Abstract
Proteins are made from 19 aa and, curiously, one N-alkylamino acid (“imino acid”), proline (Pro). Pro is thought to be incorporated by the translation apparatus at the same rate as the 19 aa, even though the alkyl group in Pro resides directly on the nitrogen nucleophile involved in peptide bond formation. Here, by combining quench-flow kinetics and charging of tRNAs with cognate and noncognate amino acids, we find that Pro incorporates in translation significantly more slowly than Phe or Ala and that other N-alkylamino acids incorporate much more slowly. Our results show that the slowest step in incorporation of N-alkylamino acids is accommodation/peptidyl transfer after GTP hydrolysis on EF-Tu. The relative incorporation rates correlate with expectations from organic chemistry, suggesting that amino acid sterics and basicities affect translation rates at the peptidyl transfer step. Cognate isoacceptor tRNAs speed Pro incorporation to rates compatible with in vivo, although still 3–6 times slower than Phe incorporation from Phe-tRNAPhe depending on the Pro codon. Results suggest that Pro is the only N-alkylamino acid in the genetic code because it has a privileged cyclic structure that is more reactive than other N-alkylamino acids. Our data on the variation of the rate of incorporation of Pro from native Pro-tRNAPro isoacceptors at 4 different Pro codons help explain codon bias not accounted for by the “tRNA abundance” hypothesis.
Keywords: non-natural amino acids, ribosomes, tRNA, EF-Tu, GTPase
N-alkylation confers special properties on amino acids and polypeptides. The N-alkyl group in Pro-tRNAPro resides directly on the nitrogen nucleophile that forms the peptide bond in translation, yet this is somehow tolerated by the translation apparatus. N-alkyl groups in polypeptides and proteins kink 3-dimensional structures, result in slow isomerization between cis and trans conformations in the case of Pro, and can improve the pharmacological properties of peptides by decreasing their degradation by proteases and increasing their membrane permeability. To investigate these special properties or endow them on translation products, there have been many studies incorporating N-alkylamino acids (“imino acids”) in translation (1–13). Small linear N-alkylamino acids (Fig. 1Upper Right), because of their similarity to Pro, were expected to incorporate well in translation, but the efficiencies were variable (1–13). Incomplete incorporations in purified translation systems (8–13) are particularly puzzling because of the lack of competing reactions, incubation times of tens of minutes, and an average codon translation time comparable with that in Escherichia coli (45 ms) (14). Here, we investigate the kinetics of incorporation of Pro and other N-alkylamino acids by combining quench-flow translation kinetics and charging of tRNAs with noncognate/unnatural amino acids (Fig. 1).
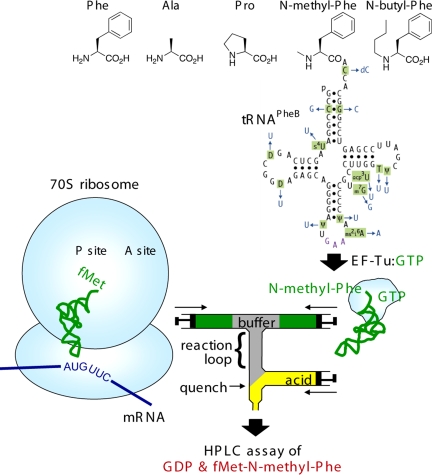
Fig. 1.
Combination of tRNAs charged with noncognate/unnatural amino acids and quench-flow translation kinetics. (Upper) The 5 amino- and N-alkylamino acids used in this study, one of which is shown charged on tRNAPheB. This synthetic, unmodified tRNA is based on natural E. coli tRNAPhe (black with purple anticodon; posttranscriptional modifications are in green), with changes in blue (11). (Lower) Aminoacyl-tRNA prebound to elongation factor Tu–GTP is mixed with ribosomes preinitiated with fMet-tRNAinitiator and then quenched rapidly with formic acid. Reactants are in green; products measured are in red.
Results
A highly flexible method for charging tRNAs with noncognate amino acids and N-alkylamino acids is to use T4 RNA ligase to join an unmodified tRNA−CA transcript to a chemically synthesized N-NitroVeratrylOxyCarbonyl (NVOC)-aminoacyl-pdCpA; the amino protecting group (NVOC) is then removed by photolysis (15). We first need to ascertain the effect of lack of nucleoside modifications and of a penultimate dC, in those aminoacyl-tRNA constructs (Fig. 1 Upper) on translation kinetics. Posttranscriptional modifications have been shown to affect marginally translation kinetics in the case of one tRNA, E. coli tRNAPheC3G-G70C, at least for N-acetyl-Phe-Phe dipeptide synthesis from poly(U) template at 5 °C, which proceeded with a codon translation time of ≈1 s (16). To assess the effects of lack of modifications also at near in vivo conditions (37 °C and codon translation times faster than 45 ms) we employ a version of our purified translation system optimized for speed and accuracy at 37 °C (17, 18). The kinetics of ribosomal synthesis of [3H]fMet-Phe-tRNAPheC3G-G70C from [3H]fMet-tRNAfMet and Phe-tRNAPheC3G-G70C, prepared by ligation and photodeprotection (termed Phe-tRNAPheB; Fig. 1) are first compared with the corresponding kinetics for the native tRNAPhe charged with Phe by Phe-tRNA synthetase. We measure the average times, τ, for both chemical reactions of codon translation, GTP hydrolysis on EF-Tu (τGTP; discussed below) and overall dipeptide synthesis (τdip), in the same reaction mixtures. The similar and short translation times for Phe-tRNAPheB and native Phe-tRNAPhe [Fig. 2 A and B, Table 1, and supporting information (SI) Table S1] show that the penultimate dC and the absence of all 10 posttranscriptional modifications in tRNAPhe (Fig. 1) have only a minor effect on dipeptide synthesis kinetics, which also validates our combination of methods.
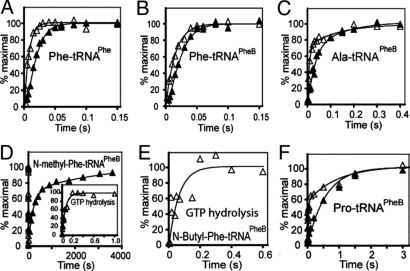
Fig. 2.
Kinetics of dipeptide synthesis with aminoacyl- and N-alkylaminoacyl-tRNASPhe. Kinetics of dipeptide synthesis (▴) from fMet-tRNAifMet and from Phe-tRNAPhe (A), Phe-tRNAPheB (B), Ala-tRNAPheB (C), N-methyl-Phe-tRNAPheB (D), N-butyl-Phe-tRNAPheB (E), and Pro-tRNAPheB (F) are shown. Kinetics of the prerequisite, faster GTP hydrolysis (▵) by EF-Tu in the same reactions are shown. The ribosome had the Phe codon UUC at the A site. All kinetics were measured at 37 °C in high-accuracy polyamine containing LS3 buffer (see Materials and Methods). Representative plots are shown for each assay.
Table 1.
Average times for dipeptide synthesis from fMet-tRNAfMet and different aminoacyl-tRNAs
| Aminoacyl-tRNA | Codon | τdip, ms | τGTP, ms | τacc,pep, ms |
|---|---|---|---|---|
| Phe-tRNAPhe | UUU | 19.6 (±2.1) | 7.8 (±0.6) | 11.9 (±2.1) |
| Phe-tRNAPhe | UUC | 18.5 (±1.7) | 7.7 (±0.6) | 10.8 (±1.8) |
| Phe-tRNABulk | UUC | 20.4 (±1.7) | 8.9 (±0.8) | 11.5 (±1.9) |
| Phe-tRNAPheB | UUC | 24.4 (±2.3) | 13.0 (±0.7) | 11.4 (±2.4) |
| N-methyl-Phe-tRNAPheB | UUC | 200,000 (±12,000) | 52.6 (±7.3) | 199,900 (±12,000) |
| N-butyl-Phe-tRNAPheB | UUC | Undetectable | 67.7 (±27) | Undetectable |
| Ala-tRNAPheB | UUC | 37.0 (±5.1) | 13.2 (±1.4) | 23.9 (±7.1) |
| Pro-tRNAPheB | UUC | 571.3 (±65) | 16.1 (±3.2) | 555 (±65) |
| Pro-tRNABulk | CCG | 62.5 (±7.6) | 12.5 (±1.4) | 50.0 (±7.7) |
| Pro-tRNABulk | CCA | 66.7 (±8.9) | 18.9 (±2.8) | 47.8 (±9.3) |
| Pro-tRNABulk | CCU | 71.4 (±9.7) | 23.8 (±3.5) | 47.6 (±10.3) |
| Pro-tRNABulk | CCC | 116.3 (±16.1) | 50.0 (±5.1) | 66.3 (±16.9) |
Data are expressed as average times for GTP hydrolysis (τGTP) and dipeptide synthesis (τdip) measured in the same reaction mixture. The average time for accommodation/peptidyl transfer is calculated as τacc,pep = τdip − τGTP. All experiments were conducted at 37 °C in high-accuracy polyamine containing LS3 buffer (see Materials and Methods); each experiment was performed at least twice.
Next, we measured the effect on translation of changing the cognate amino acid on tRNAPheB, Phe, to a much smaller, noncognate, natural amino acid, Ala. Again, the effect is minor (Fig. 2 B and C and Table 1).
In marked contrast, changing the cognate Phe to its simplest N-alkyl analog, N-methyl-Phe (Fig. 1), increases the average dipeptide synthesis time (τdip) by a factor of 8,000 to 3.5 min (Fig. 2D and Table 1). Nevertheless, virtually all N-methyl-Phe-tRNAPheB was converted to fMet-N-methyl-Phe-tRNAPheB within 30 min (Fig. 2D). Such a huge inhibitory effect of a small methyl group on the kinetics of dipeptide formation is surprising, albeit consistent with our previous inability to saturate incorporation at the third position of a tetrapeptide with this substrate (11). Mechanistic insight into the remarkably slow incorporation of N-methyl-Phe comes from measuring τGTP, the average time for GTP hydrolysis on elongation factor Tu (EF-Tu). GTP hydrolysis is greatly stimulated when the ternary complex of aminoacyl-tRNA–EF-Tu–GTP is bound to a cognate codon at the ribosomal A site. τGTP is only 4 times longer for N-methyl-Phe-tRNAPheB (Fig. 2D and Table 1) than for Phe-tRNAPheB. Further, N-methyl-Phe incorporation is strictly dependent on the aminoacyl-tRNA carrier, EF-Tu (Fig. S1), excluding the possibility of slow, direct binding of N-methyl-Phe-tRNAPheB to the A site of the ribosome. The dramatic inhibition of peptide bond formation caused by the N-methyl group (Table 1) occurs, therefore, at one or more of the translation steps subsequent to GTP hydrolysis on EF-Tu: release of EF-Tu–GDP from the aminoacyl-tRNA on the ribosome, movement of the aminoacyl-tRNA to the ribosomal peptidyltransferase site (accommodation), and the chemistry of peptide bond formation itself. The average time for these subsequent steps, τacc,pep is readily calculated from our data as τacc,pep = τdip - τGTP (Table 1).
The complete, although slow, incorporation of N-methyl-Phe into dipeptide prompted us to check the larger N-butyl-Phe (Fig. 1), whose incorporation was undetectable in the tetrapeptide system (11). Despite an incubation time of 30 min, N-butyl-Phe-tRNAPheB incorporation was undetectable (Fig. S2 and Table 1). However, its GTP hydrolysis time is similar to that of N-methyl-Phe-tRNAPheB (Fig. 2E and Table 1), again showing that the main inhibition occurs at one or more of the translation steps subsequent to GTP hydrolysis on EF-Tu. This is consistent with measurable codon-specific binding of N-butyl-Phe-tRNAPheB–EF-Tu–GTP to the ribosomal A site (11).
Given these huge inhibitions by small N-alkylations, we wondered whether Pro incorporates significantly more slowly than Phe and Ala. Indeed, incorporation of Pro from Pro-tRNAPheB into dipeptide takes ≈570 ms, 23 times slower than incorporation of Phe from Phe-tRNAPheB and 15 times slower than incorporation of Ala from Ala-tRNAPheB (Fig. 2F and Table 1). However, the GTP hydrolysis time is similar for Pro-tRNAPheB, Phe-tRNAPheB, and Ala-tRNAPheB (Fig. 2 and Table 1), indicating retardation of the accommodation/peptidyl transfer reaction (τacc,pep), as seen for N-methyl-Phe.
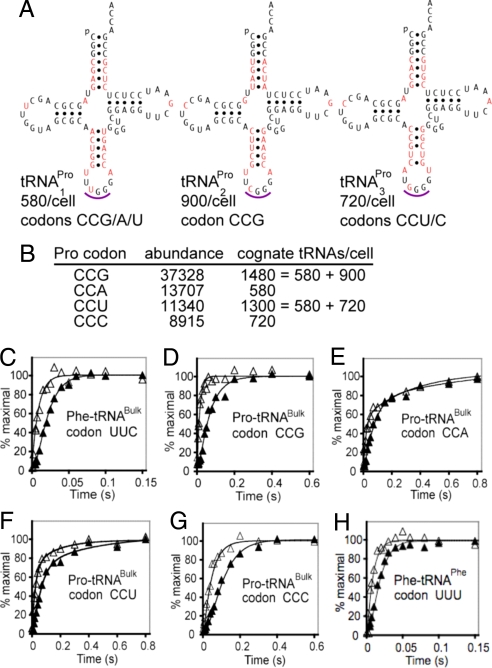
The average codon translation time (including translocation) has been estimated for 1 of the 4 Pro codons in vivo as ≈170 ms (rate of 6 s−1) at CCG in E. coli (19). Although this is substantially longer than the average in vivo codon translation time (45 ms; rate of 22 s−1) and our dipeptide synthesis times from Ala-tRNAPheB and Phe-tRNAPheB, it is 3 times shorter than our in vitro dipeptide synthesis time from Pro-tRNAPheB at UUC (which does not include the translocation time). We thus hypothesized that the faster rate for Pro in vivo might be caused by its cognate tRNA adaptors and codons (Fig. 3 A and B). To test this, we prepared natural aminoacyl-tRNA isoacceptors by charging E. coli tRNABulk substrate in situ. This substrate was first validated in our system by showing that the natural Phe-tRNAPhe substrates prepared in situ from tRNABulk and from pure tRNAPhe incorporate with very similar kinetics (Figs. 2A and 3C and Table 1). Next, tRNABulk was used as the source of the 3 natural Pro-tRNAPro isoacceptors to measure the kinetics of fMet-Pro synthesis at codon CCG. Interestingly, the dipeptide synthesis time is 62.5 ms (Fig. 3D and Table 1), 9 times shorter than for the hybrid substrate Pro-tRNAPheB and reassuringly shorter than the CCG translation time estimated in vivo. Nevertheless, the natural Pro-tRNAPro isoacceptors at CCG are still 3 times slower than natural Phe-tRNAPhe essentially because of the slower accommodation/peptidyl transfer, not GTPase reaction (Fig. 3D and Table 1). Mindful that that each of the 4 Pro codons is translated by a different subset of isoacceptors and that these isoacceptors differ significantly in sequence (73–83% homology; Fig. 3A), we also measured the Pro incorporation kinetics at the other 3 Pro codons. The kinetics at CCA and CCU are similar to CCG, but dipeptide synthesis at CCC is twice as slow because of a slower step(s) up to and including GTP hydrolysis (Fig. 3 E–G and Table 1). In contrast, translation of the synonymous codons UUC and UUU by the sole E. coli Phe-tRNAPhe isoacceptor yields virtually the same kinetics (Figs. 3H and 2A and Table 1), 6 times faster than translation by Pro-tRNA3Pro at CCC.
Fig. 3.
Kinetics of dipeptide synthesis with the native Pro-tRNAPro isoacceptors. (A) Sequences of the 3 E. coli tRNAPro isoacceptors. Nonconserved sequences are in red, and anticodons are underlined. The measured number of molecules per E. coli cell (28) and accepted codon specificities are shown (the posttranscriptional modifications have not been reported, but the Pro-1 anticodon is accepted to be modified at U34 to cmo5U) (31). (B) Lack of correlation between abundance of Pro codons in E. coli genes and measured number of cognate isoacceptor tRNAs. (C–H) Kinetics of dipeptide synthesis (▴) and accompanying GTP hydrolysis (▵) from native isoacceptors at all 6 codons for Phe and Pro. All kinetics were measured at 37 °C in high-accuracy polyamine containing LS3 buffer (see Materials and Methods). Representative plots are shown for each assay.
Discussion
We have measured the kinetics of dipeptide formation between fMet-tRNA ifMet in the P site and several different natural and unnatural aminoacyl-tRNAs under near-physiological conditions (Table 1). The rate of amino acid incorporation may vary with the identity of peptidyl-tRNA in the P site and with the length of the nascent polypeptide. However, the kinetics we observe here for native aminoacyl-tRNAs are fully compatible with the rate of bulk protein elongation in vivo, suggesting that our results are biologically relevant. Moreover, the measured times of accommodation/peptide bond formation for our 5 different amino/N-alkylamino acids (Table 1), irrespective of tRNA adaptor or codon, fit well with organic chemistry theory and experiment for nonenzymatic peptide synthesis. In solid-phase peptide synthesis, amino acids and Pro are more reactive than N-methylamino acids, which are more reactive than N-butylamino acids (20). This demonstrates, at least in organic solvent, that the increasing nucleophilicity of the respective nitrogen nucleophiles caused by the electron-releasing inductive effects is outweighed by the decreasing nucleophilicity caused by the steric effects: the N nucleophiles in Ala and Phe are less hindered than in Pro, which is less hindered than the freely rotating N in N-methyl-Phe, which is less hindered than the N in N-butyl-Phe (Fig. 1). Furthermore, amino groups are generally less basic (have lower pKa's values) in amino acids than in N-alkylamino acids, so amino acids may be less protonated and hence more nucleophilic also in aminoacyl-tRNAs on the ribosome. These correlations suggest that amino acid sterics and basicities may play a major role in governing translation incorporation rates of natural and unnatural amino acids and that the major inhibition caused by N-alkylation is at the stage of peptide bond formation, not accommodation.
Translations with N-alkylamino acids, and perhaps many other unnatural amino acids, may give low yields because the time for peptide bond formation increases to become longer than the dropoff time of ≈1 min for P site-bound, short peptidyl-tRNAs (21, 22), thereby allowing dropoff from the ribosome to occur before coupling. Decreasing dropoff experimentally may thus increase yields with unnatural amino acids (23). This could facilitate ribosomal synthesis and directed evolution of protease-resistant, membrane-permeable, peptide ligands containing multiple N-methylamino acids (23).
That Pro is the sole N-alkylamino acid in the genetic code may seem mysterious because several other N-alkylamino acids are major products of synthetic prebiotic experiments and meteorite analyses (24). However, it follows from Table 1 and our discussion above that Pro may have been selected because it is a privileged cyclic structure that forms peptide bonds much faster than linear N-alkylamino acids.
A lower reactivity of Pro compared with amino acids in peptide bond formation could explain why we find the natural tRNAPro isoacceptors to be superior adaptors for Pro (Table 1): the very special properties of Pro may require a very special design of the tRNAPro isoacceptors. The optimal tRNA design for the 19 aa may be less idiosyncratic than for Pro, so that in vivo-like incorporation rates do not always require that the amino acids are paired with their cognate tRNAs, as exemplified by the Ala incorporation from the hybrid Ala-tRNAPheB with only a 2-fold smaller rate than the Phe from the cognate Phe-tRNAPheB (Table 1). An extension of the “thermodynamic compensation” hypothesis (25–27), that evolution has optimized the pairing of each amino acid with its tRNA body for ribosomal binding and performance in translation, is therefore supported by our data in Table 1, especially for Pro.
Another hypothesis, the “uniform decoding rate” hypothesis, differs from the thermodynamic compensation hypothesis by proposing that all natural aminoacyl-tRNAs have uniform rates of incorporation in translation (26, 27). The uniform decoding rate hypothesis is supported by recent kinetic data with 10 of the 45 aminoacyl-tRNAs of E. coli at 10 codons (27), albeit at rates much smaller than those in vivo. However, the 3-fold difference in translation rate estimated for the same tRNAGlu isoacceptor at its 2 different codons in vivo (19) is an apparent exception to this hypothesis. Furthermore, our kinetic data, showing a 3- to 6-fold faster incorporation of Phe from the natural Phe-tRNAPhe isoacceptor than of Pro from the natural Pro-tRNAPro isoacceptors (Table 1), demonstrate additional exceptions to the uniform decoding rate hypothesis.
Interestingly, kinetic differences between different tRNA isoacceptors cognate to the same amino acid, here exemplified by the Pro-tRNAPro isoacceptor case (Table 1), may be a hitherto missing determinant of codon bias. The “tRNA abundance” theory of codon bias (28), which correlates tRNA isoacceptor concentrations with codon usage, breaks down for Pro codons in E. coli (Fig. 3B). Instead, codon usage for Pro correlates much better when also considering our measured incorporation times (bottom 4 τdip values in Table 1). In particular, CCC may be the rarest of the 4 Pro codons because it interacts least efficiently with a Pro-tRNA–EF-Tu–GTP ternary complex (shown by the long τGTP in Table 1). The relatively slow rate of incorporation of Pro at all 4 codons may also help explain why Pro has the second highest rate of loss of the 20 aa in evolution (29).
In conclusion, our data reveal kinetic differences of Pro and Phe incorporation from their native aminoacyl-tRNAs not expected from the uniform decoding rate hypothesis (27). In addition, our results may help explain codon bias not accounted for by the tRNA abundance hypothesis (28). The surprisingly slow accommodation/peptidyl transfer with Pro and other N-alkylamino acids fits remarkably well with organic chemistry theory. This provides a framework for understanding the rate-limiting steps in protein synthesis and improving the incorporation of unnatural amino acids.
Materials and Methods
Reagents.
We have published the preparation and characterization of the 5 N-NVOC-aminoacyl-pdCpAs (9, 11) and the purified E. coli translation system (17). Ligated tRNAs were purified by ion exchange chromatography instead of ethanol precipitation because the latter decreased maximal incorporation by severalfold (16). Buffer LS3 contains 95 mM KCl, 5 mM NH4Cl, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 30 mM Hepes (pH 7.5), 1 mM dithioerythritol, 2 mM phosphoenolpyruvate, 5 mM Mg(OAc)2, 1 mM GTP, and 1 mM ATP (30). Buffer LS3A is identical except it lacks GTP and contains 2 mM ATP, 1 μg/mL pyruvate kinase, and 0.1 μg/mL myokinase. Assuming that ATP and GTP each chelate 1 Mg2+, the free Mg2+ concentration in both buffers is 3 mM.
Kinetic Assay for Simultaneous Measurement of GTPase Activity and Dipeptide Synthesis.
Final concentrations after combining equal volumes of the 2 mixes are given. Ribosomal mix, containing 2 μM 70S ribosomes, 3 μM mRNA (sequences in Table S2), and 2 μM [3H]fMet-tRNAfMet in buffer LS3, was preincubated at 37 °C for 30 min. For precharged tRNAs, factor mix, containing 0.5 μM EF-Tu and 0.5 μM [3H]GDP in LS3A buffer, was preincubated at 37 °C for 40 min (to ensure complete conversion of GDP to GTP in solution and on EF-Tu), then 0.6 μM chemoenzymatically synthesized, photodeprotected aminoacyl-tRNA was added, and the incubation continued for 5 min. For uncharged tRNAs, factor mix, containing 0.5 μM EF-Tu, 0.5 μM [3H]GDP, and 0.1 mM Phe or Pro in LS3A buffer, was preincubated for 30 min, and then 0.1 unit/μL purified PheRS or ProRS and 0.6 μM purified tRNAPhe or tRNAPhe or tRNAPro isoacceptors within tRNABulk (20 μM and 16 μM tRNABulk respectively) were added, and the incubation continued for 10 min. Ribosomal and factor mixes were mixed rapidly at 37 °C in a quench-flow apparatus (RQF-3; KinTek Corp.), and the reaction was quenched rapidly with 50% formic acid at various times (see Fig. 1). Measured rates were independent of the exact tRNA concentration in the factor mix because the ribosomes were present in excess.
Analyses of Kinetic Assays.
Translation reactions quenched with 50% formic acid were centrifuged at 20,000 g for 15 min, and the [3H]GDP and [3H]GTP in the supernatants were quantified by Mono Q HPLC using a 0–1 M NaCl multistep gradient in 20 mM Tris (pH 7.5) and a β-RAM model 3 radio HPLC detector (IN/US Systems). The pellets containing [3H]dipeptidyl-tRNAs and unreacted [3H]fMet-tRNA were washed once with 20% formic acid to remove residual [3H]GDP/[3H]GTP and treated with KOH (to 0.5 M) at room temperature for 10 min to release the dipeptide from tRNA, and then formic acid was added (to 17%). The acid-precipitated deacylated tRNAs were then pelleted by centrifugation at 20,000 g for 15 min, and the [3H]dipeptide in the supernatant was quantified by C18 reversed-phase HPLC. Elution was as described in Fig. S2, except fMet-Ala was eluted with 0.1% trifluoroacetic acid in water, and fMet-Pro with 20% methanol/0.1% trifluoroacetic acid. All 4 dipeptides had different elution times, and these times were unaffected by the source of the elongator amino acid (free amino acid vs. N-NVOC-aminoacyl-pdCpA).
τGTP and its standard deviation were estimated with the nonlinear regression program Origin 7 (OriginLab Corp.). τdip and its standard deviation were estimated by nonlinear regression fitting to a 2-step kinetic model (18). In some cases, measured kinetics displayed double-exponential behavior, with the amplitude of the fast phase generally exceeding 70% of the total amplitude (Figs. 2 and 3). In those cases, the average times reported in Table 1 were estimated for the dominant, fast phase, representing rapid, direct binding of the aminoacyl-tRNA–EF-Tu–GTP ternary complex to the A site of the ribosome followed by GTP hydrolysis and accommodation/peptidyl transfer. The slow exponential phase represents in each case a minor, alternative pathway of dipeptide formation far too slow to be significant for the rapid protein elongation in the living cell.
Supplementary Material
Acknowledgments.
We thank Baolin Zhang and Kaweng Ieong for help with reagent preparation and Vivian Siegel for comments on the manuscript. This work was supported by National Institutes of Health grants (to V.W.C., M.E. and A.C.F.), the Swedish Research Council (M.E.) and American Cancer Society grants (to A.C.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809211106/DCSupplemental.
References
- 1.Bain JD, Wacker DA, Kuo EE, Chamberlin AR. Site-specific incorporation of non-natural residues into peptides: Effect of residue structure on suppression and translation efficiencies. Tetrahedron. 1991;47:2389–2400. [Google Scholar]
- 2.Ellman JA, Mendel D, Schultz PG. Site-specific incorporation of novel backbone structures into proteins. Science. 1992;255:197–200. doi: 10.1126/science.1553546. [DOI] [PubMed] [Google Scholar]
- 3.Chung HH, Benson DR, Schultz PG. Probing the structure and mechanism of Ras protein with an expanded genetic code. Science. 1993;259:806–809. doi: 10.1126/science.8430333. [DOI] [PubMed] [Google Scholar]
- 4.Karginov VA, et al. Probing the role of an active site aspartic acid in dihydrofolate reductase. J Am Chem Soc. 1997;119:8166–8176. [Google Scholar]
- 5.Frankel A, Millward SW, Roberts RW. Encodamers: Unnatural peptide oligomers encoded in RNA. Chem Biol. 2003;10:1043–1050. doi: 10.1016/j.chembiol.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Lummis SCR, et al. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 7.Choudhury AK, Golovine SY, Dedkova LM, Hecht SM. Synthesis of proteins containing modified arginine residues. Biochemistry. 2007;46:4066–4076. doi: 10.1021/bi062042r. [DOI] [PubMed] [Google Scholar]
- 8.Merryman C, Green R. Transformation of aminoacyl tRNAs for the in vitro selection of “drug-like” molecules. Chem Biol. 2004;11:575–582. doi: 10.1016/j.chembiol.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Tan Z, Forster AC, Blacklow SC, Cornish VW. Amino acid backbone specificity of the Escherichia coli translation machinery. J Am Chem Soc. 2004;126:12752–12753. doi: 10.1021/ja0472174. [DOI] [PubMed] [Google Scholar]
- 10.Josephson K, Hartman MC, Szostak JW. Ribosomal synthesis of unnatural peptides. J Am Chem Soc. 2005;127:11727–11735. doi: 10.1021/ja0515809. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, et al. Specificity of translation for N-alkylamino acids. J Am Chem Soc. 2007;129:11316–11317. doi: 10.1021/ja0734871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawakami T, Murakami H, Suga H. Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides. Chem Biol. 2008;15:32–42. doi: 10.1016/j.chembiol.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Subtelny AO, Hartman MC, Szostak JW. Ribosomal synthesis of N-methyl peptides. J Am Chem Soc. 2008;130:6131–6136. doi: 10.1021/ja710016v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang ST, Xu YC, Dennis P, Bremer H. mRNA composition and control of bacterial gene expression. J Bacteriol. 2000;182:3037–3044. doi: 10.1128/jb.182.11.3037-3044.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornish VW, Mendel D, Schultz PG. Probing protein structure and function with an expanded genetic code. Angew Chem Int Ed Engl. 1995;34:621–633. [Google Scholar]
- 16.Harrington KM, Nazarenko IA, Dix DB, Thompson RC, Uhlenbeck OC. In vitro analysis of translational rate and accuracy with an unmodified tRNA. Biochemistry. 1993;32:7617–7622. doi: 10.1021/bi00081a003. [DOI] [PubMed] [Google Scholar]
- 17.Pavlov MY, Freistroffer DV, MacDougall J, Buckingham RH, Ehrenberg M. Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J. 1997;16:4134–4141. doi: 10.1093/emboj/16.13.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson M, Bouakaz E, Lovmar M, Ehrenberg M. The kinetics of ribosomal peptidyl transfer revisited. Mol Cell. 2008;30:589–598. doi: 10.1016/j.molcel.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Sorensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli: The 2 glutamic acid codons GAA and GAG are translated with a 3-fold difference in rate. J Mol Biol. 1991;222:265–280. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- 20.Humphrey JM, Chamberlin AR. Chemical synthesis of natural product peptides: Coupling methods for the incorporation of noncoded amino acids into peptides. Chem Rev. 1997;97:2243–2266. doi: 10.1021/cr950005s. [DOI] [PubMed] [Google Scholar]
- 21.Karimi R, Ehrenberg M. Dissociation rates of peptidyl-tRNA from the P-site of E. coli ribosomes. EMBO J. 1996;15:1149–1154. [PMC free article] [PubMed] [Google Scholar]
- 22.Karimi R, Pavlov MY, Heurgue-Hamard V, Buckingham RH, Ehrenberg M. Initiation factors IF1 and IF2 synergistically remove peptidyl-tRNAs with short polypeptides from the P-site of translating Escherichia coli ribosomes. J Mol Biol. 1998;281:241–252. doi: 10.1006/jmbi.1998.1953. [DOI] [PubMed] [Google Scholar]
- 23.Tan Z, Blacklow SC, Cornish VW, Forster AC. De novo genetic codes and pure translation display. Methods. 2005;36:279–290. doi: 10.1016/j.ymeth.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Weber AL, Miller SL. Reasons for the occurrence of the twenty coded protein amino acids. J Mol Evol. 1981;17:273–284. doi: 10.1007/BF01795749. [DOI] [PubMed] [Google Scholar]
- 25.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 26.Wolfson AD, et al. tRNA conformity. Cold Spring Harbor Symp Quant Biol. 2001;66:185–193. doi: 10.1101/sqb.2001.66.185. [DOI] [PubMed] [Google Scholar]
- 27.Ledoux S, Uhlenbeck OC. Different aa-tRNAs are selected uniformly on the ribosome. Mol Cell. 2008;31:114–123. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong H, Nilsson L, Kurland CG. Covariation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 29.Jordan IK, et al. A universal trend of amino acid gain and loss in protein evolution. Nature. 2005;433:633–638. doi: 10.1038/nature03306. [DOI] [PubMed] [Google Scholar]
- 30.Pavlov MY, Antoun A, Lovmar M, Ehrenberg M. Complementary roles of initiation factor 1 and ribosome recycling factor in 70S ribosome splitting. EMBO J. 2008;27:1706–1717. doi: 10.1038/emboj.2008.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen P, Qian Q, Zhang S, Isaksson LA, Bjork GR. A cytosolic tRNA with an unmodified adenosine in the wobble position reads a codon ending with the noncomplementary nucleoside cytidine. J Mol Biol. 2002;317:481–492. doi: 10.1006/jmbi.2002.5435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.