Abstract
We show that arrays of nanopit structures etched in a nanoslit can control the positioning and conformation of single DNA molecules in nanofluidic devices. By adjusting the spacing, organization and placement of the nanopits it is possible to immobilize DNA at predetermined regions of a device without additional chemical modification and achieve a high degree of control over local DNA conformation. DNA can be extended between two nanopits and in closely spaced arrays will self-assemble into “connect-the-dots” conformations consisting of locally pinned segments joined by fluctuating linkers. These results have broad implications for nanotechnology fields that require methods for the nanoscale positioning and manipulation of DNA.
Keywords: nanofluidics, polymer confinement, self-assembly, DNA conformation
Controlling the on-chip organization and conformation of DNA is important for a number of interrelated nanotechnology disciplines. Nanofluidic devices for DNA analysis require methods for immobilization of DNA probes at specific points (1,2) and stretching molecules, i.e., optical mapping of the genome (3). From a “bottom-up” perspective, DNA is emerging as an important nanomaterial in its own right that can serve as a scaffold for the synthesis of nanowires (4,5), nanoparticles, and more complicated structures (6). There is a growing need for methods that can combine these self-assembly based fabrication methodologies with structures made in a clean room by using standard tools like photolithography, electron beam lithography, and reactive ion etching (7).
When the device scale falls below the characteristic size of the molecule in free solution (the radius of gyration Rg) confinement will affect DNA conformation. One striking consequence is the spontaneous stretching of DNA in 1D nanochannel devices (8–11). Confinement in 2D nanoslit devices results in suppressed DNA diffusion and modified transport properties (11,12). We introduce a type of nanostructure consisting of a nanoslit with a built-in spatial modulation of confinement created by arrays of embedded nanopits. Nanopits are square depressions in a 50- to 100-nm-deep nanoslit with a width in the range of 100–500 nm and a depth of 100 nm, as shown in Fig. 1. If the slit height is below the radius of gyration DNA will actively “feel” the nanotopography as a free-energy landscape. The more confined regions will impart a greater free energy to the DNA resulting in a spontaneous segregation of molecule contour into the nanopits. The molecule will adopt a conformation consisting of filled pits bridged by fluctuating linkers extending into the slit (Fig. 1B). Depending on the lattice spacing and nanopit size the molecule will fill a single pit or span multiple pits. In practice, this means that nanopits can be used to control where DNA is immobilized in a nanofluidic device and use DNA molecules to link specific points on a chip. For example, the nanopits might be used to connect DNA-based nanowires between electrodes in a nanofluidic system (4). Moreover, because the biotin–streptavidin bond can be used to couple DNA to a variety of additional labels or colloidal objects (14), the self-assembly of DNA in the nanopit arrays might be used to incorporate additional structure not easily obtainable via a traditional top-down fabrication approach.
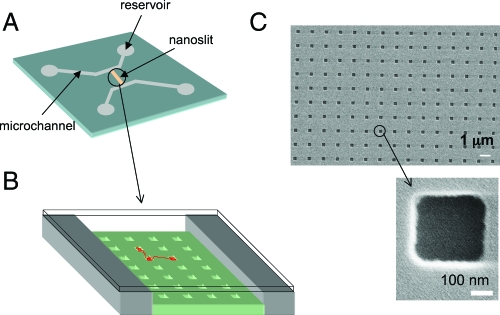
Fig. 1.
Chip design. (A) Schematic of chip design showing microchannel arms connecting the circular reservoir pads to the nanoslit (yellow). (B) An enlarged 3D view of the nanopit lattice in the nanoslit with a cartoon DNA molecule (red) spanning 3 pits. (C) SEM images of 300 × 300 nm nanopits at high and low magnification.
The nanopit lattice also serves as an ideal physics laboratory for studying the effect of nanoconfinement on polymer configuration (15). The nanopit lattice forces the polymer to adopt an effectively quantized conformation: each conformation can be characterized by N, the number of pits occupied. This simplifies the analysis; instead of attempting to extract the radius of gyration of the coil from careful image analysis, one simply counts how many pits are occupied for each frame. Moreover, the polymer conformation in the lattice is a sensitive function of the balance between the free energy of the DNA in the nanopits and the DNA in the nanoslit. Thus, the nanopit lattices can potentially be used to test scaling theories for the free energy of cavity- and slit-confined DNA (15–18). In this respect, the nanopit lattices discussed here, as they are lithographically defined, have advantages over the structures created by colloidal templating discussed in ref. 19, in particular, the ability to arbitrarily vary nanopit size and separation. Last, the nanopit-confined DNA exhibits a number of intriguing physical properties. Molecules can exhibit thermally driven fluctuations between states of different N. Long-lived metastable states can be formed by applying pressure-driven flow to stretch DNA between two nanopits. As a first step to understanding the underlying physics of this problem we introduce a free energy based on polymer-scaling theories that accounts for the net stabilization created by the filled nanopits.
Experimental Methods
The devices were fabricated on fused silica wafers (HOYA) via a 3-stage process coupling electron beam and UV contact lithography. The nanopits were defined by using electron beam lithography (JEOL) in zep520A resist (20) and then transferred to the underlying silica via CF4:CHF3 reactive ion etching (RIE). Contact UV lithography was used to expose a 450-μm-long and 50-μm-wide nanoslit in photoresist which was then etched over the nanopit array to depths of 50 and 100 nm. To introduce buffer into the nanoslit, a last UV contact lithography and etching step was used to define a 50-μm-wide microchannel 1 μm deep in 2 U-shaped arms on either side of the nanoslit leading to circular reservoirs. Loading holes were sandblasted in the reservoirs and the chip was sealed by using direct quartz-quartz bonding to 150-μm-thick fused silica coverglass (Valley Design), permitting the use of high numerical aperture oil immersion objectives. Etch depths were measured via tapping mode atomic force microscopy and a profilometer.
Experiments were conducted with λ-phage DNA (48.5 kbp, L = 16.5 μm, New England Biosciences) and T4 DNA (T4GT7 DNA, 166 kbp, L = 56.4 μm, Nippon Gene through Wako). The DNA was dyed with YOYO-1 fluorescent dye (Invitrogen) at a concentration of 1 dye molecule per every 5 base pairs. Staining the DNA increases the contour length, resulting in a λ and T4 DNA contour of 21 μm and 71.8 μm, respectively (10). The single-molecule measurements were made with a fluorescence videomicroscopy system consisting of a Nikon Eclipse TE2000 inverted microscope, 100× N.A. 1.4 oil-immersion objective coupled to an EMCCD camera (Princeton Instruments, Cascade). The chip was mounted on a chuck via o-ring seals. The chuck was designed so that pneumatic pressure could be applied to bring fresh DNA into the nanoslit from the micro loading channels. The running buffer chosen was 0.5×TBE (44.5 mM tris and borate with 1 mM EDTA). In addition, 3% 2-mercaptoethanol was added to suppress bleaching and photoknicking of DNA.
Results and Discussion
DNA in Nanopit Arrays.
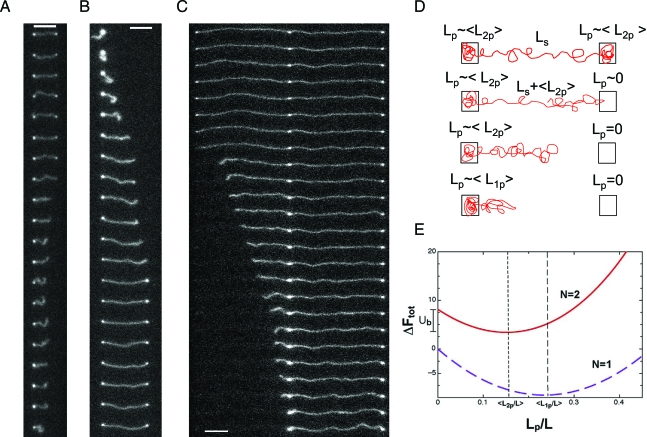
Once DNA is brought into the nanoslit from the microchannel, the molecules interact with the nanopits and spontaneously self-assemble into configurations consisting of a number of filled pits connected by fluctuating DNA linkages. The nanopits were arranged in a square lattice with a different pit-to-pit spacing, l, every 50 μm across the device so that a number of different lattice spacings could be explored in a single experiment (lattice spacings of 0.5, 1, 2, 5, 10, 15, and 20 μm were investigated). Fig. 2 shows examples of DNA molecules interacting with 300×300 nm pits, etched 100 nm deep embedded in a nanoslit 100 nm high. As the lattice spacing increases, the DNA subtends fewer pits until the molecule is completely stretched out between 2 pits. Further increase in the lattice spacing will result in single-pit occupancy. Note that, with the exception of T4 DNA in the 5-μm lattice (Fig. 2H), the DNA does not tend to extend outside the end pits. In this one exceptional case, the amount of overhanging contour was not constant but tended to fluctuate as contour was exchanged with the central linkers.
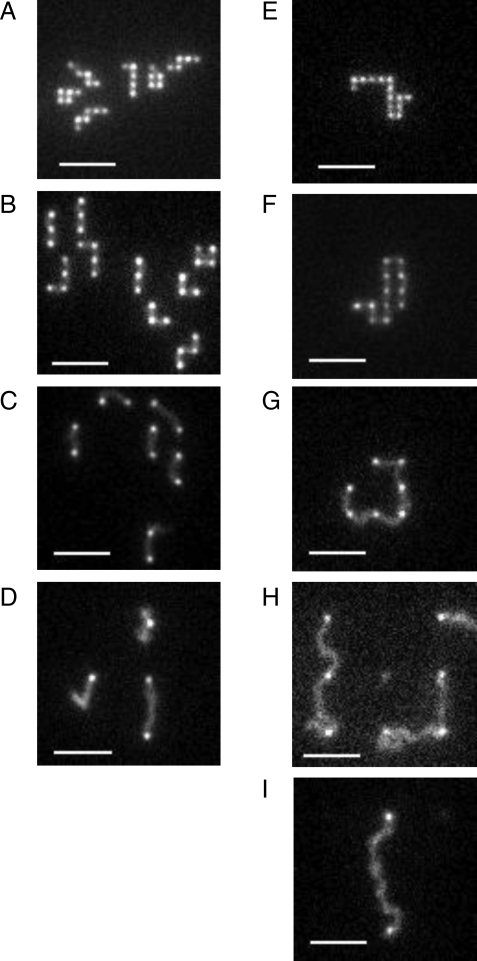
Fig. 2.
Fluorescence micrographs of λ- and T4-DNA in arrays of 300 × 300 nm nanopits etched 100 nm deep with a surrounding 100-nm-deep slit. A–D show λ-DNA in arrays with lattice spacings of, respectively, 0.5 μm, 1.0 μm, 2.0 μm, and 5.0 μm. E–I show T4-DNA in arrays with lattice spacings of, respectively, 0.5 μm, 1.0 μm, 2.0 μm, 5.0 μm, and 10.0 μm. (Scale bar, 5 μm.) Note that in D, the true equilibrium conformation is in fact the single-pit state. The stretched configuration is metastable and decays on a timescale of minutes.
In addition, the molecules are observed to undergo abrupt transitions between lattice states of varying N (see Fig. 3C–E). A natural approach to image analysis is to integrate the total intensity within boxes registered to the lattice. Each box defines a data collection channel (Fig. 3A) and an entire movie can be reduced to a set of time courses for the fluorescence of each nanopit (Fig. 3B). The integrated pit fluorescence undergoes clear jumps above baseline when the pit is filled so that a simple threshold can be applied to count the number of filled pits. The percentage of time the DNA spends in dimer, trimer, tetramer (etc.) configurations and in particular, the average pit occupancy 〈n〉, can be measured (see Fig. 5B). Increasing the lattice spacing reduces the number of accessible states. In the 2-μm lattice, λ-DNA spends the greatest time in the dimer configuration (Fig. 3E) whereas in the 0.5-μm lattice, states with N varying from 4 to 6 are of comparable probability (Fig. 3C). As the DNA tends to migrate in the slit via repeated transitions to and decays from states of higher occupancy, increasing the lattice spacing also suppresses diffusion. The λ-DNA is almost completely immobilized at lattice spacings >2μm.
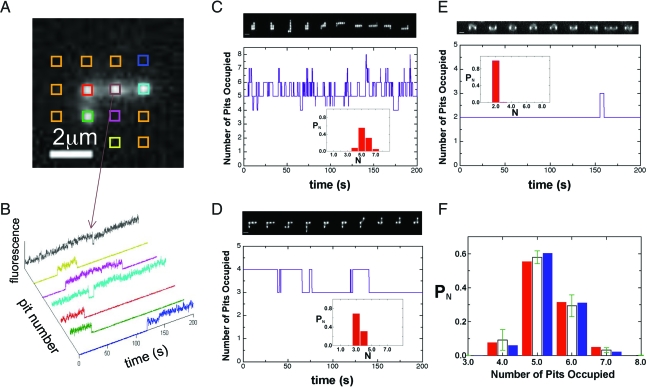
Fig. 3.
Image analysis protocol. (A) Example fluorescence image of λ-DNA in 300 × 300 nm nanopits with 1-μm spacing. The data analysis procedure involves integrating the fluorescence of boxes registered to the lattice (as shown). (B) Nanopit data channels for the molecule in A. The color of the data channels corresponds to the color of the boxes in A from which the data were obtained. The orange boxes are noise channels never occupied by the molecule during the course of the movie. A threshold can be applied to the integrated pit intensity to count the number of occupied pits as a function of time, shown in D. (C–E) The total number of filled pits as a function of time and representative fluorescence images taken 20 s apart for 300 × 300 nm nanopits with a 0.5-μm nanopit spacing (C), 1-μm nanopit spacing (D), and a 2-μm spacing (E). Insets show the histogrammed occupancy probability PN for the varying states accessed. (F) The histogrammed occupancy probability PN for the 300 × 300 nm nanopits with a spacing l = 0.5 μm. The red bars are the data for the molecule in C, the open bars with errors correspond to the mean values and standard deviations of PN for 14 molecules imaged in the 0.5-μm lattice and the blue bars are a theoretical prediction.
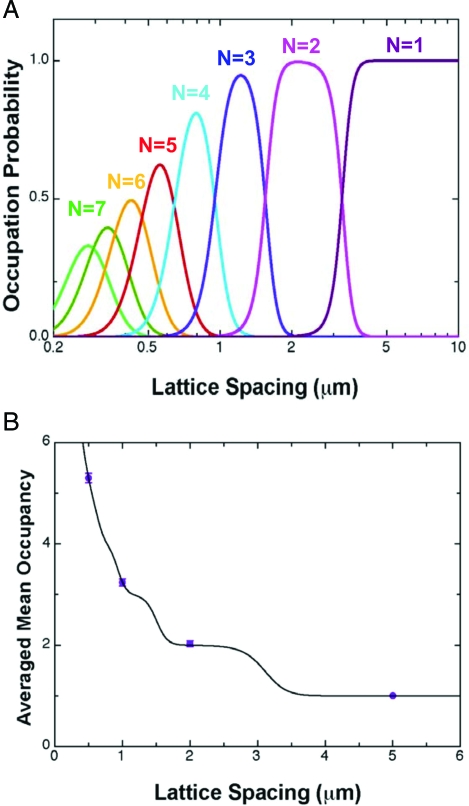
Fig. 5.
Occupancy statistics. (A) The probability of a λ-DNA molecule occupying states with N = 1–8 as a function of lattice spacing l (h = 100 nm, a = 300 nm, and d = 100 nm). (B) The averaged occupancy for λ-DNA as a function of lattice spacing l. Error bars shown, which are comparable to the size of the data points, are the standard deviation of measurements over 10 molecules.
Theory to Predict Average Nanopit Occupancy.
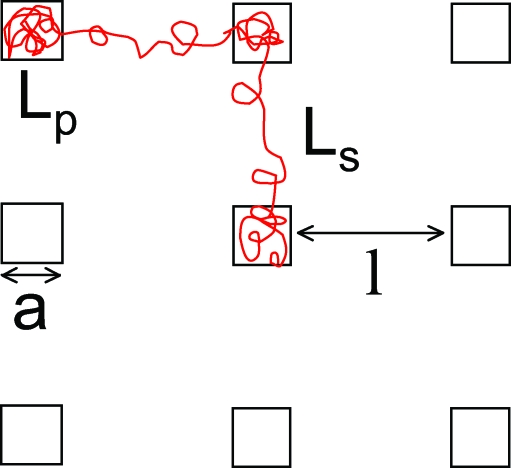
A simple model explains the observations above and predicts the DNA configurations observed in a given lattice geometry. Consider a nanopit lattice with traps that have width a, depth d, nanopit spacing l, and slit height h. The molecule has total contour length L partitioned between N filled pits and N − 1 linkers (Fig. 4). We choose not to include off-lattice diagonal linkers. While diagonal linkers are formed, due to the longer spring extension of diagonal linkers, they are suppressed relative to on-lattice linkers, as is evident from Fig. 2. Each pit contains contour length Lp (for given N the average contour stored in each pit must be equal because the pits are identical) so that the contour of each linker, Ls is given by L = NLp + (N − 1)Ls. Finally, we assume that a particular lattice configuration has reached a local equilibrium, i.e., that the timescale for transition between states of differing occupancy is greater then the relaxation time of a particular configuration.
Fig. 4.
A 2D schematic of the nanopit arrays giving the definition of the pit spacing l, pit width a, the contour per pit Lp, and contour per linker Ls. The pit depth is denoted by d and the slit height by h.
DNA spontaneously self-assembles in the nanopits because of the decrease in free energy when contour fills the (locally) less confined pit regions. Each filled pit will lower the energy by an amount Fp(Lp)−Fs(Lp), where Fp(Lp) is the confinement free energy of contour Lp if placed in the pit and Fs(Lp) is the confinement free energy of the contour Lp placed in the slit. Moreover, chain connectivity requires that DNA be stretched out a distance l between the pits to form the linkers, costing additional energy. Each linker will contribute spring energy Fspring(Ls, l) (the energy required to stretch DNA of contour Ls a distance l). The total change in free energy due to the effect of the nanopit lattice on the molecule configuration is then:
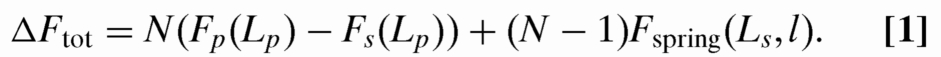 |
We can use polymer physics scaling arguments to determine how the terms in Eq. 1 depend on Lp.
The confinement free energy of DNA in the slit is proportional to the total contour in the slit. This is a property of all available theories for Fs (21–23) that follows from the extensivity of the free energy. Thus, Fs(Lp) is given by:
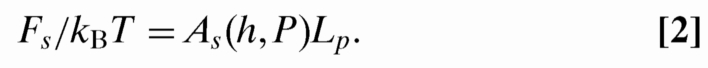 |
The constant As is a prefactor that depends on the slit height and the DNA persistence length P. The particular scaling of Fs with h depends on the ratio h/P. Brochard-Wyart suggests that for h ≫ P the confinement energy of the chain in the slit is that of an ideal polymer (22)
 |
In the regime where h ≪ P, the Odijk regime, (23). The persistence length is ≈50 nm, but there is evidence that staining with YOYO-1 increases P by roughly the same factor that the dye increases the contour length (24), so we estimate that P ≈ 64 nm.
The confinement free energy of contour Lp placed in a nanopit is given by (18)
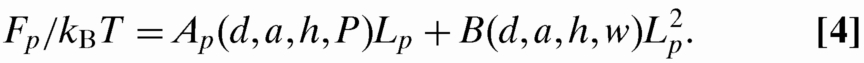 |
The first term in Eq. 4, linear in Lp, is the entropy loss for an ideal chain confined in a cavity of dimension (h + d) × a × a (18,25):
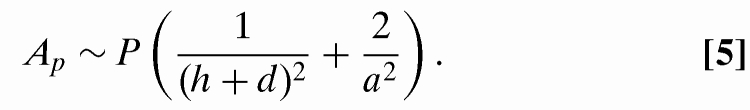 |
This term will combine with As, defined via Eq. 2, to form a constant A = As−Ap that gives the entropy gain per unit contour when a molecule is transferred from the pit into the slit. The second term in Eq. 4 arises from the excluded-volume interactions experienced by the DNA in the pit. The term scales as the number of possible binary interactions between DNA segments in the pit divided by the pit volume Vpit = (h + d)a2 (16):
 |
The quantity w is an effective chain thickness that can take into account ionic-strength-dependent electrostatic interactions enhancing the DNA self-avoidance (10,26,27). We can, in fact, prove that the excluded volume term is crucial to explaining our observations. Note that, for the case N = 1, Eq. 1 predicts that ΔFtot = Fp(Lp)−Fs(Lp) = −ALp + BLp1. If B = 0, then the free energy is minimized when the entire molecule falls into the pit, contrary to what we observe in Fig. 2D where only a portion of the molecule occupies the nanopit in the N = 1 case.
We estimate that w ≈ 9 nm in our buffer conditions (28–30). The value of weff used is obtained from the estimated ionic strength of 0.5× TBE with 3% 2-mercaptoethanol (30 mM) and reported measurements of the effective width (28). To calculate the ionic strength of 0.5×TBE, we have used the pK of Tris (8.1), boric acid (9.24) EDTA (1.99, 2.67, 6.16, 10.26), and 2-mercaptoethanol (9.6) and solved the full system of chemical equilibria (29) to obtain the concentrations of dissociated species. The EDTA is predominantly trivalent at the pH used. Ionic strength corrections to the equilibrium constants were handled via the Davies Equation (30).
Last, the N − 1 linker terms will contribute spring energy Fspring. In general, this spring energy can be obtained from integrating the force-extension curve for a worm-like chain, given to good approximation by the Marko–Siggia interpolation formula (31). The Marko–Siggia formula has been widely successful in interpreting the stretching of DNA in response to an applied force for both unconfined molecules (32) and molecules in slit-like confinement (33). The resulting spring energy is:
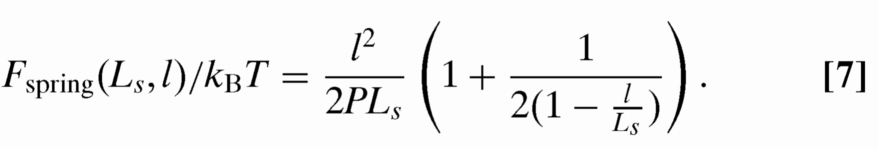 |
Note that, when l/L is small, Eq. 7 necessarily reduces to the formula for the energy of an entropic spring (21). When l/Ls is close to unity we recover the correct high force spring energy (34).
The free energy defined via Eqs. 1,2,4, and 7 can be minimized to find the equilibrium contour 〈Lp〉 placed in the nanopits as a function of N. Note that, although we do not explicitly include contour extending past the end pits (in accordance with experimental observations), the energy minimum will be obtained when there is no overhanging contour, as the linker entropy will be maximized in this case [Thermal fluctuations about the equilibrium configuration, however, can still in principle drive contour past the end pits, particularly when the central linkers are weakly extended (so that their spring constants are low).]. Inserting 〈Lp(N)〉 into Eq. 1 yields the energy of a state with N occupied pits ΔFtot(N). Using ΔFtot(N) we can evaluate the occupancy probability of each of these states in equilibrium:
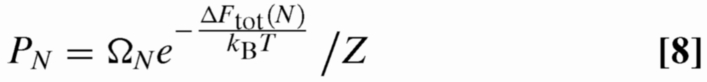 |
with Z the partition function:
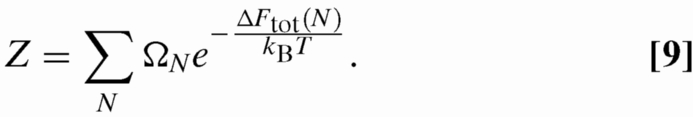 |
The degeneracy ΩN is the number of configurations possible on the lattice for N occupied pits. This is the number of translationally and rotationally unrelated self-avoiding walks with N steps on a square lattice that we obtain from ref. 35. Using PN we can then compute thermodynamically relevant parameters, for example, the average occupancy 〈n〉 = ∑N NPN.
The theoretical model predicts that a discrete spectrum of energy states ΔFtot(N) will be available to a molecule. Stable states are predicted that have both negative and positive ΔFtot. The negative states with ΔFtot/kBT < −1 can form spontaneously in equilibrium, resulting in the behavior shown in Fig. 2. In Fig. 5A we show the occupation probability predicted by Eq. 8 for λ-DNA as a function of lattice spacing. The theory predicts for λ-DNA that the molecule will tend to fluctuate between a number of neighboring states at lattice spacings <0.5 μm, will fluctuate between 3 and 2 states at l = 1–3 μm and will occupy only a single pit at l > 4μm. The average nanopit occupancy predicted by theory fits well to the experimental measurements (Fig. 5B). A least-squares fit yields A = 4.0 ± 1 μm−1 and B = 0.49 ± 0.05μm2 (errors bars give 95% confidence on parameters). Using Eqs. 3 and 5 we estimate that A = As − Ap ≈ 3.4 μm−1. Eq. 6 suggests that (we have used w ≈9 nm). Our results are thus numerically consistent with the order of magnitude predicted by theory. In addition, the fitted values of A and B can be used to predict the probability distribution PN of a molecule among states of varying N. We find that the theory yields excellent agreement with experiment in the nontrivial case of l = 0.5 μm (Fig. 3F).
Metastable States.
The states with positive ΔFtot(N) are metastable. These will not form spontaneously but can be created by the application of an external force. Once formed, these states will remain stable over a timescale τ as the cost of pit-to-pit contour transfer creates a significant energy barrier preventing the immediate decay of the state. For example, in the device with 300×300 nm nanopits, h = 100 nm and l = 5 μm dimer states do not form spontaneously. It is possible, however, to transform a N = 1 state into a N = 2 state by applying a small pulse of pressure to stretch the molecule out between the pits and then releasing the pressure once the molecule fills the second pit. Once formed for this particular geometry, dimer states will last for ≈ 1 − 2 min and then decay back to N = 1 (Fig. 6A). Although it is not possible to form strongly stretched long-lived metastable states with a 100-nm-deep slit, decreasing h increases the energy barrier and allows for greater stretching and longer-lived states. Decreasing the slit height to 50 nm, for example, it is possible to stretch λ-DNA out between 2 pits spaced 10 μm apart (Fig. 6B). This extension is comparable to that observed in 100×100 nm nanochannels (9). This stretched state lasted >3 min, relevant for optical mapping applications. Furthermore in the 50-nm device we were able to stretch T4-DNA between 3 pits spaced 20 μm apart (Fig. 6C) although this state decayed to the dimer configuration after 2.0 min.
Fig. 6.
Metastable conformations. (A) The decay of a λ-DNA dimer to a monomer state for h = 100 nm, a = 300 nm, d = 100 nm, and l = 5 μm. Time runs from top to bottom with each image 0.24 s apart. The dimer state existed for about 50 s (only a portion of the movie around the decay event is shown). (B) The formation of a λ-DNA dimer state in a slit with h = 50 nm, a = 300 nm, d = 100 nm, and l = 10 μm. Time runs from top to bottom with each image 0.24 s apart. These states last in excess of 3 min. (C) The decay of a T4-DNA trimer state to a dimer with h = 50 nm, a = 300 nm, d = 100 nm, and l = 20 μm. Time runs from top to bottom with each image 0.45 s apart. The trimer state existed for 2 min before decaying. (D) A schematic illustration of the decay of a dimer state. Initially the molecule is suspended between 2 pits. The contour in each pit Lp fluctuates around the average 〈L2p〉 for the dimer configuration. A thermal fluctuation then empties the right pit and the linker recoils to the remaining filled nanopit which gains contour. Eventually, the molecule is left in a monomer state with the remaining nanopit containing contour 〈L1p〉. (E) The free energy as a function of nanopit contour Lp for a dimer and monomer state with a = 300 nm, d = 100 nm, and l = 5 μm (corresponding to the situation in A). The nanopit empties when a fluctuation drives Lp to the origin, corresponding to crossing the energy barrier Ub. Once Lp = 0, the molecule will recoil to the remaining nanopit and reach the monomer conformation (corresponding to the lower curve). (Scale bar, 5 μm.)
Coupling Kramers theory of barrier crossing to Eq. 1, it is possible to estimate the timescale τ for decay from a state with occupancy N = 2 to N = 1 (the same arguments can be extended to decays from states of larger N). Consider a fluctuation that drives all contour Lp from the pit to the adjoining linker (Fig. 6D). Once the contour has been completely drained from the pit, the transition to a lower-energy state will occur immediately as the adjoining linker recoils to the pit that remains filled. This scenario can be modeled as a Kramers problem for escape from a minimum at Lp = 〈Lp〉 in the potential U(Lp) ≡ ΔFtot(N = 2,Lp)/kBT to a perfectly absorbing barrier at Lp = 0 (Fig. 6E). The escape rate for such a problem can be estimated from (36)
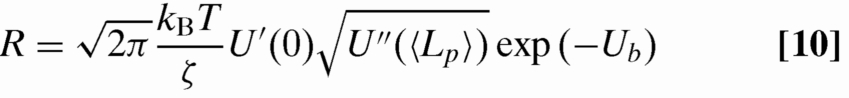 |
with ζ the friction factor, Ub the barrier height, U′(0) the derivative of U(Lp) with respect to Lp at Lp = 0 and U″(〈Lp〉) the second derivative at Lp = 〈Lp〉.
In principle the decay process could occur over a number of different paths, depending in detail how the contour draining from the pit is partitioned between the linker and the remaining pit. For simplicity, we will choose the following scenario: all of the contour in the pit being emptied is transferred to the linker before the linker recoils. This scenario minimizes the energy barrier for pit-to-pit contour transfer, for as long as the linker is extended between the 2 pits, increasing the contour in the linker will lower the spring energy, while placing the contour in the remaining pit will tend to enhance self-exclusion interactions via Eq. 4. We can estimate the energy barrier by computing the energy cost for emptying a single pit, taking into account as well the decrease in spring energy created by the increase in contour of the linker:
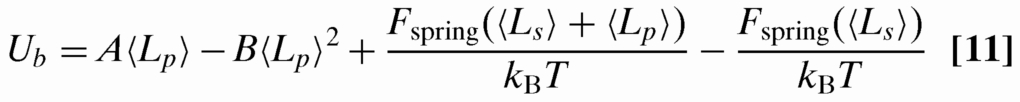 |
The derivatives U′(0) and U″(〈Lp〉) can be computed directly from ΔFtot. We estimate ζ as being on order of a Kuhn segment times viscosity 2Pη. This theory yields decay times τ on order of a minute for λ-DNA with h = 100 nm and l = 5 μm, approximately comparable to the experimental values.
Conclusion
We have demonstrated that embedded nanopit arrays can be used to control the positioning and conformation of DNA in nanoslits. In addition we have derived a free energy that can account for the number of nanopits occupied by λ-DNA in lattices of 300×300 nm nanopits. In the future this work should be extended to study the effect of varying nanopit size and, in particular, thermal fluctuations of the contour stored in each pit.
Acknowledgments.
We thank Peixiong Shi for assistance with ebeam lithography at the Danchip clean room facility and Derek Stein for helpful comments and discussion on this work. This work was supported by Danish Research Council for Technology and Production Sciences Grant 274-05-0375.
Footnotes
The authors declare no conflict of interest.
References
- 1.Moller R, Csaki A, Kohler JM, Fritzsche W. DNA probes on chip surfaces studied by scanning force microscopy using specific binding of colloidal gold. Nucleic Acids Res. 2000;28:e91. doi: 10.1093/nar/28.20.e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blow N. Microfluidics: in search of a killer application. Nat Methods. 2007;4:665–670. [Google Scholar]
- 3.Lin J, et al. Whole-genome shotgun optical mapping of deinococcus radiodurans. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 4.Braun E, Eichen Y, Sivan U, Ben-Yoseph G. DNA-templated assembly and electrode attachment of a conducting silver wire. Nature. 1998;391:775–778. doi: 10.1038/35826. [DOI] [PubMed] [Google Scholar]
- 5.Gu Q, et al. DNA nanowire fabrication. Nanotechnology. 2006;17:R14–R25. [Google Scholar]
- 6.Yan H, Park S, Finkelstein G, Reif J, LaBean T. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 7.Klinov D, Atlasov K, Kotlyar A, Dwir B, Kapon E. DNA nanopositioning and alignment by electron-beam-induced surface chemical patterning. Nano Lett. 2007;7:3583–3587. doi: 10.1021/nl072177y. [DOI] [PubMed] [Google Scholar]
- 8.Tegenfeldt JO, et al. The dynamics of genomic-length DNA molecules in 100-nm channels. Proc Natl Acad Sci USA. 2004;101:10979–10983. doi: 10.1073/pnas.0403849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reisner W, et al. Statics and dynamics of single DNA molecules confined in nanochannels. Phys Rev Lett. 2005;94:196101. doi: 10.1103/PhysRevLett.94.196101. [DOI] [PubMed] [Google Scholar]
- 10.Reisner W, et al. Nanoconfinement-enhanced conformational response of single DNA molecules to changes in ionic environment. Phys Rev Lett. 2007;99:058302. doi: 10.1103/PhysRevLett.99.058302. [DOI] [PubMed] [Google Scholar]
- 11.Reccius CH, Mannion JT, Cross JD, Craighead HG. Compression and free expansion of single DNA molecules in nanochannels. Phys Rev Lett. 2005;95:268101. doi: 10.1103/PhysRevLett.95.268101. [DOI] [PubMed] [Google Scholar]
- 12.Stein D, Heyden FHJ, Koopmans W, Dekker C. Pressure-driven transport of confined DNA polymers in fluidic channels. Proc Natl Acad Sci USA. 2006;103:15853–15858. doi: 10.1073/pnas.0605900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balducci A, Mao P, Han J, Doyle P. Double-stranded DNA diffusion in slitlike nanochannels. Macromolecules. 2006;39:6273–6281. [Google Scholar]
- 14.Zimmermann RM, Cox EC. DNA stretching on functionalized gold surfaces. Nucleic Acids Res. 1994;22:492–497. doi: 10.1093/nar/22.3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jun S, Thirumalai D, Ha BY. Compression and stretching of a self-avoiding chain in cylindrical nanopores. Phys Rev Lett. 2008;101:138101. doi: 10.1103/PhysRevLett.101.138101. [DOI] [PubMed] [Google Scholar]
- 16.Cacciuto A, Luijten E. Self-avoiding flexible polymers under spherical confinement. Nano Lett. 2006;6:901–905. doi: 10.1021/nl052351n. [DOI] [PubMed] [Google Scholar]
- 17.Sakaue T, Raphael E. Polymer chains in confined spaces and flow-injection problems: some remarks. Macromolecules. 2006;39:2621–2628. [Google Scholar]
- 18.Sakaue T. Semiflexible polymer confined in closed spaces. Macromolecules. 2007;40:5206–5211. [Google Scholar]
- 19.Nykypanchuk D, Strey H, Hoagland D. Single molecule visualizations of polymer partitioning within model pore geometries. Macromolecules. 2005;38:145–150. [Google Scholar]
- 20.Ed Rai-Choudhury P. SPIE Handbook of Microlithography, Micromachining and Microfabrication. Vol. 1. Bellingham, WA: SPIE; 1997. Section 2.7.2.4. [Google Scholar]
- 21.de Gennes PG. Scaling Concepts in Polymer Physics. Ithaca, NY: Cornell Univ Press; 1979. [Google Scholar]
- 22.Brochard-Wyart F, Tanaka T, Borghi N, de Gennes PG. Semiflexible polymers confined in soft tubes. Langmuir. 2005;21:4144–4148. doi: 10.1021/la0474114. [DOI] [PubMed] [Google Scholar]
- 23.Burkhardt T. Free energy of a semiflexible polymer in a tube and statistics of a randomly-accelerated particle. J Phys A. 1997;30:L167–L172. [Google Scholar]
- 24.Quake S, Babcock H, Chu S. The dynamics of partially extended single molecules of DNA. Nature. 1997;388:151–154. doi: 10.1038/40588. [DOI] [PubMed] [Google Scholar]
- 25.Doi M, Edwards SF. The Theory of Polymer Dynamics. New York, NY: Oxford Univ Press; 1986. [Google Scholar]
- 26.Rybenkov V, Cozzarelli N, Vologodskii A. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc Natl Acad Sci USA. 1993;90:5307–5311. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh CC, Balducci A, Doyle P. Ionic effects on the equilibrium dynamics of DNA confined in nanoslits. Nano Lett. 2008;8:1683–1688. doi: 10.1021/nl080605+. [DOI] [PubMed] [Google Scholar]
- 28.Rybenkov V, Cozzarelli N, Vologodskii A. Proc Natl Acad Sci USA. 1993;90:5307. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li A, Qi L, Shih H, Marx K. Biopolymers. 1996;38:367. doi: 10.1002/(sici)1097-0282(199603)38:3<367::aid-bip9>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 30.Perrin D, Dempsey B. Buffers for pH and Metal Ion Control. London: Chapman and Hall; 1974. [Google Scholar]
- 31.Marko J, Siggia E. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 32.Bustamante C, Smith S, Liphardt J, Smith D. Single-molecule studies of DNA mechanics. Curr Opin Struct Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- 33.Bakajin OB, et al. Electrohydrodynamic stretching of DNA in confined environments. Phys Rev Lett. 1998;80:2737–2740. [Google Scholar]
- 34.Wilhelm J, Frey E. Radial distribution function of semiflexible polymers. Phys Rev Lett. 1996;77:2581–2584. doi: 10.1103/PhysRevLett.77.2581. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan D, Aynechi T, Voelz V, Kuntz I. Information content of molecular structures. Biophys J. 2003;85:174–190. doi: 10.1016/S0006-3495(03)74464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLeish T. In: Soft Condensed Matter Theory in Mol Cell Biol. Poon WCK, Andelman D, editors. New York: Taylor and Francis; 2006. pp. 123–135. [Google Scholar]