Abstract
The selectins are cell adhesion proteins that must resist applied forces to mediate leukocyte tethering and rolling along the endothelium and have 2 conformational states. Selectin–ligand bond dissociation increases only modestly with applied force, and exhibits catch bond behavior in a low-force regime where bond lifetimes counterintuitively increase with increasing force. Both allosteric and sliding–rebinding models have emerged to explain catch bonds. Here, we introduce a large residue into a cleft that opens within the lectin domain to stabilize the more extended, high-affinity selectin conformation. This mutation stabilizes the high-affinity state, but surprisingly makes rolling less stable. The position of the mutation in the lectin domain provides evidence for an allosteric pathway through the lectin domain, connecting changes at the lectin–EGF interface to the distal binding interface.
Keywords: cell rolling, mechanochemistry, vasculature
In an inflammatory response, leukocytes in the bloodstream first tether and roll along the endothelium, then arrest and extravasate through the vessel wall to the site of infection or injury (1, 2). The ability of leukocytes to tether and roll over the wide range of shear stresses experienced in the vasculature is mediated by cell surface-displayed lectins referred to as the selectins. P-selectin is expressed on activated endothelium and platelets, and its primary ligand P-selectin glycoprotein ligand 1 (PSGL-1), which is expressed on leukocytes (2). The processes of cell tethering, rolling, and arrest have all been reproduced by using in vitro flow chambers.
Rolling through selectins is unusually stable to changes in the concentration of ligand on the substrate and the wall shear stress. As wall shear stress increases, rolling velocity increases much less. One factor contributing to the mechanical stability of rolling through selectins is the relatively moderate increase in the off rates for selectin–ligand bonds as the force experienced by the bond is increased (3–5). Unique to the selectins among leukocyte adhesion molecules is the observation of a shear threshold effect for cell tethering and rolling adhesion. At low shear stresses few cells tether and rollingly adhere, whereas above a threshold shear stress (or shear), many cells tether and roll. With increasing shear, more and more cells tether and roll, until a peak is reached, beyond which increasing shear results in decreased numbers of rolling cells. This effect was first demonstrated for L-selectin (6) and later for P- and E-selectin (7). Measurements of the number of bonds between a rolling cell and the substrate have provided an explanation for these effects (8). More bonds are present being a rolling cell and the substrate at high shear than low shear. The shear threshold occurs at the shear where the number of bonds between the cell and the substrate is close to 1. Thus, as wall shear stress increases, the higher rate of breakage of individual selectin–ligand bonds is largely compensated by the formation of a larger number of selectin–ligand bonds between the cell and the substrate, suggesting that the rate of bond formation increases with increasing shear (8).
Most receptor–ligand bonds are slip bonds that decrease in lifetime as tensile force increases on the receptor–ligand complex. In contrast, catch bonds increase in lifetime under a range of applied forces. Experimental evidence for the existence of catch bonds came through atomic force microscopy experiments examining the interaction between P-selectin and PSGL-1 (9). With increasing force, the lifetime of these bonds first increased, and then at higher forces, decreased. Furthermore, with the observation of 2 distinct kinetic pathways of P-selectin dissociation from its ligands by using a biomembrane force probe, Evans et al. (10) developed a mathematical model in which force altered the propensity for dissociation through the 2 different pathways.
Two distinct states of a selectin have been defined by crystallography (11). Selectins contain from N to C terminus a ligand-binding lectin domain, an epidermal growth factor-like (EGF) domain, multiple short consensus repeat (SCR) domains, a transmembrane domain, and a short cytoplasmic domain. P-selectin crystallized with and without a bound PSGL-1 ligand fragment showed 2 different conformations that differed in the angle between the lectin and EGF domains, and here are termed extended and bent, respectively. It should be pointed out that the bent conformation crystallized in the absence of ligand can also bind ligand, as shown by soaking ligand into preformed crystals (11). The conformation crystallized in the presence of ligand is more extended, because the distance between the ligand binding site and the C terminus of the EGF domain, where tensile force would be applied in vivo, is greater than in the bent conformation.
Extended protein conformations are favored by applied tensile force, and if the extended conformation is higher affinity, this provides a mechanism for explaining catch-bond behavior (12–14). A mutation introduced into the lectin–EGF interface designed to favor the extended, ligand-bound conformation of P-selectin does indeed exhibit a higher affinity for ligand in solution, and when expressed on the cell surface results in slower cell rolling velocities and increased resistance to shear stress (13). This finding provides support for the role of a conformational change in selectin function. Mutations at the lectin–EGF interface in L-selectin also altered its rolling behavior (13).
A second model for catch-bond behavior was recently advanced to explain the increased lifetime of selectin–ligand interactions under applied force. The “sliding-rebinding” model is based on cell rolling data with wild-type and mutant L-selectin with the Asn138Gly substitution in the EGF domain at the interface with the lectin domain (15, 16). The model describes the lectin–EGF interface as a hinge region with 2 preferred orientations, with flexibility at the hinge influenced by residue 138 (15, 16). In this model, extension is not proposed to change the conformation of the ligand binding site. Instead, extension tilts the interface to align with the direction of force application. It is further proposed that the ligand does not bind to a unique binding site in the lectin domain, and that after slipping out of one binding site, the ligand could bind to many further, overlapping binding sites that are distributed over the lectin domain surface in the direction of the pulling force, and would also have the opportunity to subsequently rebind to sites from which it had previously slipped out. Thus, as force is applied, multiple opportunities to reorient and rebind prolong the association time of the bond (15, 16).
Here, rather than altering P-selectin at the lectin–EGF interface, we mutate P-selectin at a point of conformational change within the lectin domain, to demonstrate that subtle motions within the lectin domain are an important part of selectin conformational change and function. The data support models that invoke a simple 2-state conformational equilibrium in selectins, rather than the sliding-rebinding model.
Results
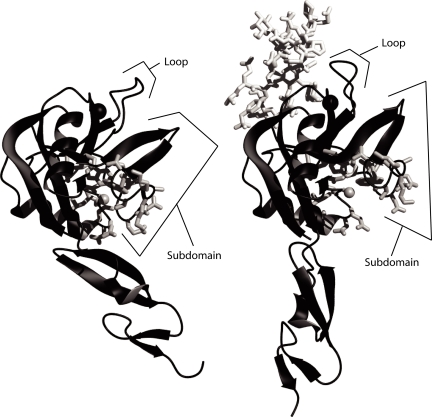
Visual inspection of the crystal structures of P-selectin crystallized in the absence (Fig. 1Left) or presence (Fig. 1 Right) of ligand (11) reveals a lobe in the lectin domain that reorients to bind ligand, such that a cleft “opens” within the lectin domain. This cleft is not at the lectin–EGF interface, or near the residues involved in binding ligand. For a residue with a small side chain and a Cα-Cβ vector pointing into the cleft, replacement with a larger residue would be expected to favor the extended conformation of P-selectin. The design module of Rosetta (17) was used to scan for an appropriate mutation. Alanine 28 was chosen to be replaced with a histidine. For most substitutions of residue 28 larger than alanine, no rotamer could be found that would be free of steric clashes in the bent conformation. In contrast, rotamers could be found that would be free of steric clashes in the extended conformation of P-selectin.
Fig. 1.
Opening of a cleft in the lectin domain in the extended selectin conformation. (Left) P-selectin in the unliganded conformation (PDB ID code 1G1Q). (Right) P-selectin in the liganded, extended conformation (1G1S). In each structure, the Cβ atom of Ala-28 in the cleft is shown as a gray sphere, and other side chains that line the cleft are shown as white sticks. The divalent cation in the ligand binding site is shown as a larger black sphere. Molecules are shown as ribbons in identical orientations after superposition on the lectin domain.
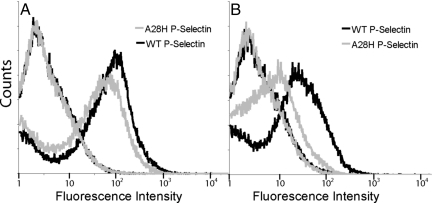
P-selectin containing the lectin, EGF, and first 2 short consensus repeat (SCR) domains was displayed on the yeast surface as a fusion to α-agglutinin, which is covalently linked to the yeast cell wall. Expression levels of A28H P-selectin on yeast were comparable to wild-type P-selectin levels based on antibody binding to the SCR domains of P-selectin (Fig. 2A). However, significantly lower staining of the A28H mutant is seen when an anti-lectin domain antibody is used (Fig. 2B). A similar decrease in G3 mAb reactivity was previously observed when a glycan wedge was introduced into the lectin–EGF domain interface to stabilize the extended conformation of P-selectin (13).
Fig. 2.
P-selectin expression on yeast. Yeast with plasmids encoding wild-type P-selectin or point mutant A28H P-selectin were labeled with anti-P-selectin antibodies and visualized with phycoerythrin-conjugated secondary antibodies. In both A and B, the left-most curves are uninduced yeast cultures, and the right-most curves are postinduction yeast cultures. (A) Antibody directed at the short consensus repeats of P-selectin (AC1.2). (B) Antibody directed at the lectin domain of P-selectin (G3).
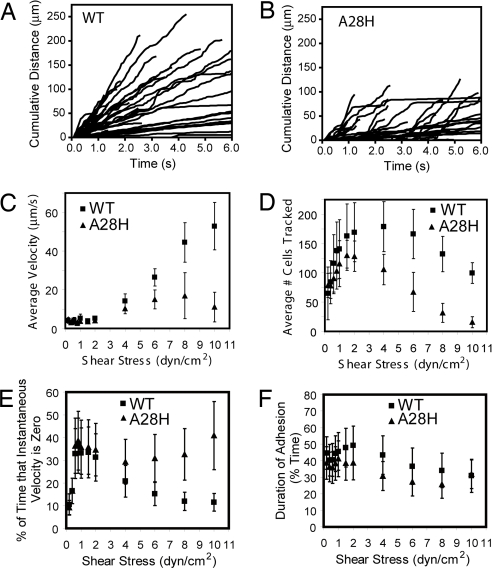
Tethering and rolling of yeast cells expressing wild-type and A28H P-selectin on a PSGL-1 substrate were evaluated in a parallel wall chamber (Fig. 3). Rolling velocities of mutant and wild-type cells were similar at lower shear, but mutant cells rolled more slowly at higher wall shear stresses (Fig. 3C), including at 4 dyn/cm2 (Fig. 3 A and B). This observation of slower rolling can arise by two mechanisms. First, rolling through A28H could be less stable. As shear stress increases, less stable rolling would result in cells detaching more often and at lower shear stresses. Because there is a distribution in the amount of P-selectin expressed per cell, it is possible that a subpopulation of cells with on average higher expression of P-selectin would remain bound at higher shear stresses, resulting in the observation of slower rolling at higher shear. This is consistent with the observation that rolling through A28H P-selectin is less stable, as described below. Second, the rolling mediated through A28H P-selectin could be slower solely due to the mutation; however, we do not have any information that allows us to discriminate between the first and second possibilities.
Fig. 3.
Yeast rolling profiles. (A and B) Cumulative distance rolled of tracked cells at 4 dyn/cm2 for wild-type P-selectin and A28H P-selectin, respectively. Each line represents a single cell. (C) Average rolling velocity ± SD, calculated from the average rolling velocities determined from each of 9 separate trials. See Materials and Methods for more detail. (D) Average number of cells tracked at each shear stress. Densities of cells infused to the flow chamber were comparable. (E) Quantitation of rolling stability. Of the 6 seconds tracked at each shear, percentage of time that the instantaneous velocity was zero. (F) Another measure of rolling stability. Of the 6 s tracked, percentage of time that cells were adherent (rolling or stopped but attached) as opposed to detaching and no longer being tracked in the 6-s window.
Importantly, rolling through A28H P-selectin is less stable than rolling through wild-type P-selectin, as shown in several ways in Fig. 3 D–F. The average number of cells tracked at each shear is shown in Fig. 3D. The cells infused to the flow chamber were at comparable concentrations for each experiment. At low shear stresses fewer cells are tracked, as cells in flow continue to attach with increasing shear. Beyond a given shear stress, fewer cells are tracked as they begin to detach. Wild-type cells attached at higher shear, as reflected by the peak number of cells tracked at 4 dyn/cm2 for wild-type as opposed to 1.5 dyn/cm2 for the A28H mutant. Also, the resistance to detachment was greater for wild-type cells. At 10 dyn/cm2 compared with 4 dyn/cm2, the percentage of cells remaining bound was 55% for wild-type and 15% for mutant (Fig. 3D). Both of these observations indicate greater rolling stability through wild-type P-selectin.
The tracks of cells displaying A28H P-selectin exhibit more plateaus or “steps” during rolling than cells displaying wild-type P-selectin (Fig. 3 A and B). This is quantitated in Fig. 3E as the amount of time that cells spend with an instantaneous velocity of zero. This again demonstrates that rolling through the mutant protein is less smooth and hence less stable. This can also be seen in terms of the amount of time that cells are adherent (whether stationary or rolling) relative to the total time of 6 s that cells were tracked at each shear. Cells were tracked as long as they were adherent for at least 0.5 s of the observation period, whether they tethered to the substrate during the 6-s observation period, or detached before the end of this period. Despite large standard deviations in the population of rolling cells, Fig. 3F shows that cells rolling through A28H P-selectin are more likely to be adherent for a shorter period.
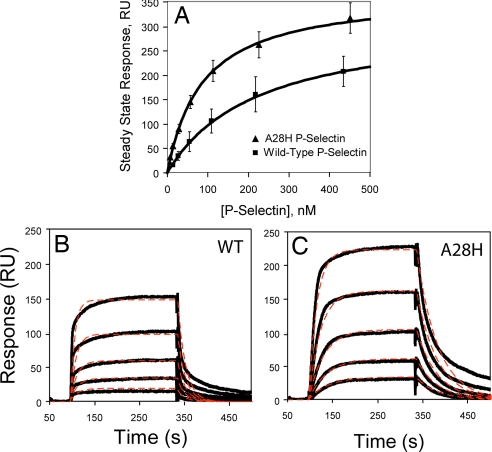
To understand the differences in binding properties that contribute to altered rolling behavior, surface plasmon resonance was used. Purified WT or A28H P-selectin fragments containing the lectin, EGF, and SCR domains 1 and 2 were injected over immobilized PSGL-1-Fc. Steady-state analysis (Fig. 4A) revealed a higher binding affinity of 87 ± 4 nM for the A28H P-selectin compared with 224 ± 7 nM for wild-type P-selectin.
Fig. 4.
Surface plasmon resonance binding responses of wild-type P-selectin and A28H P-selectin. (A) Steady-state response levels fit to a 1:1 binding isotherm. (B and C) Global fits of the data (thick lines) to a 1:1 binding event with single exponential on and off rates (thin dashed lines).
Qualitatively, the kinetic profiles reveal during the dissociation phase a slower koff for A28H P-selectin (solid lines, Fig. 4 B and C). Although the steady-state response levels are reasonably well captured by a 1:1 binding model, certain features of the association and dissociation phases are not reproduced (Fig. 4 B and C, dashed lines). A model in which two equilibrated conformations (bent and extended) are capable of binding and dissociating from the chip surface with differing kinetics results in too many highly correlated fitting parameters, and reliable values for these parameters could not be extracted. Furthermore, heterogeneity in the glycosylation or sulfation of PSGL-1 could contribute to heterogeneity in on and off rates (11). Therefore, the main features of the P-selectin interaction with PSGL-1 are summarized by fitting the steady-state responses and the kinetics to a simple 1:1 model (Table 1). Estimates from both approaches are within the same range, and show a 3- to 4-fold higher affinity of A28H mutant compared with wild-type. The values for WT P-selectin interactions with PSGL-1 are consistent with previous observations (11, 13).
Table 1.
Affinity and kinetics
| P-selectin | Steady state |
Kinetic binding model |
||
|---|---|---|---|---|
| Kd, nM | Kd, nM | ka, M−1s−1 | kd, s−1 | |
| WT | 224 ± 7 | 288 ± 292 | 1.4 (±1.0) x105 | 0.04 ± 0.03 |
| A28H | 87 ± 4 | 64 ± 29 | 2.9 (±1.3) x105 | 0.019 ± 0.003 |
Discussion
Swapping portions of domains between E-, L-, and P-selectins identified the EGF domain as being capable of drastically altering selectin function (18–20). Later, the same changes in function were seen by changing a single residue (position 138) in the lectin domain-proximal portion of the EGF domain of L-selectin (13, 15). It was also shown that introducing a glycan wedge to favor a more obtuse, extended-like angle between the lectin and EGF domains would bring about similar a functional change in P-selectin (13).
The profound effects on selectin function of mutations at the lectin–EGF domain interface have led to two mutually exclusive structural models to explain the coupling between ligand binding and the orientation between the lectin and EGF domains. The sliding-rebinding model does not propose that extension is linked to a conformational change in the lectin domain. Instead, extension is proposed to promote mechanical stability by enabling force to align the lectin–ligand interface so that multiple distinct but overlapping ligand binding sites can be occupied along the force-induced unbinding pathway (15, 16). In contrast, the allostery model proposes that there is a single ligand binding site that can exist in high- and low-affinity conformations, and that the conformation at the ligand binding site is coupled through a network of atomic interactions to the orientation between the lectin and EGF domains (13).
The most significant differences in the crystal structures of the lectin and EGF domains of P-selectin unbound and bound to a glycosulfopeptide (11) are the reorientation of the lectin and EGF domains relative to one another, the opening of a cleft in the lectin domain that alters the environment around Ala-28, and the movement of a loop (residues 83–89) toward the ligand binding site, which provides additional binding surface to interact with ligand (Fig. 1). The work presented here focuses on a position within the cleft that opens in the ligand-bound structure.
Alanine 28 is ideally positioned within the lectin domain to assess whether conformational change within the lectin domain is propagated to the ligand binding site. Alanine 28 is buried in the bent conformation of P-selectin, but is exposed in the extended conformation. This position is not in the pivot region between the lectin and EGF domains, nor is it contacting ligand. The mutation of alanine to histidine serves to fill the space observed in the extended, ligand-bound conformation of P-selectin. Filling this space with a larger residue is expected to create steric clashes in the bent conformation and therefore favor the extended conformation even in the absence of ligand or applied force.
Based on differential antibody staining, the mutation does alter the conformation of the lectin domain. The mutation results in decreased binding by mAb G3 to the lectin domain, with no effect on binding by a mAb to the SCR domains. Ala-28 cannot be part of the G3 epitope, because it has no solvent exposure in the bent P-selectin conformation that binds this antibody best. This suggests that the conformation of the lectin domain has indeed been altered due to the A28H point mutation. A similar decrease in G3 mAb reactivity was previously observed when a glycan wedge was introduced into the lectin–EGF domain interface to stabilize the extended conformation of P-selectin (13). Therefore, effects of two different mutations, one designed to force the angle between the lectin and EGF domains to resemble the extended conformation, and another designed to prohibit the lectin domain from adopting the bent configuration, both have similar effects on antibody binding.
Furthermore, the A28H mutation, as predicted, results in a higher affinity for ligand. This strongly suggests that the change at Ala-28 propagates through the subdomain above alanine 28 to the loop (residues 83–89) to affect the ligand binding interface. Favoring extension with a glycan wedge also increased affinity for ligand (13). It is reasonable to suggest that the change at Ala-28 is propagated to the lectin–EGF interface region as well, favoring the extended orientation of the lectin and EGF-like domains.
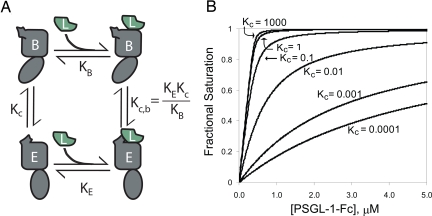
The findings that simple changes at the lectin–EGF interface can alter selectin function, and that function can be disrupted by altering the ability of the selectin to adopt crystallographically observed conformations, suggest a simple model for the equilibrium between two distinct selectin conformations, with different affinities for ligand (Fig. 5). We do not claim that our data uniquely support this model or that the model is novel; indeed it is a recapitulation of classical laws of allostery and mass action. However, given the large amount of attention devoted recently to the sliding-rebinding theory (15, 16), we believe it is important to clearly state the alternative model. A single equilibrium constant Kc describes the ratio of the two unbound conformations. In this model of conformational equilibria, the observed binding isotherm will fit a simple 1:1 binding interaction. However, the binding constant that fits this curve will actually reflect the underlying equilibria depicted in Fig. 5 with the following relationship:
where KB is the affinity of ligand for the bent conformation, KE is the affinity of ligand for the extended conformation, and Kc is the equilibrium between the two unbound conformations. A change in Kc to favor the higher-affinity conformation increases the observed binding affinity (Fig. 5B).
Fig. 5.
Model of a conformational equilibrium between 2 distinct conformations of the selectins. (A) Equilibrium between 2 distinct conformations of P-selectin in solution whereby each conformation is capable of binding ligand with different affinities. (B) Effect of shifting only the equilibrium (Kc) between the 2 unliganded conformations on an observed binding curve.
We interpret our results in the context of this model. The steady-state SPR data revealed a higher binding affinity for the mutant P-selectin as compared with the wild-type protein. Kinetic measurements showed an increase in kon and a decrease in koff. Further, koff/kon yielded a Kd in agreement with the steady-state data and a similar estimate of the increase in affinity of the mutant. The increased affinity and change in a lectin domain epitope is consistent with the properties seen when P-selectin is designed to favor the extended conformation by introducing a glycan wedge (13). Higher affinity induced by mutation corresponds to an increase in the conformational equilibrium constant in absence of ligand, Kc, in our model. It is important to note that the previous study altered the lectin–EGF interface, whereas here the mutation is within the lectin domain, removed from the interface and removed from the ligand binding site. Our results suggest that a mutation in the allosteric pathway between the lectin/EGF inferface and the ligand binding site can increase affinity for ligand.
The A28H mutant rolls less stably. Within the context of the model of conformational equilibria, the less stable rolling may be reflecting an impaired ability to interconvert between conformations, i.e., lower rates of equilibration between conformations. Although having higher affinity for ligand, and a higher on rate, cells bearing the mutant receptor nonetheless bound to substrate less efficiently, and were more susceptible to detachment. Further, rolling was less smooth, with longer pauses during rolling seen in the mutant than wild-type cells. It should be emphasized that our data on ligand binding and dissociation kinetics do not provide information on the rate with which selectins equilibrate between the bent and extended conformations. However, our finding that a selectin mutant with more favorable binding properties mediates less stable rolling leads us to speculate that the mutant shows a decrease in the rate of equilibration between conformational states, and that the rate at which selectins interconvert between bent and extended conformations may be an important component of the rolling mechanism.
The model presented here can be applied to the glycan wedge mutation in P-selectin (13), and the A28H mutation described here. This model can also be applied to mutants of L-selectin that introduce the N138G mutation in the EGF domain (13, 15, 18–20). Previously, the N138G mutation has been suggested to increase flexibility of the lectin–EGF interface, in support of the sliding-rebinding model. However, this mutation could shift the equilibrium to a more extended conformation (i.e., increase Kc in Fig. 5) by altering the equilibrium for conformational change at the interface between the lectin and EGF domain. Furthermore, in both the A28H and glycan wedge P-selectin mutants, flexibility of the lectin domain with respect to the EGF domain is expected to decrease. However, increases in affinity for ligand are observed. The sliding-rebinding model posits no linkage between the EGF domain and affinity for ligand in the absence of force, and no conformational change in the lectin domain that alters ligand binding affinity. Our results are inconsistent with the sliding-rebinding model, and support the hypothesis that the conformation at the EGF–lectin interface is allosterically transmitted through the lectin domain to the ligand binding site.
Evans et al. (10) presented a model that considers two different bound-state configurations to explain the observation of two dissociation pathways when the selectin–ligand interaction is forced apart. In that model, two distinct bound-state configurations are considered to be in equilibrium, either of which can dissociate under applied force. The ratio between the two bound states in the absence of force is represented by Φo in their model and is equivalent to Kc,b (Fig. 5A). As force increases, Φ changes so that the more slowly rupturing configuration is more populated. Evans et al. did not propose a structural correlate for the two bound-state configurations, or mechanism by which force would alter Φ. Here, we propose that the two bound-state configurations with fast and slow rupture correspond to ligand bound to the bent and extended selectin conformations, respectively. The structural details of this model are included in an accompanying article (21). In addition to conformational equilibrium (Kc,b between the two bound-state configurations), we have included the conformational equilibrium Kc between the unbound selectins (Fig. 5A). We propose that the structures seen in the absence of force, i.e., in the crystals of bent E-selectin soaked with sialyl LewisX, or in the crystals of extended P-selectin formed in the presence of ligand, are close to the structures of the bound-state configurations in the presence of applied force. Although not consistent with all data (10), the idea is extremely attractive that the observation of two dissociation pathways in the presence of force is a natural consequence of the underlying equilibria in the absence of force.
Thomas et al. (22, 23) have developed a model similar to that of Evans et al. (10) to describe the experimental data collected on the bacterial adhesion protein FimH. They have applied an allosteric-like model to FimH whereby changes at the interface of two domains are propagated to the ligand binding site to affect binding properties, resulting in a resistance to unbinding under applied force (22, 24). It has been proposed that a similar allosteric model might also apply to the selectins (23). Another example is provided by the integrin I domains where the case has been made that structurally disparate adhesion molecules have conformational states in which the more extended state is higher affinity, and that force-induced extension may be a general mechanism by which force can act as an allosteric effector in the mechanochemistry of adhesion molecules (14).
Materials and Methods
Detailed methods are found in supporting information (SI) Text. In brief, the design module of Rosetta was used to examine substitutions to every residue except Cys in the lectin and EGF domains of P-selectin and find those that would destabilize the bent but not the extended conformation. Wild-type and A28H mutant P-selectin fragments containing the lectin, EGF, and sushi domains 1 and 2 were expressed in HEK293S GnT1− cells and purified by using C-terminal tags. The same segments were displayed on yeast covalently linked through their C termini to the cell wall. Yeast cells were infused into a parallel wall flow chamber with PSGL-1-Fc adsorbed on the lower wall by using a syringe pump and subjected to incremental increases every 10 s in wall shear stress. Video images were captured through an inverted microscope. Custom macros in ImagePro 6.0 were used to track individual, nonbudding yeast cells from frame to frame. Track data were exported into Excel and used to determine number of cells and cell velocities. Three runs through shear stress increases were performed for 3 separate yeast cultures, resulting in 9 trials for both wild-type and A28H P-selectin. Surface plasmon resonance on a BIACore 3000 used P-selectin concentrations from 5 nM to 0.5 μM injected over PSGL-1-Fc surfaces.
Supplementary Material
Acknowledgments.
We thank Drs. Larry Shapiro, Wendy Thomas, and William Somers for reviewing this manuscript. This work was supported by National Institutes of Health Grant HL-48675 and a Kirschstein National Research Service Award.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810620105/DCSupplemental.
References
- 1.Butcher EC. Leukocyte-endothelial cell recognition: Three (or more) steps to specificity and diversity. Cell. 1991;67:1033–1036. doi: 10.1016/0092-8674(91)90279-8. [DOI] [PubMed] [Google Scholar]
- 2.Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: The multi-step paradigm. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 3.Alon R, Hammer DA, Springer TA. Lifetime of the P-selectin: Carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature. 1995;374:539. doi: 10.1038/374539a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Alon R, Fuhlbrigge RC, Springer TA. Rolling and transient tethering of leukocytes on antibodies reveal specializations of selectins. Proc Natl Acad Sci USA. 1997;94:3172–3177. doi: 10.1073/pnas.94.7.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Chateau M, Chen S, Salas A, Springer TA. Kinetic and mechanical basis of rolling through an integrin and novel Ca2+-dependent rolling and Mg2+-dependent firm adhesion modalities for the α4β7-MAdCAM-1 interaction. Biochemistry. 2001;40:13972–13979. doi: 10.1021/bi011582f. [DOI] [PubMed] [Google Scholar]
- 6.Finger EB, et al. Adhesion through L-selectin requires a threshold hydrodynamic shear. Nature. 1996;379:266–269. doi: 10.1038/379266a0. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence MB, Kansas GS, Kunkel EJ, Ley K. Threshold levels of fluid shear promote leukocyte adhesion through selectins (CD62L,P,E) J Cell Biol. 1997;136:717–727. doi: 10.1083/jcb.136.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S, Springer TA. An automatic braking system that stabilizes leukocyte rolling by an increase in selectin bond number with shear. J Cell Biol. 1999;144:185–200. doi: 10.1083/jcb.144.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marshall BT, et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 10.Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc Natl Acad Sci USA. 2004;101:11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Somers WS, Tang J, Shaw GD, Camphausen RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLeX and PSGL-1. Cell. 2000;103:467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- 12.Konstantopoulos K, Hanley WD, Wirtz D. Receptor-ligand binding: ‘catch’ bonds finally caught. Curr Biol. 2003;13:R611–R613. doi: 10.1016/s0960-9822(03)00529-3. [DOI] [PubMed] [Google Scholar]
- 13.Phan UT, Waldron TT, Springer TA. Remodeling of the lectin/EGF-like interface in P- and L-selectin increases adhesiveness and shear resistance under hydrodynamic force. Nat Immun. 2006;7:883–889. doi: 10.1038/ni1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Astrof NS, Salas A, Shimaoka M, Chen JF, Springer TA. Importance of force linkage in mechanochemistry of adhesion receptors. Biochemistry. 2006;45:15020–15028. doi: 10.1021/bi061566o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lou J, et al. Flow-enhanced adhesion regulated by a selectin interdomain hinge. J Cell Biol. 2006;174:1107–1117. doi: 10.1083/jcb.200606056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lou J, Zhu C. A structure-based sliding-rebinding mechanism for catch bonds. Biophys J. 2007;92:1471–1485. doi: 10.1529/biophysj.106.097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 18.Kansas GS, Spertini O, Stoolman LM, Tedder TF. Molecular mapping of functional domains of the leukocyte receptor for endothelium, LAM-1. J Cell Biol. 1991;114:351–358. doi: 10.1083/jcb.114.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kansas GS, et al. A role for the epidermal growth factor-like domain of P-selectin in ligand recognition and cell adhesion. J Cell Biol. 1994;124:609–618. doi: 10.1083/jcb.124.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dwir O, Kansas GS, Alon R. An activated L-selectin mutant with conserved equilibrium binding properties but enhanced ligand recognition under shear flow. J Biol Chem. 2000;275:18682–18691. doi: 10.1074/jbc.M001103200. [DOI] [PubMed] [Google Scholar]
- 21.Springer TA. Structural basis for selectin mechanochemistry. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0810784105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Bacterial adhesion to target cells enhanced by shear force. Cell. 2002;109:913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 23.Thomas W. For catch bonds, it all hinges on the interdomain region. J Cell Biol. 2006;174:911–913. doi: 10.1083/jcb.200609029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas W, et al. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys J. 2006;90:753–764. doi: 10.1529/biophysj.105.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.