C, 32P incorporation into PKD or CREB was quantified by PhosphorImager. The data are from the same experiment depicted in A; similar results were obtained in a separate experiment. D, PKD1-dependent syntide-2 phosphorylation was examined in triplicate according to “Experimental Procedures”; the data represent the results of a single experiment, with identical results obtained in two separate experiments.
Syntide-2 kinase assays were performed in a similar manner, with 0.36 mg/ml syntide-2 included as the PKD1 substrate. Assays were terminated by adding 30 μl of 350 mm phosphoric acid followed by centrifugation at 14,000 × g for 10 min. 50 μl of each supernatant was spotted onto phosphocellulose filter papers (P-81), dropped into 75 mm phosphoric acid, washed (three times for 5 min), dried, and counted for radioactivity.
RESULTS
KD-PKD1 Is Phosphorylated in Trans at Ser916—Cardiomyocytes that heterologously overexpress KD-PKD1 at levels ∼8-fold higher than endogenous PKD1 were treated with PMA (a direct activator of PKD and phorbol ester-sensitive PKC isoforms) as an initial strategy to resolve PKD1 phosphorylation via an obligate intramolecular autophosphorylation reaction from PKD1 phosphorylations in trans by some other cellular Ser/Thr kinase. PKD1 phosphorylation was tracked by immunoblot analysis with a PSSA that recognizes phosphorylation at Ser916 and two PSSAs that recognize phosphorylation at the activation loop. We used a PSSA from Cell Signaling Technology (Cell Signaling Technology) that is designated anti-PKD-Ser(P)744/Ser(P)748. This PSSA was raised against a peptide phosphorylated on serines equivalent to Ser744 and Ser748 in rodent PKD1. However, the Rozengurt laboratory has reported that this PSSA primarily recognizes PKD1-Ser744 phosphorylation. We also used an Abcam anti-PKD1-Ser(P)748 PSSA (nomenclature based upon rodent sequence) that preferentially recognizes PKD1 phosphorylation at Ser748 (5). Preliminary experiments validated the specificity of this reagent, showing that the Abcam anti-PKD1-Ser(P)748 PSSA specifically recognizes PMA-dependent phosphorylation of WT-PKD1, but it does not recognize the PMA-activated PKD1-S744A/S748A or PKD1-S744E/S748E enzymes (data not shown). Conditions were optimized so that phosphorylation of both endogenous PKD and the KD-PKD1 transgene could be detected in a single experiment by varying gel exposure.
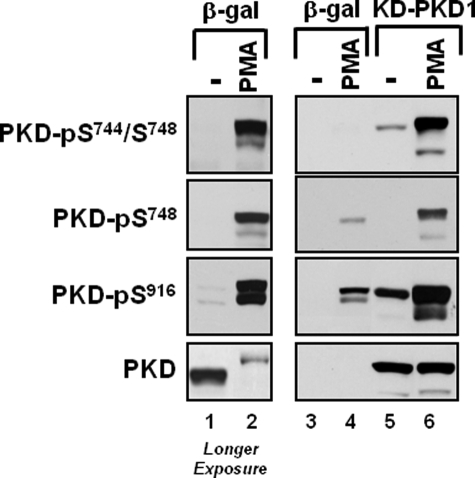
Fig. 1 shows that PMA treatment leads to a robust increase in endogenous PKD phosphorylation that is recognized by the Cell Signaling Technology anti-PKD1-Ser(P)744/Ser(P)748, the Abcam anti-PKD1-Ser(P)748, and the anti-PKD1-Ser(P)916 PSSAs (lanes 1 and 2). The PMA-activated PKD1 enzyme also migrates more slowly in SDS-PAGE. Heterologously overexpressed KD-PKD1 also is recognized by the anti-PKD-Ser(P)744/Ser(P)748 and anti-PKD1-Ser(P)916 PSSAs in resting cardiomyocytes (Fig. 1, lane 5). This cannot be dismissed as an effect of these PSSAs to recognize KD-PKD1 in a phosphorylation-independent manner at high levels of transgene overexpression, because anti-PKD1-Ser(P)744/Ser(P)748 and anti-PKD1-Ser(P)916 immunoreactivity increases further (to a level that exceeds immunoreactivity in basal Ad-KD-PKD1 cultures or PMA-treated Ad-β-gal cultures) when Ad-KD-PKD1 cultures are treated with PMA (Fig. 1, lane 6). Because KD-PKD1 cannot execute a cis autophosphorylation reaction, KD-PKD1 must be phosphorylated at Ser744/Ser748 and Ser916 in trans by an endogenous cellular Ser/Thr kinase.
FIGURE 1.
PMA increases KD-PKD1-Ser916 phosphorylation in cardiomyocytes. Cardiomyocytes were infected with Ad-β-gal or Ad-KD-PKD1 and then challenged with vehicle or 300 nm PMA. Cell extracts were subjected to immunoblotting for PKD protein and PKD phosphorylation. Lanes 1 and 2 depict a longer exposure time for the data presented in lanes 3 and 4, so that the phosphorylation of endogenous PKD can be detected in Ad-β-gal cultures. Results were replicated in three separate experiments. It should be noted that the anti-PKD-Ser(P)744/Ser(P)748 PSSA used in this and subsequent figures preferentially recognizes PKD1 phosphorylation at Ser744 (numbering based upon rodent sequence, corresponding to human PKD1-Ser738). Ser748 phosphorylation is tracked with the anti-PKD-Ser748 PSSA.
A comparison of KD-PKD1 phosphorylation at individual activation loop sites also is revealing. Fig. 1 shows that the Cell Signaling Technology anti-PKD1-Ser(P)744/Ser(P)748 PSSA detects a low level of KD-PKD1 phosphorylation in resting cardiomyocytes; KD-PKD1-Ser744/Ser748 phosphorylation is markedly increased by PMA. In contrast, the anti-PKD1-Ser(P)748 PSSA detects KD-PKD1 in PMA-treated, but not resting, cardiomyocytes. Although anti-PKD1-Ser(P)748 immunoreactivity is higher in PMA-treated Ad-KD-PKD1 cultures than in PMA-treated Ad-β-gal cultures, the increment in anti-PKD1-Ser(P)748 immunoreactivity because of KD-PKD1 overexpression is quite modest, given the high level of transgene overexpression (Fig. 1, compare lanes 6 and 4). These results provided the first hint that PKD1 phosphorylation at Ser744 and Ser748 might be controlled via distinct molecular mechanisms (and that an autophosphorylation mechanisms is required for optimal PKD1 phosphorylation at Ser748 but not Ser744).
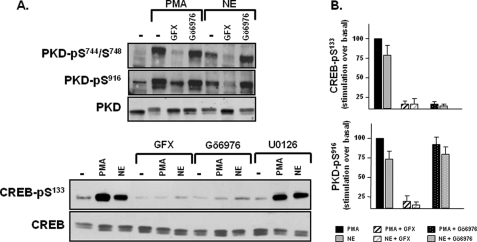
Gö6976 Does Not Block PKD-Ser916 Phosphorylation in Vivo in Cardiomyocytes—PKD-Ser916 phosphorylation mechanisms also were interrogated with pharmacologic inhibitors. We recently reported that PKD is a PKCδ-activated enzyme that functions downstream from α1-adrenergic receptors as a CREB-Ser133 kinase in cardiomyocytes (6). Fig. 2A replicates some of the findings from the previous study, showing that the α1-adrenergic receptor agonist norepinephrine (NE) and PMA promote PKD phosphorylation at Ser744/Ser748 and Ser916, and this is associated with an increase in CREB phosphorylation at Ser133. NE- and PMA-dependent CREB-Ser133 phosphorylation is inhibited by GF109203X, a PKC inhibitor that prevents PKC-dependent PKD activation (but does not directly inhibit PKD activity). NE- and PMA-dependent CREB-Ser133 phosphorylation also is inhibited by Gö6976, a PKD inhibitor that does not inhibit nPKC isoforms. In contrast, whereas previous studies demonstrated that NE and PMA activate p90 ribosomal S6 kinase via a MEK-ERK-dependent pathway that is abrogated by the MEK inhibitor U0126, and p90 ribosomal S6 kinase has been implicated as a CREB-Ser133 kinase in many cell types, NE- and PMA-dependent CREB-Ser133 phosphorylation is not blocked by U0126 in cardiomyocytes (Fig. 2A) (6).
FIGURE 2.
Gö6976 blocks PKD-dependent CREB-Ser133 phosphorylation without blocking PKD1 phosphorylation at Ser916. Cardiomyocytes were pretreated for 45 min with GF109203X (GFX), Gö6976, or U0126 (each at 5 μm) prior to stimulation with NE (1 μm) or PMA (300 nm) as indicated. Cell extracts were subjected to immunoblot analysis for PKD and CREB phosphorylation, with immunoblot analysis for PKD and CREB protein expression provided as a loading control. A depicts immunoblotting data that are representative of results obtained in four separate experiments on different culture preparations. Results from the group of experiments are expressed as stimulation over basal, normalized to the stimulatory response to PMA, and quantified in B (n = 4).
Fig. 2 shows that the PKC inhibitor GF109203X prevents agonist-dependent PKD phosphorylation at Ser744/Ser748 and Ser916, consistent with the notion that PKD activation is initiated by PKC-dependent PKD-Ser744/Ser748 phosphorylation mechanism that increases PKD activity and leads to subsequent PKD autophosphorylation at Ser916.Gö6976 does not block NE- or PMA-dependent PKD-Ser744/Ser748 phosphorylation, as predicted by prevailing models that attribute PKD-Ser744/Ser748 phosphorylation to an nPKC isoform; nPKC isoforms are not inhibited by Gö6976 (6). However, Fig. 2 provides surprising evidence that a biologically relevant concentration of Gö6976, which inhibits PKD-dependent trans-phosphorylation of CREB at Ser133, does not inhibit PKD-Ser916 phosphorylation. This result was unanticipated; we had planned to use immunoblotting for PKD-Ser916 phosphorylation as a convenient method to assess the efficacy of PKD inhibition by Gö6976. The divergent controls for PKD-Ser916 and CREB-Ser133 phosphorylation identified in our study argue that PKD-Ser916 phosphorylation does not necessarily provide a valid read-out of PKD activity in Gö6976-treated cardiomyocytes.
In Vitro Kinase Assays Identify Distinct ATP Requirements for PKD1 Autophosphorylation at Ser916 and PKD1 Phosphorylation of Heterologous Substrates—Because Gö6976 acts as a competitive inhibitor of ATP, we reasoned that a PKD1-Ser916 autophosphorylation reaction might be relatively resistant to inhibition by Gö6976 if its ATP requirement was low, relative to the ATP requirement for PKD-dependent phosphorylation of CREB. We compared the ATP requirements for PKD1 autophosphorylation at Ser916 and PKD1 phosphorylation of target substrates in trans to test this hypothesis. In vitro kinase assays were performed in parallel with either CREB or cTnI as substrate to examine PKD1 activity toward two physiologically relevant substrates. Immunoblot analysis was performed with PSSAs that track PKD1-Ser916 autophosphorylation, PKD1-dependent CREB-Ser133 may contribute to PKD1 activation loop autophosphorylation (5), PKD1 autophosphorylation also was examined using the Cell Signaling Technology anti-PKD-Ser(P)744/Ser(P)748 PSSA that preferentially recognizes phosphorylation at Ser744 and the Abcam anti-PKD-Ser(P)748 PSSA.
Fig. 3A shows that PKD1 executes autophosphorylation reactions at the activation loop (at both Ser744 and Ser748) and at Ser916 with markedly different ATP requirements. PKD-Ser916 autophosphorylation is detected at 1 μm ATP; the threshold concentration for PKD-Ser916 autophosphorylation is ∼0.3 μm (data not shown). PKD-Ser916 autophosphorylation does not increase at higher ATP concentrations, although the anti-PKD-Ser(P)916 PSSA detects a pronounced phosphorylation-dependent mobility shift as the ATP concentration increases to 6–66 μm. PKD autophosphorylations at Ser744 or Ser748 are not detected at 1 μm ATP; PKD1-Ser744 and Ser748 autophosphorylations are detected at 6 μm ATP and maximal at 66 μm ATP. These results suggest that the ATP requirement for PKD autophosphorylation at Ser916 is considerably lower than the ATP requirement for PKD autophosphorylation at Ser744 or Ser748. However, the apparent differences in PKD1 autophosphorylation at Ser916 versus Ser744/Ser748 in Fig. 3A could be a feature of the immunoblotting method, if the PSSAs used in our study recognize individual phosphorylation reactions with markedly different affinities. Therefore, the in vitro kinase assays were performed with buffers containing [32P]ATP, and 32P incorporation into PKD1 was quantified by PhosphorImager (a highly sensitive detection method that is not undermined by the uncertainties inherent in immunoblotting with a panel of PSSAs). Fig. 3B shows that overall PKD1 autophosphorylation (detected as 32P incorporation into PKD1) increases in a dose-dependent manner as the ATP concentration is increased from 6 to 200 μm. The observation that 1 μm ATP leads to maximal PKD1-Ser916 phosphorylation but only a very low level of 32P incorporation into PKD1 provides unambiguous evidence that PKD1-Ser916 phosphorylation is via a mechanism that is distinct from the mechanism that controls autophosphorylation at other sites in the enzyme.
The ATP requirements for PKD1 phosphorylation of CREB and cTnI were examined in parallel, to determine whether substrate phosphorylation correlates with overall PKD1 autophosphorylation (detected as 32P incorporation into the protein) or PKD1 autophosphorylation at Ser916. The immunoblotting studies in Fig. 3A show that 1 μm ATP does not support PKD1-dependent CREB-Ser133 or cTnI-Ser23/Ser24 phosphorylation. PKD1 induces a dose-dependent increase in CREB phosphorylation as the ATP concentration increases from 6 to 66 μm; cTnI phosphorylation is detected at 20 μm and is maximal at 300 μm. Fig. 3B shows that overall CREB phosphorylation (detected as 32P incorporation into CREB) also increases in a dose-dependent manner as the ATP concentration increases from 6 to 200 μm. The observation that PKD1 substrate phosphorylation correlates with overall PKD1 autophosphorylation (measured as 32P incorporation into PKD1), but not with PKD1-Ser916 autophosphorylation, indicates that the Ser916-phosphorylated form of PKD1 is not necessarily active toward target substrates.
A PKD1 autophosphorylation reaction that requires only limiting levels of ATP is predicted to be relatively refractory to inhibition by Gö6976. This prediction was tested directly by comparing Gö6976 inhibition of PKD1 autophosphorylation at Ser916 (detected by immunoblot analysis) with Gö6976 inhibition of overall PKD1 autophosphorylation (detected as 32P incorporation into PKD1 by PhosphorImager). Fig. 4A (bottom) shows that 32P incorporation into PKD1 is quite low at 1 μm ATP; 32P incorporation into PKD1 increases progressively (in parallel with an increase in PKD-Ser744/Ser748 phosphorylation; Fig. 4A, top) as the ATP concentration is increased to 66 μm. At 66 μm ATP, 32P incorporation into PKD1 and PKD1-Ser744/Ser748 autophosphorylation are inhibited in a dose-dependent manner by Gö6976; a high level of inhibition is evident at 1–5 μm Gö6976. In contrast, PKD1-Ser916 autophosphorylation is similar at 1, 6, and 66 μm ATP, and it is relatively resistant to inhibition by Gö6976. 1–5 μm Gö6976 does not block PKD1-Ser916 autophosphorylation at 66 μm ATP. PKD-Ser916 autophosphorylation at 6 μm ATP is blocked by Gö6976 but only at a high inhibitor concentration. An effect of lower Gö6976 concentrations to inhibit PKD1-Ser916 autophosphorylation is detected only when the ATP concentration is reduced to 1 μm.
We next examined whether PKD1-Ser916 autophosphorylation remains relatively refractory to inhibition by Gö6976 when kinase assays are performed with a heterologous substrate such as cTnI. Fig. 4B shows that PKD1-Ser744/Ser748 autophosphorylation and PKD1 phosphorylation of cTnI are detected at 66 μm ATP and at a much lower level at 6 μm ATP. These phosphorylation reactions (that are impaired when the ATP concentration is reduced to 6 μm) are effectively inhibited by 1–5 μm Gö6976. In contrast, PKD1-Ser916 autophosphorylation is blocked only by a high Gö6976 concentration when assays are performed with 6 μm ATP. Similar results were obtained in assays with CREB as substrate (data not shown). Collectively, these studies provide novel evidence that the ATP requirements for PKD1 autophosphorylation at the activation loop (Ser744 and Ser748) and PKD1 trans-phosphorylation of heterologous substrates such as cTnI or CREB are considerably higher than the ATP requirement for PKD1-Ser916 autophosphorylation.
PKD1 autophosphorylation can be mediated either by a cis intramolecular reaction or a intermolecular phosphorylation in trans. GFP-tagged WT-PKD1 and HA-tagged KD-PKD1 (which have different mobilities and can be resolved by SDS-PAGE) were heterologously overexpressed in COS-7 cells, immunoprecipitated, and subjected to in vitro kinase assays with varying amounts of ATP to determine whether the ATP requirements for the cis WT-PKD1-Ser916 autophosphorylation reaction and WT-PKD1 phosphorylation of KD-PKD1 (an obligate trans-phosphorylation) can be distinguished. In vitro kinase assays were performed with buffers containing [32P]ATP to examine whether the KD-PKD1 enzyme preparation might contain contaminants with kinase activity. Fig. 5 shows that assays with KD-PKD1 do not lead to any 32P incorporation into KD-PKD1 or possible co-precipitating proteins; KD-PKD-Ser916 phosphorylation also is not detected by immunoblot analysis under these conditions. In contrast, kinase assays with the WT-PKD1 preparation results in the appearance of a single prominent radiolabeled band that co-migrates with the WT-PKD1 enzyme; no contaminating enzymes or substrates are detected in the WT-PKD1 preparation. WT-PKD1-Ser916 autophosphorylation is detected in assays with 1 μm ATP. WT-PKD1 also phosphorylates KD-PKD1 at Ser916 in trans, but this is detected only when the ATP concentration is increased to 6–66 μm. The failure to detect an intermolecular KD-PKD1 phosphorylation by WT-PKD1 at 1 μm ATP suggests that the very efficient WT-PKD1-Ser916 autophosphorylation at 1 μm ATP is mediated by a cis intramolecular autocatalytic reaction.
Autophosphorylation of PKD1-S744E/S748E and PKD1-S744A/S748A Enzymes—The in vitro controls of PKD1 activity identified in our studies conflict with two of the major prevailing assumptions regarding the mechanisms that contribute to PKD1 activation. First, conventional models view PKD1 phosphorylation as a hierarchical process that is initiated by a phosphorylation reaction at the activation loop that primes the enzyme for a subsequent autophosphorylation at Ser916. Our results showing that PKD1 executes a Ser916 autophosphorylation reaction at 1 μm ATP, under conditions that do not support (and do not require) activation loop autophosphorylation, are at odds with this formulation. Second, although there is evidence that activation loop phosphorylation is required for PMA-dependent PKD1 activation in a cellular context, previous studies with a PKD1-S744A/S748A mutant were interpreted as evidence that activation loop phosphorylation is not required for in vitro PKD1 activity (21). This conclusion is surprising, based upon a substantial literature that implicates activation loop phosphorylation as a modification that is essential for the catalytic competence of other Ser/Thr kinases. Our observation that PKD1 activity toward target substrates such as CREB and cTnI increases in parallel with an increase in PKD1 activation loop autophosphorylation (Fig. 3) provided the rationale to revisit studies that consider a possible role for activation loop phosphorylation in the control of PKD1 catalytic activity.
We performed in vitro kinase assays with WT-PKD1, PKD1-S744E/S748E, and PKD1-S744A/S748A to directly examine the role of activation loop phosphorylation in the mechanism leading to PKD1-Ser916 autophosphorylation and PKD1 phosphorylation of target substrates in trans. Fig. 6A shows that WT-PKD1, PKD1-S744E/S748E, and PKD1-S744A/S748A were heterologously overexpressed in HEK293 cells at similar levels. PKD1-S744E/S748E is recovered from resting HEK293 cells as a constitutively Ser916-phosphorylated enzyme that undergoes little-to-no further increase in Ser916 phosphorylation when subjected to in vitro kinase assays (Fig. 6B). However, PKD1-S744E/S748E executes a prominent in vitro autophosphorylation reaction that is detected as an electrophoretic mobility shift in SDS-PAGE (Fig. 6B) and an increase in 32P incorporation by PhosphorImager (Fig. 6D). Because PKD1-S744E/S748E harbors activation loop phosphomimetic substitutions and PKD1-S744E/S748E is constitutively phosphorylated at Ser916, the electrophoretic mobility shift and increased 32P incorporation must denote PKD1 autophosphorylation elsewhere in the protein. We used a mutagenesis approach to map the structural requirements for the band shift. Fig. 6C shows that a PH domain deletion mutant (PKD1-ΔPH) undergoes a prominent band shift as a result of the in vitro kinase assay, whereas a PKD1 N-terminal truncation mutant lacking the C1 domain (PKD1-ΔC1) does not. These results indicate that the band shift is because of an autophosphorylation reaction that maps to (or is dependent upon) the N terminus of the enzyme. Of note, PKD1 autophosphorylation sites have been mapped to Ser205/Ser208 and Ser219/Ser223 in the C1A/C1B interdomain (22). Although these sites are deleted in the PKD1-ΔC1 mutant, additional studies showed that S205A/S208A- and S219A/S223A-substituted forms of PKD1 undergo pronounced mobility shifts during in vitro kinase assays (data not shown). These results suggest that PKD1 autophosphorylates at yet additional sites; efforts to map the autophosphorylation sites(s) underlying the mobility shift are ongoing.
Fig. 6, B and E, shows that PKD1-S744E/S748E is a robust CREB and cTnI kinase. Maximal CREB-Ser133 and cTnI-Ser23/Ser24 phosphorylation by immunoblot analysis (Fig. 6B) or 32P incorporation into CREB or cTnI (by PhosphorImager analysis, Fig. 6E) is considerably higher in assays with PKD1-S744E/S748E than in assays with WT-PKD1. Of note, a S744E/S748E substitution leads to an increase in PKD1 autophosphorylation measured as 32P incorporation into PKD1 and enhanced PKD1 phosphorylation of heterologous substrates in trans, but it does not result in increased PKD1-Ser916 autophosphorylation.
Fig. 6B shows that the PKD1-S744E/S748E enzyme promotes CREB-Ser133 phosphorylation at 1–6 μm ATP, under conditions that do not lead to a detectable increase in CREB-Ser133 phosphorylation by WT-PKD1. Although these results could suggest that activation loop Ser → Glu substitutions lower the ATP requirement for CREB phosphorylation by PKD1, a Ser → Glu substitution that increases overall CREB-Ser133 phosphorylation might lead to an apparent left shift of the curve by bringing a subthreshold CREB-Ser133 phosphorylation reaction at 1–6 μm ATP into the detectable range. To avoid a result that might be an artifact of the detection method, CREB phosphorylation also was tracked by Phosphor-Imager analysis. Fig. 6, E and F, shows that activation loop Ser → Glu substitutions increase overall 32P incorporation into CREB and also decrease the ATP requirement for CREB phosphorylation by PKD1; this left shift in the dose-response curve for ATP is most evident in Fig. 6F, where the data are normalized for differences in maximal WT-PKD1 and PKD1-S744E/S748E activity.
A PKD1-S744E/S748E mutant with a reduced ATP requirement should be relatively refractory to inhibition by Gö6976. This prediction was tested by comparing the dose-response curves for Gö6976 inhibition of WT-PKD1 and PKD1-S744E/S748E. Fig. 7A shows that WT-PKD1 executes autophosphorylation reactions at the activation loop (Ser748) and at Ser916 and that Gö6976 selectively prevents PKD1 autophosphorylation at Ser748 but not Ser916. WT-PKD1 also phosphorylates CREB and cTnI; this is detected as an increase in anti-CREB-Ser(P)133 and anti-cTnI-Ser(P)23/Ser(P)24 immunoreactivity by Western blot analysis (Fig. 7A) and an increase in 32P incorporation by PhosphorImager analysis (Fig. 7B). By either measure, WT-PKD1 phosphorylation of CREB or cTnI is abrogated by 5 μm Gö6976. PKD1-S744E/S748E is recovered as a constitutively Ser916-phosphorylated enzyme that is not recognized by the anti-PKD1-Ser(P)748 antibody. PKD1-S744E/S748E promotes a high level of CREB-Ser133 and cTnI-Ser23/Ser24 phosphorylation (Fig. 7A). Gö6976 inhibits PKD1-S744E/S748E phosphorylation of CREB and cTnI, without blocking PKD1-S744E/S748E-Ser916 autophosphorylation. However, the effect of Gö6976 to inhibit PKD1-S744E/S748E activity requires high inhibitor concentrations, ∼4-fold higher than the Gö6976 concentration sufficient to inhibit WT-PKD1 (Fig. 7B). These results expose a potential limitation of PKD1 inhibitors that act by competing for ATP binding; the efficacy of these compounds will be influenced by the activation state of (and post-translational modifications on) the enzyme.
Studies of the PKD1-S744A/S748A mutant also were revealing. Fig. 6B shows that the PKD1-S744A/S748A enzyme autophosphorylates at Ser916, similar to WT-PKD1. However, PKD1-S744A/S748SA incorporates relatively little 32P; it does not band shift, and it is a very ineffective CREB-Ser133 or cTnI-Ser23/Ser24 kinase when subjected to in vitro kinase assays with PMA (Fig. 6B, D, and E). These results provide direct evidence that PKD1 does not require a phosphorylation or negative charge in the activation loop to execute an autophosphorylation reaction at Ser916. The results also suggest that a phosphorylation or negative charge in the activation loop is required for PKD1 activity toward target substrates. Although this conclusion resonates with prevailing concepts regarding the mechanisms regulating the activity of various other Ser/Thr kinases, it is at odds with results published by the Rozengurt laboratory showing that a S744A/S748A substitution does not interfere with in vitro PKD1 activity (21). Because conclusions from the Rozengurt laboratory were based upon in vitro kinase assays performed with a different lipid cofactor (PDBu, rather than PMA) and a peptide (syntide-2) rather than a protein substrate, we performed additional kinase assays to reconcile discrepancies between this and the previous study.
Fig. 8 shows that WT-PKD1 executes similar robust autophosphorylation reactions, detected as either Ser916 phosphorylation (Fig. 8A) or 32P incorporation into the enzyme (Fig. 8B), when in vitro kinase assays are performed with either PDBu, PMA, or dextran sulfate (a compound used by the Rozengurt laboratory as a potent activator of PKD1 (21)). WT-PKD1 activity toward syntide-2 also is increased by lipid cofactors and dextran sulfate (Fig. 8D). In contrast, the magnitude of the lipid cofactor-dependent increase in WT-PKD1-dependent CREB phosphorylation is quite modest when compared with the massive dextran sulfate-induced increase in CREB kinase activity. These results suggest that the controls of WT-PKD1 activity toward peptide and protein substrates might differ.
Fig. 8A shows that the PKD1-S744A/S748A mutant autophosphorylates at Ser916 when kinase assays are performed with lipid cofactors or dextran sulfate. Although basal 32P incorporation into PKD1-S744A/S748A is reduced relative to basal 32P incorporation into WT-PKD1, lipid cofactor-dependent increases in 32P incorporation into WT-PKD1, and PKD1-S744A/S748A enzymes are quite similar (Fig. 8B). However, the absolute level of lipid-dependent syntide-2 kinase activity for the PKD1-S744A/S748A mutant was quite low compared with WT-PKD1 (Fig. 8D). Moreover, the PMA-activated PKD1-S744A/S748A enzyme is autophosphorylated at Ser916, but it does not act as a CREB kinase. However, the PKD1-S744A/S748A enzyme exhibits a band shift, and PKD1-S744A/S748A becomes an effective CREB and syntide-2 kinase (with activity comparable with WT-PKD1), when assays are performed in the presence of dextran sulfate. These results indicate that activation loop Ser → Ala substitutions interfere with PKD1 activation by lipids, but they do not lead to structural changes that inherently disable the enzyme.
PKD1-Ser748 Autophosphorylation Is a Hierarchical Process That Requires a Negative Charge at Ser916—Having demonstrated that PKD1 does not require a phosphorylation reaction (or negative charge) at the activation loop to autophosphorylate at Ser916, the final set of studies used a mutagenesis strategy to examine whether Ser916 autophosphorylation exerts a reciprocal effect to regulate PKD1 activation loop autophosphorylation and/or PKD1 activity. Fig. 9A shows that the PKD1-S916A enzyme undergoes an autophosphorylation reaction that is detected as an electrophoretic mobility shift and an increase in immunoreactivity for the Cell Signaling Technology anti-PKD1-Ser(P)744/Ser(P)748 PSSA, similar to WT-PKD1. However, 32P incorporation into PKD1-S916A (at maximal ATP) is reduced by 37.8 ± 6%, compared with 32P incorporation into WT-PKD1 (n = 3, Fig. 9B). Although this presumably is attributable to the loss of Ser916 as a phosphoacceptor site, our studies expose an additional effect of the S916A substitution to induce a secondary defect in PKD1 autophosphorylation at Ser748. Additional studies show that WT-PKD1 and PKD1-S916A enzymes elicit similar increases in CREB and cTnI phosphorylation, measured as increased CREB-Ser133 and cTnI-Ser23/Ser24 phosphorylation by immunoblot analysis (Fig. 9A) or 32P incorporation into CREB and cTnI by PhosphorImager analysis (Fig. 9B). These results indicate that a PKD1-Ser748/Ser916 autophosphorylation defect does not impair in vitro PKD1 activity toward protein substrates.
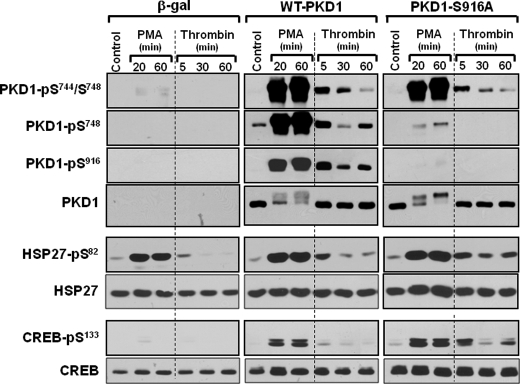
WT-PKD1 and PKD1-S916A were heterologously overexpressed in HEK293 cells to determine whether Ser916 phosphorylation also regulates PKD1-Ser748 phosphorylation and PKD1 activity in vivo in a cellular context. Fig. 10 shows that PMA treatment leads to a similar mobility shift and increase in Ser744/Ser748 phosphorylation for WT-PKD1 and PKD-S916A enzymes. In contrast, PMA treatment increases Ser748 phosphorylation on the WT-PKD1 enzyme, but PMA-dependent Ser748 phosphorylation is markedly reduced in the PKD1-S916A enzyme. The S916A substitution also influenced PKD1 activation by thrombin, an agonist for the G protein-coupled protease-activated receptor. Thrombin triggers similar increases in WT-PKD1 and PKD1-S916A phosphorylation at Ser744/Ser748; thrombin-dependent Ser744/Ser748 is maximal at 5 min and wanes when the stimulation interval is prolonged to 30–60 min. In contrast, thrombin induces biphasic increases in WT-PKD1 phosphorylation at Ser748 and Ser916. Phosphorylation at these sites increases at 5 min and wanes as the stimulation interval is increased to 30 min. However, a secondary increase in PKD1-Ser748/Ser916 phosphorylation is detected when treatment with thrombin is prolonged to 60 min. The effect of thrombin to promote PKD1-Ser748 phosphorylation (at all time points) is abrogated by the S916A substitution.
FIGURE 10.
The PKD1-S916A mutant exhibits a Ser748 phosphorylation defect and a prolonged signaling response in HEK293 cells. WT-PKD1, PKD1-S916A, and β-galactosidase were heterologously overexpressed in HEK293 cells that were then treated with vehicle, 300 nm PMA, or 1 unit/ml thrombin for the indicated intervals. Cell extracts were subjected to immunoblot analysis for PKD1, CREB, and HSP27 phosphorylation and protein expression. Results are representative of data obtained in three separate experiments.
The final series of studies examined whether the S916A substitution influences in vivo PKD1 activity toward physiologically relevant substrates such as CREB and HSP27. Fig 10 shows that PMA and thrombin increase CREB-Ser133 and HSP27-Ser82 phosphorylation in control β-galactosidase-overexpressing cultures. PMA induces a sustained increase in HSP27-Ser82 phosphorylation that persists for at least 60 min, whereas thrombin-dependent HSP27-Ser82 phosphorylation is maximal at 5 min and wanes when treatment with thrombin is prolonged to 60 min. WT-PKD1 overexpression increases the magnitude, without changing the kinetics, of the thrombin-dependent HSP27-Ser82 and CREB-Ser133 phosphorylation responses. In contrast, thrombin-dependent CREB-Ser133 and HSP27-Ser82 phosphorylation persists for at least 60 min in PKD1-S916A overexpressing cultures. Thrombin treatment for 60 min leads to a 6.2 ± 0.5-fold higher CREB-Ser133 phosphorylation and a 3.1 ± 0.03-fold higher HSP27-Ser82 phosphorylation, in PKD1-S916A overexpressing cultures than in WT-PKD1 overexpressing cultures (n = 3, p < 0.05). Collectively, these data indicate that a PKD1-S916A substitution leads to a defect in PKD1 autophosphorylation at Ser748 and that PKD1-Ser916 and/or Ser748 autophosphorylation plays a heretofore unrecognized role to terminate the cellular PKD1 signaling response.
DISCUSSION
PKD1 has recently emerged as a signaling enzyme that is activated by many physiologically important stimuli and contributes to growth, survival, and functional responses in many cell types. PKD1 activity is tightly regulated in vivo by phosphorylation at two highly conserved serine residues in the catalytic domain activation loop. Current dogma holds that PKD1 activation loop phosphorylation is mediated by PKC and is followed by PKD1 autophosphorylation at Ser916. Studies reported herein challenge several aspects of this dogma, showing the following: 1) PKD1 executes an autophosphorylation reaction at the activation loop; 2) PKD1-Ser748 autophosphorylation is a hierarchical process that requires prior phosphorylation at Ser916; and 3) PKD1 autophosphorylation at Ser916 is mediated by a unique catalytic mechanism that proceeds at exceedingly low ATP concentrations, does not require prior PKD1 phosphorylation at Ser744/Ser748, and is not necessarily accompanied by increased PKD1 activity toward heterologous substrates. We also show that PKD1-Ser916 phosphorylation can be mediated by either an intra- or inter-molecular reaction, further undermining the conclusion that PKD1-Ser916 autophosphorylation provides a valid readout of the phosphorylation state of that particular PKD1 molecule.
Although PKD-Ser916 autophosphorylation is generally touted as a valid surrogate measurement of PKD activity, discrepancies between PKD1-Ser916 phosphorylation and measurements of PKD activity are quite common in the literature (16–18). These inconsistencies are not readily reconciled by established models of PKD1 activation and have generally been ignored. This study presents a series of observations that challenge the prevailing assumption that PKD-Ser916 autophosphorylation provides a valid surrogate measure of PKD activity under all experimental conditions. First, we show that KD-PKD1 (which by definition cannot undergo Ser916 autophosphorylation) is recovered from cardiomyocytes as a Ser916-phosphorylated enzyme. Of note, KD-PKD1-Ser916 trans-phosphorylation by a GF109203X-sensitive signaling pathway also was previously identified in B lymphocytes (23) and may be a common feature of cells with relatively high endogenous PKC/PKD activity. Second, we show that Gö6976 effectively blocks PKD-dependent phosphorylation of cTnI or CREB, but this is not associated with a coordinate inhibition of PKD1-Ser916 autophosphorylation. Although this observation was initially surprising, it is interesting to note that Gö6976 is widely used to implicate PKD in various signaling responses, but experimental evidence showing that Gö6976 inhibits agonist-dependent PKD-Ser916 autophosphorylation is conspicuously absent from the literature. Third, we show that WT-PKD1 autophosphorylates at Ser916, but WT-PKD1 does not phosphorylate heterologous substrates when in vitro kinase assays are performed with very low ATP concentrations. Fourth, we show that the PKD1-S744A/S748A mutant autophosphorylates at Ser916, but it does not phosphorylate heterologous substrates such as CREB or cTnI. Collectively, these results provide clear evidence that PKD1 autophosphorylation at Ser916 is a superbly efficient reaction that proceeds under conditions that do not support PKD autophosphorylation at other sites or PKD1 phosphorylation of heterologous substrates; these results argue that PKD1-Ser916 autophosphorylation does not necessarily provide a valid surrogate measure of PKD1 activation in all cellular contexts.
PKD is rapidly phosphorylated at Ser744/Ser748 in response to various physiologic stimuli in many cell types. This rapid increase in PKD-Ser744/Ser748 phosphorylation has been ascribed to a nPKC-dependent mechanism based upon a large number of studies showing that PKD1-Ser744/Ser748 phosphorylation is effectively blocked by PKC inhibitors (that do not directly inhibit PKD activity). However, it is important to note that there is only very limited direct in vitro experimental evidence that nPKC isoforms phosphorylate PKD1 at Ser744 and/or Ser748 (24). A model that implicates PKCs as obligatory PKD1 activation loop kinases also is difficult to reconcile with studies from the Rozengurt laboratory showing that the PKD1-ΔPH mutant is recovered from GF109203X-treated COS-7 cells as a constitutively active Ser744/Ser748-phosphorylated enzyme (25, 26). These results suggest that PKD1-ΔPH-Ser744/Ser748 phosphorylation is mediated by a PKC-independent mechanism. In fact, the Rozengurt laboratory has recently revisited this issue and demonstrated that PKD1-ΔPH activation loop phosphorylation can be mediated by an autocatalytic reaction (5). This study provides direct evidence that the PKD1 activation loop is phosphorylated via an autocatalytic mechanism that may contribute to the physiological control of PKD1 activity when PKC isoforms are down-regulated.
Current concepts regarding the mechanisms that regulate PKD1 activation loop phosphorylation are based almost exclusively on studies that used the anti-PKD1-Ser(P)744/Ser(P)748 PSSA. The notion that this reagent tracks phosphorylation primarily at Ser744, and that the controls and consequences of PKD1 phosphorylation Ser748 may differ, has not been considered. This study exploits the properties of a recently developed PSSA that specifically recognizes PKD1 phosphorylation at Ser748 to expose striking differences in the mechanisms and consequences of PKD1-Ser744 and -Ser748 phosphorylation. First, cell-based studies show that heterologously overexpressed KD-PKD1 displays robust phosphorylation in trans at Ser744, but catalytically inactive PKD1 displays only a low level of Ser748 phosphorylation. These results suggest that in vivo PKD1-Ser748 phosphorylation is primarily via a cis autophosphorylation reaction that is defective in the catalytically inactive enzyme. Second, this study presents novel evidence that Ser748 autophosphorylation requires a prior priming autophosphorylation at Ser916; the S916A substitution leads to a PKD1-Ser748 phosphorylation defect, without any changes in Ser744 phosphorylation in vitro and in vivo in agonist-activated HEK293 cells. These results provide strong evidence that Ser748 phosphorylation is mediated by an autocatalytic reaction (rather than a trans-phosphorylation by PKC). These results suggest that the PKD1 C terminus, and particularly the Ser916 autophosphorylation site, plays a heretofore unrecognized role to structure the kinase core for some aspect of its catalytic function. Although this mode of regulation is novel for PKD1, it is well described for cAMP-dependent protein kinase and PKC, where phosphorylation at the extreme C terminus tethers the C terminus away from the active site and stabilizes the enzyme in a conformation that favors catalysis (27, 28). However, Ser916/Ser748 autophosphorylation defects do not lead to gross abnormalities in PKD1 autophosphorylation at other sites (such as Ser744 or the sites elsewhere in the protein that underlie the electrophoretic mobility shift) or PKD1 phosphorylation of heterologous substrates such as CREB or cTnI. Rather, cell-based studies identify a biphasic thrombin-dependent Ser916/Ser748 phosphorylation response and a more prolonged PKD1 signaling response when Ser916/Ser748 autophosphorylation is prevented (by the S916A substitution). These results suggest autophosphorylation reactions at Ser916 and/or Ser748 function to limit the duration of the PKD1 signaling response; these results emphasize that studies that use Ser916 autophosphorylation as a marker of PKD1 activation may be misleading.
Whereas the distinct controls and consequences for activation loop Ser744 and Ser748 phosphorylation identified in this study have not previously been noted for mammalian PKD family enzymes, they resemble features recently described for the Caenorhabditis elegans PKD enzymes, DKF-1 and DKF-2 (29–31). The DKF-1 activation loop sequence contains a single phospho-acceptor site at QFRKT588, corresponding to Ser748 in PKD1. Thr588 phosphorylation is via a PKC-independent mechanism and plays a dual role to mediate the PMA-dependent increase in DKF-1 activity and to tag DKF-1 for ubiquitinylation and proteasomal degradation (29, 30). The 925SFRRS929 activation loop sequence in the other C. elegans PKD enzyme DKF-2 is identical to the 744SFRRS748 sequence in PKD1 (31). Phosphorylation reactions at Ser925 and Ser929 in DKF-2 have been attributed to a PKC-dependent mechanism, with Ser925 phosphorylation implicated in the PMA-dependent mechanism that increases DKF-2 activity and Ser929 phosphorylation implicated as a modification that terminates the DKF-2 signaling response. The results of this study, showing that phosphorylation reactions at Ser744 and Ser748 play functionally distinct roles to regulate either the magnitude or the kinetics of the PKD1 signaling response, suggest that these features have been evolutionally conserved in the mammalian PKD1 enzyme.
This study implicates Ser744 phosphorylation as a modification that is required for PKD1 activity toward heterologous substrates such as CREB, cTnI, and syntide-2. Although these results resonate with considerable literature that implicates activation loop phosphorylation as a modification that controls the catalytic function of other Ser/Thr kinases, the results are at odds with studies from the Rozengurt laboratory that have been interpreted as evidence that the in vitro kinase activities of PKD1-S744A/S748A and WT-PKD1 are indistinguishable. Of note, the discrepancies between this and the previous study are readily attributed to differences in assay conditions and methods of data analysis. Studies from the Rozengurt laboratory focused exclusively on PKD1 phosphorylation of the peptide substrate syntide-2; results for lipid cofactor-dependent enzyme activity were normalized to correct for any differences in basal PKD1 versus PKD1-S744A/S748A activity. Studies reported herein show that lipid cofactor-dependent increments in PKD1-S744A/S748A and WT-PKD1 syntide-2 kinase activities appear comparable when the data are expressed in this manner. However, the normalized data obscure the marked reduction in the absolute level of lipid-dependent syntide-2 kinase activity that results from a S744A/S748A substitution. Moreover, studies reported herein show that a S744A/S748A substitution abrogates lipid-dependent activity toward protein substrates such as CREB and cTnI, exposing an important potential fallacy of studies that rely exclusively on kinase assays with model peptides to interrogate mechanisms that control PKD1 phosphorylation of physiologically relevant substrates.
Finally, this study shows that PKD1 executes a series of phosphorylation reactions with distinct ATP requirements. PKD1 undergoes an intramolecular autophosphorylation at Ser916 at a very low ATP concentration; PKD1 autophosphorylation at other sites and PKD1 phosphorylation of target substrates are detected only at considerably higher ATP concentrations. Although intracellular ATP levels are sufficiently high to support all PKD1 phosphorylation reactions under physiologic conditions, these differences in ATP requirements could become relevant in ischemia, where a fall in ATP might lead to a selective defect in the phosphorylation of target substrates such as cTnI and CREB with little-to-no associated defect in PKD-Ser916 autophosphorylation (a modification that is required for PKD1 interactions with PDZ domain-containing binding partners and PKD1 targeting within the cell). Our studies showing that post-translational modifications such as activation loop phosphorylation lower the ATP requirement for PKD1-dependent phosphorylation of heterologous substrates also may have clinical implications; our results raise the concern that the “therapeutic window” for ATP-competitive inhibitor compounds may vary according to the activation status of the PKD1 enzyme. These results emphasize an inherent limitation of kinase inhibitors that compete with ATP at the binding pocket and the importance of developing novel compounds that act through different mechanisms.
Acknowledgments
We acknowledge technical assistance from Ms. Talia Saal.
This work was supported, in whole or in part, by National Institutes of Health Grant HL 77860 (USPHS NHLBI). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PKD1, protein kinase D1; PKD, protein kinase D; cTn, cardiac troponin; CREB, cAMP-response element-binding protein; DKF-1, D kinase family-1; DKF-2, D kinase family-2; ERK, extracellular signal-regulated kinase; GFP, green fluorescent protein; HDAC5, histone deacetylase 5; HSP27, heat shock protein 27; KD-PKD1, kinase-dead PKD1; MBP, maltose-binding protein; MEK,; mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; NE, norepinephrine; PDBu, phorbol 12,13-dibutyrate; PH, pleckstrin homology; PSSA, phosphorylation site-specific antibody; PKC, protein kinase C; nPKC, novel PKC isoforms (δ, ε, η, and θ); PMA, phorbol 12-myristate 13-acetate; PS, phosphatidylserine; WT-PKD1, wild-type PKD1; HA, hemagglutinin.
References
- 1.Rozengurt, E., Rey, O., and Waldron, R. T. (2005) J. Biol. Chem. 280 13205-13208 [DOI] [PubMed] [Google Scholar]
- 2.Avkiran, M., Rowland, A. J., Cuello, F., and Haworth, R. S. (2008) Circ. Res. 102 157-163 [DOI] [PubMed] [Google Scholar]
- 3.Nishikawa, K., Toker, A., Wong, K., Marignani, P. A., Johannes, F. J., and Cantley, L. C. (1998) J. Biol. Chem. 273 23126-23133 [DOI] [PubMed] [Google Scholar]
- 4.Hausser, A., Storz, P., Link, G., Stoll, H., Liu, Y. C., Altman, A., Pfizenmaier, K., and Johannes, F. J. (1999) J. Biol. Chem. 274 9258-9264 [DOI] [PubMed] [Google Scholar]
- 5.Jacamo, R., Sinnett-Smith, J., Rey, O., Waldron, R. T., and Rozengurt, E. (2008) J. Biol. Chem. 283 12877-12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozgen, N., Obreztchikova, M., Guo, J., Elouardighi, H., Dorn, G. W., Wilson, B. A., and Steinberg, S. F. (2008) J. Biol. Chem. 283 17009-17019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johannessen, M., Delghandi, M. P., Rykx, A., Dragset, M., Vandenheede, J. R., Van Lint, J., and Moens, U. (2007) J. Biol. Chem. 282 14777-14787 [DOI] [PubMed] [Google Scholar]
- 8.Fielitz, J., Kim, M. S., Shelton, J. M., Qi, X., Hill, J. A., Richardson, J. A., Bassel-Duby, R., and Olson, E. N. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 3059-3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vega, R. B., Harrison, B. C., Meadows, E., Roberts, C. R., Papst, P. J., Olson, E. N., and McKinsey, T. A. (2004) Mol. Cell. Biol. 24 8374-8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison, B. C., Kim, M. S., van Rooij, E., Plato, C. F., Papst, P. J., Vega, R. B., McAnally, J. A., Richardson, J. A., Bassel-Duby, R., Olson, E. N., and McKinsey, T. A. (2006) Mol. Cell. Biol. 26 3875-3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haworth, R. S., Cuello, F., Herron, T. J., Franzen, G., Kentish, J. C., Gautel, M., and Avkiran, M. (2004) Circ. Res. 95 1091-1099 [DOI] [PubMed] [Google Scholar]
- 12.Cuello, F., Bardswell, S. C., Haworth, R. S., Yin, X., Lutz, S., Wieland, T., Mayr, M., Kentish, J. C., and Avkiran, M. (2007) Circ. Res. 100 864-873 [DOI] [PubMed] [Google Scholar]
- 13.Doppler, H., Storz, P., Li, J., Comb, M. J., and Toker, A. (2005) J. Biol. Chem. 280 15013-15019 [DOI] [PubMed] [Google Scholar]
- 14.Evans, I. M., Britton, G., and Zachary, I. C. (2008) Cell. Signal. 20 1375-1384 [DOI] [PubMed] [Google Scholar]
- 15.Matthews, S. A., Rozengurt, E., and Cantrell, D. (1999) J. Biol. Chem. 274 26543-26549 [DOI] [PubMed] [Google Scholar]
- 16.Brandlin, I., Hubner, S., Eiseler, T., Martinez-Moya, M., Horschinek, A., Hausser, A., Link, G., Rupp, S., Storz, P., Pfizenmaier, K., and Johannes, F. J. (2002) J. Biol. Chem. 277 6490-6496 [DOI] [PubMed] [Google Scholar]
- 17.Storz, P., Doppler, H., and Toker, A. (2004) Mol. Pharmacol. 66 870-879 [DOI] [PubMed] [Google Scholar]
- 18.Celil, A. B., and Campbell, P. G. (2005) J. Biol. Chem. 280 31353-31359 [DOI] [PubMed] [Google Scholar]
- 19.Rybin, V. O., Guo, J., Sabri, A., Elouardighi, H., Schaefer, E., and Steinberg, S. F. (2004) J. Biol. Chem. 279 19350-19361 [DOI] [PubMed] [Google Scholar]
- 20.Oancea, E., Bezzerides, V. J., Greka, A., and Clapham, D. E. (2003) Dev. Cell 4 561-574 [DOI] [PubMed] [Google Scholar]
- 21.Iglesias, T., Waldron, R. T., and Rozengurt, E. (1998) J. Biol. Chem. 273 27662-27667 [DOI] [PubMed] [Google Scholar]
- 22.Hausser, A., Link, G., Bamberg, L., Burzlaff, A., Lutz, S., Pfizenmaier, K., and Johannes, F. J. (2002) J. Cell Biol. 156 65-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez-Ruiloba, L., Cabrera-Poch, N., Rodriguez-Martinez, M., Lopez-Menendez, C., Martin Jean-Mairet, R., Higuero, A. M., and Iglesias, T. (2006) J. Biol. Chem. 281 18888-18900 [DOI] [PubMed] [Google Scholar]
- 24.Storz, P., Doppler, H., and Toker, A. (2004) Mol. Cell. Biol. 24 2614-2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iglesias, T., and Rozengurt, E. (1998) J. Biol. Chem. 273 410-416 [DOI] [PubMed] [Google Scholar]
- 26.Waldron, R. T., Rey, O., Zhukova, E., and Rozengurt, E. (2004) J. Biol. Chem. 279 27482-27493 [DOI] [PubMed] [Google Scholar]
- 27.Edwards, A. S., and Newton, A. C. (1997) J. Biol. Chem. 272 18382-18390 [DOI] [PubMed] [Google Scholar]
- 28.Knighton, D. R., Zheng, J. H., Ten Eyck, L. F., Ashford, V. A., Xuong, N. H., Taylor, S. S., and Sowadski, J. M. (1991) Science 253 407-414 [DOI] [PubMed] [Google Scholar]
- 29.Feng, H., Ren, M., Wu, S. L., Hall, D. H., and Rubin, C. S. (2006) J. Biol. Chem. 281 17801-17814 [DOI] [PubMed] [Google Scholar]
- 30.Feng, H., Ren, M., and Rubin, C. S. (2006) J. Biol. Chem. 281 17815-17826 [DOI] [PubMed] [Google Scholar]
- 31.Feng, H., Ren, M., Chen, L., and Rubin, C. S. (2007) J. Biol. Chem. 282 31273-31288 [DOI] [PubMed] [Google Scholar]