Abstract
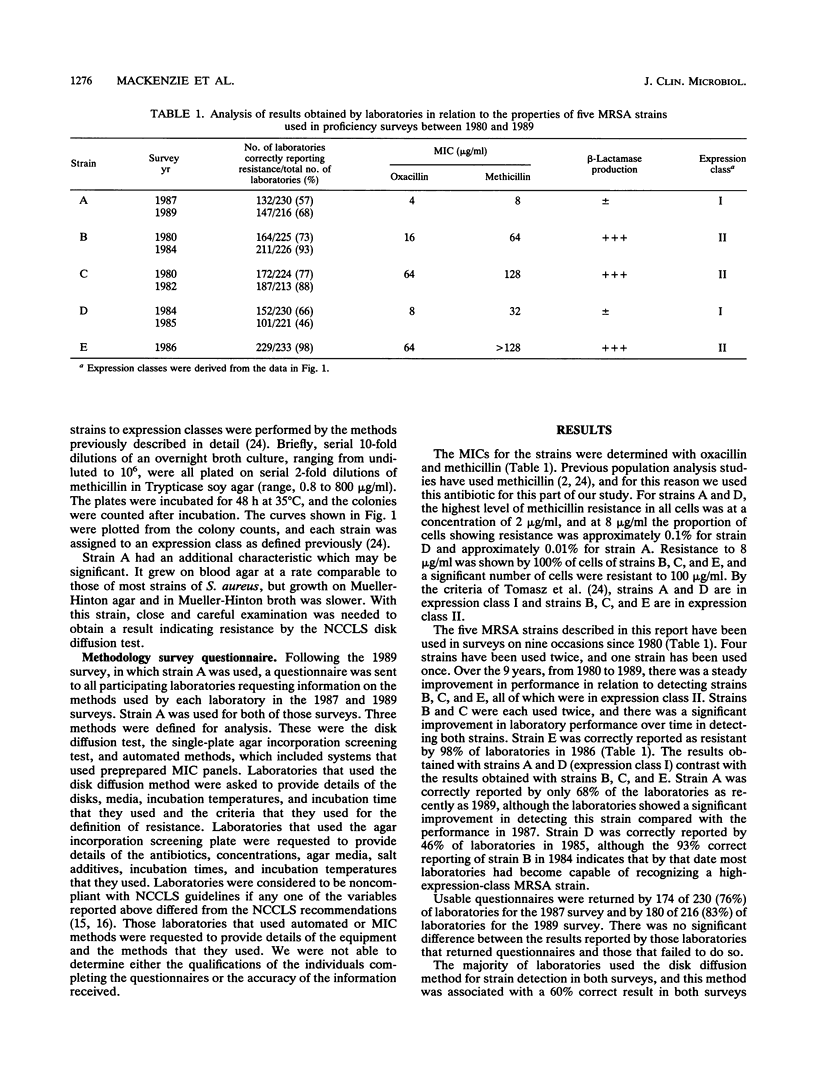
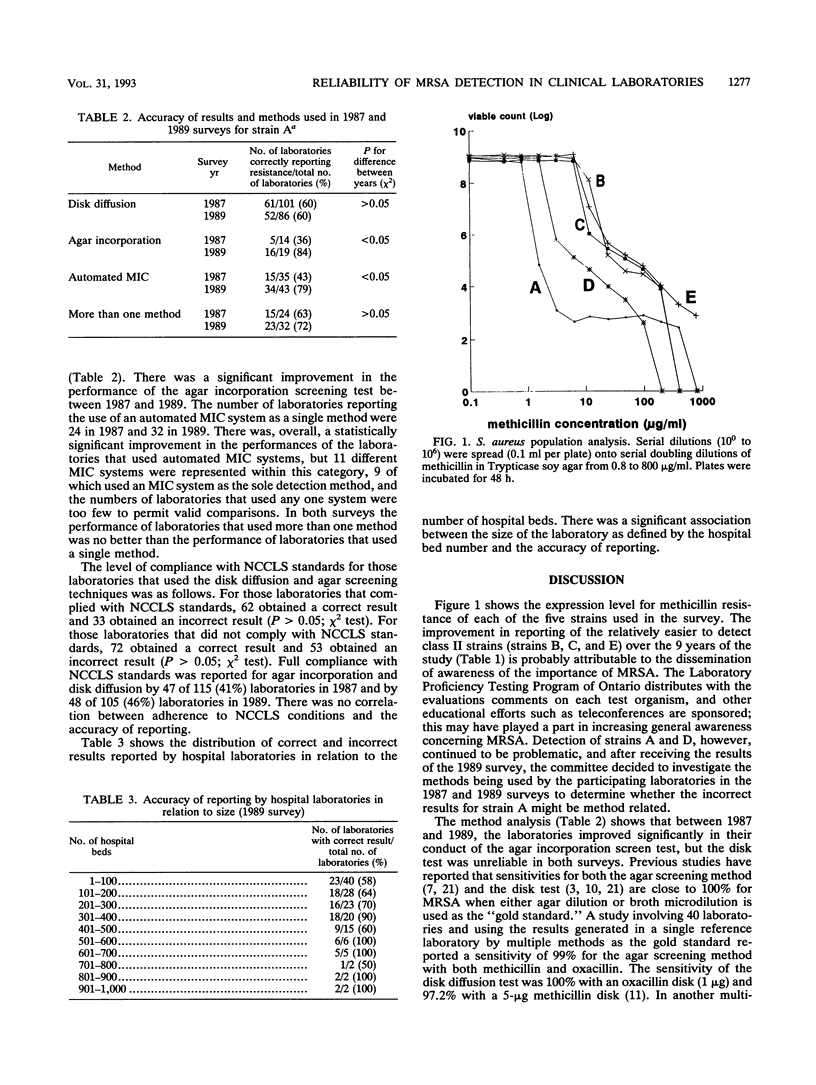
We report the results of a province-wide quality control program in which five methicillin-resistant Staphylococcus aureus strains were circulated to all Ontario laboratories (hospital, private, and public health laboratories) on nine occasions between 1980 and 1989. The level of expression of methicillin resistance in each of the isolates was determined by performing viable colony counts on serial dilutions of methicillin in agar, and each isolate was assigned to an expression class according to previous published criteria (A. Tomasz, S. Nachman, and H. Leaf, Antimicrob. Agents Chemother. 35:124-129, 1991). Over this time there was an improvement in the performance of laboratories in the recognition of three strains that were relatively easy to detect (strains B, C, and E). These strains were of expression class II, and 98% of laboratories reported correct identifications in 1986. Performance in identifying two strains (strains A and D) of expression class I remained poor. Strain A was circulated in two surveys in 1987 and 1989, and laboratories were sent a questionnaire requesting details of the methods used in those two surveys. The methods used by the laboratories were classified into three categories: disk diffusion, single-plate screening by agar incorporation, and automated methods, which included premanufactured MIC panels. Between the 1987 and 1989 surveys, there was no change in the performance of the disk diffusion test (60% correct on both occasions), but there was improvement in the sensitivity of the agar incorporation test (36% correct in 1987 and 84% correct in 1989) and in automated methods (43% correct in 1987 and 79% correct in 1989). Over a decade, there was overall improvement in the performance of laboratories in detecting easy-to-detect strains, but there were difficulties in detecting organisms of low expression class, and an organism of very low expression class should be designated as a control organism for routine testing of methicillin-resistant s. aureus isolates.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldridge K. E., Janney A., Sanders C. V., Marier R. L. Interlaboratory variation of antibiograms of methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains with conventional and commercial testing systems. J Clin Microbiol. 1983 Nov;18(5):1226–1236. doi: 10.1128/jcm.18.5.1226-1236.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Strässle A., Gustafson J. E., Kayser F. H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992 Jul;36(7):1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce J. M. Reevaluation of the ability of the standardized disk diffusion test to detect methicillin-resistant strains of Staphylococcus aureus. J Clin Microbiol. 1984 Jun;19(6):813–817. doi: 10.1128/jcm.19.6.813-817.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumfitt W., Hamilton-Miller J. Methicillin-resistant Staphylococcus aureus. N Engl J Med. 1989 May 4;320(18):1188–1196. doi: 10.1056/NEJM198905043201806. [DOI] [PubMed] [Google Scholar]
- Chambers H. F. Methicillin-resistant staphylococci. Clin Microbiol Rev. 1988 Apr;1(2):173–186. doi: 10.1128/cmr.1.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudron P. E., Jones D. L., Dalton H. P., Archer G. L. Evaluation of laboratory tests for detection of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. J Clin Microbiol. 1986 Nov;24(5):764–769. doi: 10.1128/jcm.24.5.764-769.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbarth C. J., Chambers H. F. Methicillin-resistant staphylococci: detection methods and treatment of infections. Antimicrob Agents Chemother. 1989 Jul;33(7):995–999. doi: 10.1128/aac.33.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S. L., Freedy P. K. Variation in the abilities of automated, commercial, and reference methods to detect methicillin-resistant (heteroresistant) Staphylococcus aureus. J Clin Microbiol. 1984 Sep;20(3):494–499. doi: 10.1128/jcm.20.3.494-499.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindler J. A., Warner N. L. Effect of source of Mueller-Hinton agar on detection of oxacillin resistance in Staphylococcus aureus using a screening methodology. J Clin Microbiol. 1987 Apr;25(4):734–735. doi: 10.1128/jcm.25.4.734-735.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. N., Barry A. L., Gardiner R. V., Packer R. R. The prevalence of staphylococcal resistance to penicillinase-resistant penicillins. A retrospective and prospective national surveillance trial of isolates from 40 medical centers. Diagn Microbiol Infect Dis. 1989 Sep-Oct;12(5):385–394. doi: 10.1016/0732-8893(89)90108-9. [DOI] [PubMed] [Google Scholar]
- Lindberg M., Sjöström J. E., Johansson T. Transformation of chromosomal and plasmid characters in Staphylococcus aureus. J Bacteriol. 1972 Feb;109(2):844–847. doi: 10.1128/jb.109.2.844-847.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal L. K., Thornsberry C. The role of beta-lactamase in staphylococcal resistance to penicillinase-resistant penicillins and cephalosporins. J Clin Microbiol. 1986 May;23(5):832–839. doi: 10.1128/jcm.23.5.832-839.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Wakefield D. S., Hammons G. T., Massanari R. M. Variation from standards in Staphylococcus aureus susceptibility testing. Am J Clin Pathol. 1987 Aug;88(2):231–235. doi: 10.1093/ajcp/88.2.231. [DOI] [PubMed] [Google Scholar]
- Ryffel C., Kayser F. H., Berger-Bächi B. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 1992 Jan;36(1):25–31. doi: 10.1128/aac.36.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., McDougal L. K. Detecting methicillin resistance in Staphylococcus aureus by polymerase chain reaction. Antimicrob Agents Chemother. 1992 Jul;36(7):1585–1586. doi: 10.1128/aac.36.7.1585-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry C., McDougal L. K. Successful use of broth microdilution in susceptibility tests for methicillin-resistant (heteroresistant) staphylococci. J Clin Microbiol. 1983 Nov;18(5):1084–1091. doi: 10.1128/jcm.18.5.1084-1091.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokue Y., Shoji S., Satoh K., Watanabe A., Motomiya M. Comparison of a polymerase chain reaction assay and a conventional microbiologic method for detection of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992 Jan;36(1):6–9. doi: 10.1128/aac.36.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Drugeon H. B., de Lencastre H. M., Jabes D., McDougall L., Bille J. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP 2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob Agents Chemother. 1989 Nov;33(11):1869–1874. doi: 10.1128/aac.33.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Nachman S., Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991 Jan;35(1):124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]


