Abstract
The small helical protein BBL has been shown to fold and unfold in the absence of a free energy barrier according to a battery of quantitative criteria in equilibrium experiments, including probe-dependent equilibrium unfolding, complex coupling between denaturing agents, characteristic DSC thermogram, gradual melting of secondary structure, and heterogeneous atom-by-atom unfolding behaviors spanning the entire unfolding process. Here, we present the results of nanosecond T-jump experiments probing backbone structure by IR and end-to-end distance by FRET. The folding dynamics observed with these two probes are both exponential with common relaxation times but have large differences in amplitude following their probe-dependent equilibrium unfolding. The quantitative analysis of amplitude and relaxation time data for both probes shows that BBL folding dynamics are fully consistent with the one-state folding scenario and incompatible with alternative models involving one or several barrier crossing events. At 333 K, the relaxation time for BBL is 1.3 μs, in agreement with previous folding speed limit estimates. However, late folding events at room temperature are an order of magnitude slower (20 μs), indicating a relatively rough underlying energy landscape. Our results in BBL expose the dynamic features of one-state folding and chart the intrinsic time-scales for conformational motions along the folding process. Interestingly, the simple self-averaging folding dynamics of BBL are the exact dynamic properties required in molecular rheostats, thus supporting a biological role for one-state folding.
Keywords: downhill folding, folding landscape, landscape topography, protein dynamics
Theory asserts that protein folding kinetics can be described as diffusion on a low dimensional free energy surface obtained by projecting the hyperdimensional energy landscapes of proteins into one or a few suitable order parameters (1, 2). The overall topography of such free energy surface and the conformational motions guiding folding are not resolvable by classical folding kinetics, but could be probed by time-resolved experiments of downhill folding (3). The difficulty resides in identifying examples of downhill folding relaxations. Stretched exponential decays and kinetic memory effects are not reliable signatures because they require the downhill free energy landscape to be rugged (4), and can also originate from other sources (5). Recently, downhill folding has been pursued by reengineering the fast-folding λ-repressor to accelerate folding with either mutations (6, 7) or stabilizing cosolvents (8). Approach to the barrierless regime was correlated with the emergence of an additional faster kinetic relaxation interpreted as the downhill decay from a vanishing barrier top (7). From these experiments, a folding speed limit of ≈2.5 μs at 340 K was proposed for λ-repressor. This time-scale is close to the recent upper limit estimate of N/100 μs (9) for λ-repressor [N = 80] residues.
A powerful alternative would be to measure conformational dynamics on a folding free energy surface that remains barrierless at all conditions (one-state or global downhill folding) (10). One-state folding produces characteristic thermodynamic behavior that can be used for its identification (11). Building upon this idea, the 40-residue helical protein BBL has been found to exhibit one-state folding thermodynamics according to a battery of quantitative experimental criteria: (i) probe-dependent equilibrium unfolding (12); (ii) complex coupling between denaturing agents (13); (iii) characteristic DSC thermogram (14); (iv) gradual melting of secondary structure (15); (v) heterogeneous atom-by-atom unfolding behaviors spanning the entire unfolding process (16); and (vi) generalized baseline crossings in fits to global two-state models (17). BBL equilibrium unfolding has also been investigated in molecular simulations, ranging from off-lattice models with Go potentials (18–22) to replica exchange molecular dynamics (REMD) simulations in explicit solvent (23, 24). All simulations indicate that BBL crosses very minimal folding barriers, if any. In REMD simulations the downhill noncooperative equilibrium behavior is maintained for different variants and/or experimental conditions (24), whereas simulations with Go potentials produce slight variations in face of the differences in the contact maps (21).
However, the dynamics of global downhill folding remain largely unexplored experimentally, and thus its distinctive features are still unclear. In contrast to common assumption, single exponential decays are compatible with barrierless relaxation processes (5, 11, 19). However, the probe-dependent equilibrium of one-state folding should correspond exactly with probe dependent amplitudes in relaxation experiments. Recently, it has been proposed that relaxation times may also exhibit probe dependence in rough downhill free energy surfaces (25). Coarse-grained off-lattice simulations have shown flat or even inverted chevron plots (i.e., plots of folding relaxation rate versus chemical denaturant concentration) for one-state folding (19). Calculations on perfectly smooth downhill free energy surfaces show similar flattening for folding barriers <3 RT, but the plot remains V-shaped even down in the global downhill limit (26). However, a completely flat chevron plot has been recently reported for the villin headpiece subdomain (27), a protein that folds in few microseconds over an ≈2 RT barrier (26, 28).
Here, we set out to measure protein conformational dynamics of BBL as model case of one-state folding. To this end, we perform nanosecond temperature-jump experiments probing end-to-end distance by FRET and backbone structure by IR absorption. From detailed analysis of these multiprobe experiments we demonstrate quantitative compliance with the one-state folding scenario. At the same time we are able to rule out alternative folding models involving free energy barrier crossings, such as classical two-state and three-state kinetic schemes. Our results provide useful guidelines for the interpretation of dynamic experiments in or near the downhill folding regime. Furthermore, they reveal that the critical signatures of one-state folding dynamics reside in the amplitudes of multiprobe experiments. Finally, we take advantage of these experimental results to extract the time-scales of folding motions and probe the overall topography of the folding free energy surface.
Results
BBL folding has been investigated in variants with slight differences in its unstructured tails that do not change the overall equilibrium folding behavior (29). In this work, we employ our original variant previously termed Naf-BBL, and Naf-BBL labeled at the C terminus with a dansyl group for FRET measurements. The term BBL is used from hereafter for simplicity.
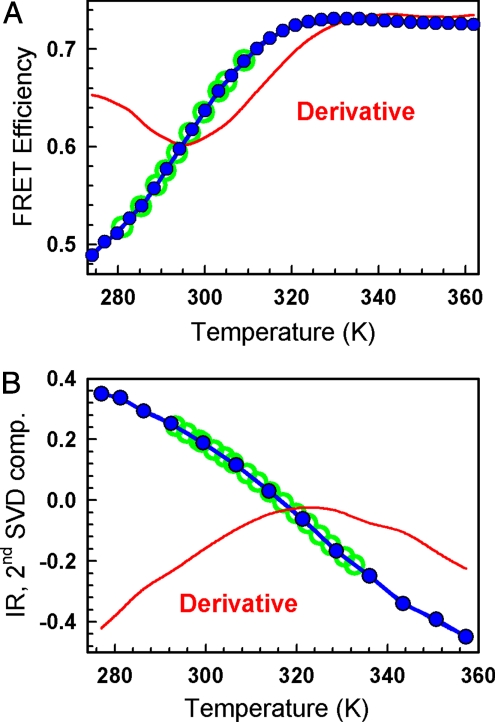
Fig. 1A shows FRET changes during the equilibrium thermal unfolding of dansyl-labeled BBL. The FRET probes (R0 ∼2.5 nm, assuming that k2 = 2/3) are placed at the protein ends, which in the native structure point in opposite directions lying at the farthest protein distance of ≈2.5 nm (12)(e.g., see rightmost BBL structure in Fig. 4). Upon thermal unfolding the FRET efficiency goes from 0.5 (consistent with the native structure) to ≈0.72, sensing the randomized reorientations of the 2 BBL α-helices upon disruption of the protein core, as observed by NMR (16) and REMD simulations (24). The experiment shows that the end-to-end distance decreases upon unfolding with Tm ≈295 K (indicated by the derivative extremum). A similar equilibrium experiment monitoring backbone structure by IR (mostly the α-helices for BBL) is displayed in Fig. 1B. It is apparent from the IR derivative that BBL α-helices melt at much higher temperature with Tm ≈ 325 K. Therefore, tertiary packing and backbone structure develop at different stages of BBL folding process (see refs. 12 and 16 for more details). These equilibrium experiments that report on the two main structural features linked to protein folding, and display very different apparent Tm, lay the foundations for the multiprobe laser T-jump experiments.
Fig. 1.
Equilibrium unfolding of BBL. (A) FRET-monitored equilibrium thermal unfolding. Blue circles, experimental data; red curve, derivative; green circles, final temperatures for the FRET T-jump experiments. (B) FTIR-monitored equilibrium thermal unfolding. Blue circles, amplitude of the second singular value decomposition (SVD) component of the matrix of amide I band infrared spectra of BBL versus temperature; red curve, derivative; green circles, final temperatures for the IR T-jump experiments.
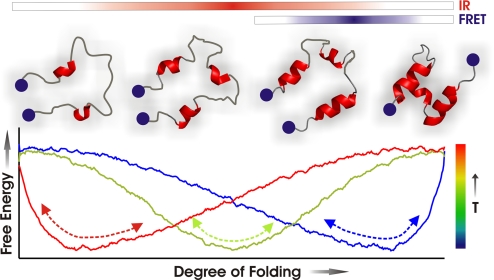
Fig. 4.
Diagram of one-state downhill folding dynamics in BBL.
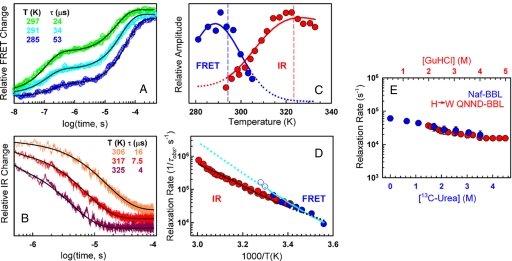
For the FRET probe we performed nanosecond T-jump experiments with final temperatures in the range of 280–310 K because the FRET changes are maximal in this region (green circles in Fig. 1A). In these experiments we used dansyl-labeled BBL at 60 μM and low salt, ensuring completely reversible folding-unfolding within this temperature range (29). The T-jump relaxations showed 2 components (Fig. 2A). The fast component is an exponential decrease in end-to-end distance (increased FRET efficiency) with a relaxation time (τobs) of ≈100 ns. At low temperatures, where BBL is mostly folded, the amplitude is very small, but it increases linearly with temperature dominating the overall relaxation at >300 K (see Fig. S1A) up to 320 K, where the overall FRET signal disappears following the equilibrium curve (Fig. 1A). This nanosecond process is equivalent in rate (Fig. S1B) and signal to the temperature-induced hydrophobic collapse relaxation of acid denatured BBL (30). The slower component is another exponential decrease in end-to-end distance occurring in tens of μs (Fig. 2A). The amplitude exhibits a maximum at ≈290 K, and then quickly decreases (blue circles in Fig. 2C) following the overall trend of the equilibrium FRET derivative (red curve in Fig. 1A). The amplitude extremum occurs at slightly lower temperatures because the equilibrium FRET change also includes the fast component amplitude, which increases linearly in this range.
Fig. 2.
Time-resolved experiments. (A) FRET relaxation decays. Colors indicate temperature. Black curves are biexponential fits. (Inset) Table showing final temperatures and relaxation times for the microsecond phase. Increases in the vertical scale correspond to increases in FRET efficiency. (B) As in A, but for IR relaxation decays. Black curves are single-exponential fits; decreases in the vertical scale correspond to decreases in the absorbance at 1,632 cm−1. (C) Amplitude of the microsecond relaxation. Blue circles, FRET time-resolved experiments; blue curve, fit to FRET data; red circles, IR time-resolved experiments; red curve, fit to IR data. Dashed lines show the extremum in the equilibrium derivative (Fig. 1). (D) Arrhenius plot of the FRET (blue circles) and IR (red circles) relaxation rate, and fits to the FRET (continuous cyan curve) and IR data (continuous dark red curve). Blue open circles correspond to FRET experiments with marginal amplitude for the microsecond phase. Dotted lines show FRET (cyan) and IR (dark red) relaxation rate extrapolations by the fitted model. (E) Relaxation rate at 298 K versus [13C-urea] for Naf-BBL measured by IR T-jump (blue circles and scale) and versus [guanidinium chloride] for H->W QNND-BBL (31) (red circles and scale). The two scales are shifted 0.5 M to overlay the datasets at their denaturation midpoints.
Time-resolved IR measurements were performed from 293 to 333 K (final temperature) where the changes in IR signal upon T-jumps are larger (open green circles in Fig. 1B). The experiments were carried out in non-dansylated BBL at ≈2.5 mM, a concentration that results in entirely reversible unfolding transitions for this protein (29). The IR relaxation follows a microsecond single exponential decrease in α-helix content (decrease in absorbance at 1,632 cm−1 corresponding to the α-helix band) (Fig. 2B). The amplitude (red circles in Fig. 2C) tracks the derivative of the equilibrium unfolding curve, indicating that it accounts for the whole conformational change in equilibrium.
Although the observation windows accessible to time-resolved FRET and IR overlap only partially, it is apparent from Fig. 2 A and B that τobs for the lone IR and the microsecond FRET process follow a monotonic trend with temperature. This is best observed plotting the relaxation rate (1/τobs) versus the inverse temperature (filled circles in Fig. 2D). Fig. 2D highlights a common τobs for backbone structure formation and the change in end-to-end distance associated to protein core disruption (i.e., slow FRET phase), which are thus controlled by concerted motions. Moreover, it demonstrates that the dansyl label does not significantly perturb BBL folding dynamics, as was shown for the thermodynamics in ref. 29. Another important observation is the significant temperature dependence of the microsecond phase, which accelerates by 2 orders of magnitude (i.e., from 120 to 1.3 μs) in only 55 K. Finally, at >300 K, there are hints of a slight accelerating trend in the FRET over the IR data (open blue circles in Fig. 2D). This effect, however, is within experimental error given the minimal amplitude remaining at these temperatures.
Fig. 2E (blue circles) shows 1/τobs for the single IR relaxation at room temperature versus urea concentration, measured using 13C-urea to shift the high absorbance of urea in the amide I region. 1/τobs decreases very slightly at increasing urea with a maximal decrease of ≈3-fold (i.e., ≈RT) at the 3 M urea denaturation midpoint. The relaxation could not be measured at >3.5 M urea because at such concentrations even 13C-urea is too absorptive at 1,632 cm−1. However, it is apparent that the behavior is very similar to that of a mutated longer BBL variant at high ionic strength (31) (H->W QNND-BBL, red circles in Fig. 2E). The two experiments use different chemical denaturants, and the mutated QNND-BBL variant is more stable in their conditions (i.e., midpoint at ≈3.5 M guanidinium chloride). Nevertheless, the folding relaxation rate of the 2 BBL variants is almost identical at their respective midpoints, and similarly insensitive to chemical denaturant, demonstrating that they exhibit the same dynamics.
A critical issue when interpreting kinetic experiments is whether the data provides sufficient information to rule some of the alternative models out. What realistic models could be invoked to interpret the BBL data? The observation of 2 FRET phases immediately draws attention to a classical three-state kinetic scheme [i.e., unfolded (U), intermediate (I), and folded (F)] (32). However, we found that the three-state model is incapable of reproducing the data in any of its versions (see SI Text, Fig. S2, and Tables S1 and S3). This assertion can be justified with a few concrete points. Due to the large difference in time-scales, the three possible kinetic schemes (on-pathway, off-pathway, and triangular) produce essentially the same results. The fast phase corresponds to the reequilibration between I and one end state, and the slow phase to the reequilibration of the other end state with everything else. The FRET nanosecond phase increases in amplitude with temperature (Fig. S1A), which implies that the fast equilibrium must be I-U. The lack of nanosecond IR phase requires the IR signal to be equal for U and I. Therefore, the amplitude of the slow IR phase reports the depopulation of F whereas the slow FRET phase does the same for F and I combined. Because F alone must depopulate at lower temperature by definition, the three-state model demands that the maximum in the IR amplitude occurs at lower temperature than that of the FRET microsecond phase. This is the exact opposite of the experimental observation (Fig. 2C). We can thus rule out a three-state mechanism for BBL.
By setting the preexponential to agree with the fast rate [i.e., ≈1/(200 ns)], the three-state model becomes equivalent to a two-state model (i.e., F-U) in which one of the states changes structurally with temperature (from the nanosecond phase amplitude it must be U here) in a downhill relaxation that sets the preexponential for the slower microsecond phase (Fig. S3 and Table S2). However, this version of the three-state model is equally inconsistent with the FRET and IR amplitudes. In an alternative two-state model, we could assume that the nanosecond FRET phase corresponds to a nonfolding relaxation process (see below). Under this interpretation the equilibrium differences between FRET and IR result from the FRET signal being a composite of the two processes, and the microsecond phase is the true two-state folding. This model is again incompatible with the data because the FRET and IR microsecond phase amplitudes should have the maximum in exactly the same position (Fig. S4) while experimentally they are ≈35 K apart (Fig. 2C). Furthermore, the predicted amplitudes for the two FRET phases are reversed in trend relative to experiment (Fig. S4C).
The third possibility is that we are indeed resolving the dynamics of one-state folding plus a nanosecond FRET process orthogonal to the folding reaction coordinate. The nanosecond process then reports on the same end-to-end relaxation observed in acid denatured BBL (30). The increasing amplitude with temperature would reflect the larger amplitude of motion of the tails as the protein becomes progressively more unstructured. The microsecond phase observed by FRET and IR would correspond to the one-state unfolding process showing exponential decays (Fig. 2 A and B), probe-independent rate (Fig. 2D), probe dependent amplitudes (and thus probe-dependent equilibrium) (Fig. 2C), and a flat still V-shaped rate versus urea plot (Fig. 2E).
To determine whether one-state folding can explain all of the data quantitatively, we performed a detailed analysis with a one-dimensional free energy surface model. The model (see SI Text) uses the property nativeness (n, or the probability of finding residues in native conformation) as reaction coordinate, and has been successful describing the kinetics of two-state and ultrafast folding proteins (26). In this model the absence or presence of folding barrier (and its magnitude) is determined by how steeply the stabilization enthalpy decays as a function of nativeness. This decay is assumed exponential in n with steepness determined by the exponent kΔH (see SI Text). kΔH is thus a fitting parameter that determines the fraction of stabilization enthalpy at the intercept with the conformational entropy contribution to the free energy, reflecting the ratio between non-local and local contributions.
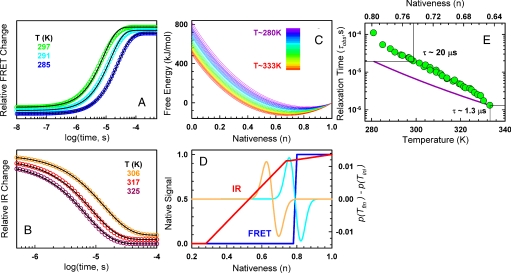
Remarkably, the one-dimensional model fitted very well the BBL time-resolved data with only 5 floating physical parameters (3 determining the smooth free energy surface and 2 for the diffusion coefficient; see Experimental Procedures) and idealized representations of the FRET and IR signals as a function of n that require 4 additional fitting parameters (blue and red curves in Fig. 3D). This set of fitting parameters is still 2 fewer than those required for the simplest three-state model (see SI Text). Fig. 2C shows that the model does indeed reproduce the large differences between FRET (blue curve) and IR (red curve) relaxation amplitudes together with the relaxation rate versus temperature curves (Fig. 2D, cyan for FRET and dark red for IR). The simulations of the FRET and IR relaxations exhibit quasi-perfect exponential decays (Figs. 3 A and B), as observed experimentally. Furthermore, the model even predicts the apparent speed up in the FRET relaxation at >300 K (dotted cyan line in Fig. 2D) hinted by the FRET data not used in the fits (open blue circles). The resulting one-dimensional free energy surfaces are globally downhill, with a minimum shifting from order to disorder as temperature rises (Fig. 3C). At the highest probed temperature (333 K) the surface minimum is at n = 0.64 because the unfolding process is still incomplete (see Fig. 1B). The minimum is predicted to shift down to n = 0.5 in strongly unfolding conditions (373 K), consistently with the residual structure in thermally unfolded BBL (16).
Fig. 3.
Quantitative analysis of time-resolved experiments. (A) FRET relaxation decays produced by the fitted model. As before, color code indicates the temperature, black curves are single exponential fits to the predicted decays. Note that the predicted relaxation corresponds only to the microsecond phase. The nanosecond phase is orthogonal to the folding process and thus is not accounted for by the model. (B) As in A, but for IR relaxations. (C) One-dimensional free-energy surfaces produced by the fitted model. The color code signifies temperature according to the scale shown in Inset. (D) Representation of how the FRET (blue) and IR (red) signals depend on the degree of nativeness, and examples of population redistributions after experimental temperature jumps: (cyan) T-jump to 294 K (orange) T-jump to 333 K. (E) Relaxation time of BBL as a function of temperature (lower scale) and the position of the free energy surface minimum on the reaction coordinate (top scale). Green circles, combined FRET and IR data; purple line, calculation for 40 kJ/mol activation energy using the value at 333 K as reference.
This quantitative analysis also indicates that the BBL folding free energy surface must be single welled and rather smooth. The model is incapable of reproducing the dynamic experiments when even a marginal folding barrier (≈1.5 RT) is enforced during the fitting procedure. Neither can the experiments be reproduced with a rough downhill free energy surface (Fig. S5A), which would be analogous to a folding scenario characterized by a downhill ladder of partly structured intermediates separated by local barriers
Therefore, the multiprobe folding dynamics observed in BBL could be summarized with the sketch shown in Fig. 4, in which the one-dimensional folding free energy surface exhibits a global minimum (single well) that shifts from native (blue profile) to unfolded (red profile) as temperature rises. FRET is mostly sensitive to the separation of the two helices, which occurs early in the continuous unfolding process (blue bar in Fig. 4; blue in Fig. 3D). IR monitors the backbone conformation unwinding more gradually and at higher temperature (red bar in Fig. 4, red in Fig. 3D). This sequence of structural events have also been observed in recent REMD simulations of BBL unfolding (24). If the surface approximates a harmonic well and is smooth (peaks and troughs below RT in Fig. 4), the relaxation after a small perturbation is exponential with relaxation time of the form:
where n is an order parameter, ω is the curvature of the single well, and D is the diffusion coefficient. To a first approximation, τobs is probe-independent. However, the observed decay ultimately depends on the diffusive path as detected by the probe. This normally small effect causes the apparent FRET speed up at >300 K because this probe is mostly insensitive to high temperature population redistributions (e.g., compare blue and orange curves in 3D). At those temperatures the FRET signal clips most of the true relaxation making it seemingly faster (dotted cyan line in 2D). It is also apparent from Eq. 1 that the τobs for BBL one-state folding may depend on temperature and on the position of the single well along the folding process (i.e., the value of the order parameter), which also depends on temperature.
Discussion
Our results shed much needed light onto the folding dynamics of BBL and the one-state folding scenario. From the multiprobe time-resolved experiments we can rule out that BBL folds over a free energy barrier, or via an intermediate, or through a ladder of partially folded intermediates separated by local barriers. Subsequent analysis with a noncommittal free energy surface model indicates that BBL folding dynamics are globally downhill. As in the previous thermodynamic analysis (12), these conclusions emerge directly from a quantitative analysis of the experiments. This critical requirement of calibrating phenomenological models with experimental data has been surprisingly ignored in recent work in which a ≈2–3 RT folding barrier was claimed for BBL based on calculations with a phenomenological model that does not reproduce the experiments quantitatively (33).
On another front, a variety of extraneous factors have been proposed in recent literature as sources of our previous equilibrium results in BBL: artifactual interactions through fluorescent labels, partial histidine protonation, excessive trimming of the unstructured tails, and partial destabilization at low ionic strength (see for example ref. 31). Our time-resolved experiments contradict all these arguments. We observe the same relaxation time in BBL variants with and without the dansyl label. We find that BBL behaves as a global downhill folder both in equilibrium and dynamically at pH 7.0, where its 2 histidines are not protonated. The chevron plot of our BBL variant [with a naftyl-alanine (29)] is very similar to the one reported by the Fersht group on a mutant of the longer QNND-BBL variant, which does not carry the naftyl-alanine and was studied at high ionic strength. Moreover, at the chemical midpoint, where a putative folding barrier would be maximal, the relaxation times are nearly identical (see Fig. 2E). The comparison highlights the little sensitivity of BBL's folding relaxation rate to factors that affect its overall stability. This is expected for a one-state folding scenario in which changes in stability tilt the free energy surface, but are not directly propagated to a diffusive relaxation rate.
According to Eq. 1, the time-scale changes shown in Fig. 3E for BBL result directly from the dependence of D on temperature (bottom scale) and reaction coordinate (top scale). At 333 K the position of the BBL one-state minimum (n = 0.65) (Fig. 3E) matches typical values for the top of folding barriers (26). Because this temperature is similar to typical temperatures in fast-folding experiments, the 1.3 μs of BBL at 333 K could be seen as a folding speed limit to compare with other fast-folding proteins. This interpretation is consistent with the N/100 μs estimate of Eaton and coworkers (9), and in perfect agreement with the ≈2.5-μs relaxation reported on λ-repressor (6) (1.3 μs × 80λ-repressor/40BBL ≈ 2.6 μs). Likewise, applying this speed limit to the 5 μs folding time for 35-residue villin (34) would indicate a marginal barrier of 1.5 RT, in very good agreement with independent barrier estimates (26, 28).
The data in Fig. 3E correspond to an apparent activation energy for D of ≈60 kJ/mol (see Fig. S6). In principle, D could change with temperature due to changes in water viscosity and steric hindrance during peptide bond rotations [together ≈24 kJ/mol (35)], steric dewetting (36), and the roughness in the energy landscape that is smoothed out by the free energy projection. The latter could also make D decrease along the reaction coordinate (2) and produce superArrhenius temperature dependence (37). In a recent empirical analysis, the temperature dependence for D at the top of the barrier has been estimated as ≈1kJ/mol per residue (26) (i.e., 40 kJ/mol for BBL, see purple line in Fig. 3E). The remaining 20 kJ/mol in BBL could originate from a position dependent D. A simple calculation (see Fig. S6) shows that, to be consistent with our experimental results, D cannot decrease >2.5 times between the two extreme positions of the BBL one-state minimum (n = 0.64 and n = 0.8). This effect can therefore account for an additional ≈12 kJ/mol. The rather low temperatures that we access for BBL hint some superArrhenius curvature(e.g., <290 K in Fig. 3E), which could also increase the apparent activation energy.
This discussion brings us to another interesting issue. We show here that diffusive global-downhill folding relaxations can be exponential and with probe-independent rates even when D changes along the reaction coordinate. The reason is that for one-state folding the temperature perturbation results in very little displacement in the reaction coordinate (see Fig. 3C). For example, linear extrapolation of a 2.5 decrease in D between n = 0.65 and n = 0.8 would result in a factor of ≈10 for a downhill relaxation from the fully unfolded state (n ≈ 0.3) down to the native structure. Such relaxation would be highly stretched and probe-dependent.
Finally, our results could have important implications for the biological function of BBL. One-state folding has been postulated as a mechanism to achieve molecular rheostats (12). In particular, BBL could act as a molecular oscillator coordinating the action of the 3 enzymatic steps performed by the oxo-glutarate reductase complex, and/or as a recoiling mechanism for the swinging arm of this complex (15). These two mechanisms require self-averaging conformational dynamics on a smooth single folding well exactly as we observe here for BBL.
Experimental Procedures
Equilibrium Experiments.
FRET experiments were performed at pH 7.0 with 20 μM protein and as described in ref. 12. For FTIR experiments Naf-BBL samples were prepared with 2.5 mM protein in 99.9% 2H2O at pH 7.0 (see SI Text).
Time-Resolved Experiments.
Time-resolved FRET experiments were performed at pH 7.0 with 60 μM protein, using our custom-built fluorescence laser T-jump apparatus as described in ref. 30. Time-resolved IR experiments were performed on a custom-built version of the instrument by Feng Gai and coworkers (38) (see SI Text).
Fitting of Experimental Data to the Theoretical Model.
For the analysis of BBL data with the free energy surface model (see SI Text), we fixed the heat capacity parameters to: ΔCp,res = 58 J/(mol·K) per residue and kΔCp = 4.3 (26). The 3 fitted parameters that determine the temperature dependent free energy surface are: ΔSres = 61 J/(mol·K·res), κΔH = −0.78, and ΔH385 = 72 kJ/(mol·res). It is important to emphasize that these parameters do not have any macroscopic meaning in the global downhill folding regime. They are just phenomenological parameters that reproduce the required shape for the free energy surface. The fitted parameters for the diffusion coefficient are D0 = 680 n2·s−1 and Ea = 60 kJ/mol.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grant RO1-GM066800, National Science Foundation Grant MCB-0317294, and Marie Curie Excellence Award MEXT-CT-2006-042334.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802986106/DCSupplemental.
References
- 1.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein-folding—a synthesis. Proteins Struct Funct Genet. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 2.Socci ND, Onuchic JN, Wolynes PG. Diffusive dynamics of the reaction coordinate for protein folding funnels. J Chem Phys. 1996;104:5860–5868. [Google Scholar]
- 3.Eaton WA. Searching for “downhill scenarios” in protein folding. Proc Natl Acad Sci USA. 1999;96:5897–5899. doi: 10.1073/pnas.96.11.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plotkin SS, Onuchic JN. Understanding protein folding with energy landscape theory part II: Quantitative aspects. Quarterly Reviews in Biophysics. 2002;35:205–286. doi: 10.1017/s0033583502003785. [DOI] [PubMed] [Google Scholar]
- 5.Hagen SJ. Probe-dependent and nonexponential relaxation kinetics: Unreliable signatures of downhill protein folding. Proteins Struct Funct Bioinf. 2007;68:205–217. doi: 10.1002/prot.21342. [DOI] [PubMed] [Google Scholar]
- 6.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 7.Yang WY, Gruebele M. Rate-temperature relationships in lambda-repressor fragment lambda(6–85) folding. Biochemistry. 2004;43:13018–13025. doi: 10.1021/bi049113b. [DOI] [PubMed] [Google Scholar]
- 8.Yang WY, Gruebele M. Folding λ-repressor at its speed limit. Biophys J. 2004;87:596–608. doi: 10.1529/biophysj.103.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubelka J, Hofrichter J, Eaton WA. The protein folding “speed limit”. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Muñoz V. Conformational dynamics and ensembles in protein folding. Ann Rev Biophys Biomol Struct. 2007;36:395–412. doi: 10.1146/annurev.biophys.36.040306.132608. [DOI] [PubMed] [Google Scholar]
- 11.Muñoz V. Thermodynamics and kinetics of downhill protein folding investigated with a simple statistical mechanical model. Int J Quantum Chem. 2002;90:1522–1528. [Google Scholar]
- 12.Garcia-Mira MM, Sadqi M, Fischer N, Sanchez-Ruiz JM, Muñoz V. Experimental identification of downhill protein folding. Science. 2002;298:2191–2195. doi: 10.1126/science.1077809. [DOI] [PubMed] [Google Scholar]
- 13.Oliva FY, Muñoz V. A simple thermodynamic test to discriminate between two-state and downhill folding. J Am Chem Soc. 2004;126:8596–8597. doi: 10.1021/ja048309w. [DOI] [PubMed] [Google Scholar]
- 14.Muñoz V, Sanchez-Ruiz JM. Exploring protein-folding ensembles: A variable-barrier model for the analysis of equilibrium unfolding experiments. Proc Natl Acad Sci USA. 2004;101:17646–17651. doi: 10.1073/pnas.0405829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naganathan AN, Doshi U, Fung A, Sadqi M, Muñoz V. Dynamics, energetics, and structure in protein folding. Biochemistry. 2006;45:8466–8475. doi: 10.1021/bi060643c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadqi M, Fushman D, Muñoz V. Atom-by-atom analysis of global downhill protein folding. Nature. 2006;442:317–321. doi: 10.1038/nature04859. [DOI] [PubMed] [Google Scholar]
- 17.Sadqi M, Fushman D, Muñoz V. Structural biology—Analysis of protein-folding cooperativity—Reply. Nature. 2007;445:E17–E18. doi: 10.1038/nature05644. [DOI] [PubMed] [Google Scholar]
- 18.Zuo GH, Wang J, Wang W. Folding with downhill behavior and low cooperativity of proteins. Proteins Struct Funct Bioinf. 2006;63:165–173. doi: 10.1002/prot.20857. [DOI] [PubMed] [Google Scholar]
- 19.Knott M, Chan HS. Criteria for downhill protein folding: Calorimetry, chevron plot, kinetic relaxation, and single-molecule radius of gyration in chain models with subdued degrees of cooperativity. Proteins Struc Funct Bioinf. 2006;65:373–391. doi: 10.1002/prot.21066. [DOI] [PubMed] [Google Scholar]
- 20.Prieto L, Rey A. Influence of the native topology on the folding barrier for small proteins. J Chem Phys. 2007;127:175101. doi: 10.1063/1.2780154. [DOI] [PubMed] [Google Scholar]
- 21.Cho SS, Weinkman P, Wolynes PG. Origins of barriers and barrierless folding in BBL. Proc Natl Acad Sci USA. 2008;105:118–123. doi: 10.1073/pnas.0709376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Badasyan A, Liu Z, Chan HS. Probing possible downhill folding native contact topology likely places a significant constraint on the folding cooperativity of proteins with approximately 40 residues. J Mol Biol. 2008;384:512–530. doi: 10.1016/j.jmb.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Li W, Wang J, Qin M, Wang W. All-atom replica exchange molecular simulation of protein BBL. Proteins Struct Funct Bioinf. 2008;72:1038–1047. doi: 10.1002/prot.22001. [DOI] [PubMed] [Google Scholar]
- 24.Pitera JW, Swope WC, Abraham FF. Observation of non-cooperative folding thermodynamics in simulations of 1BBL. Biophys J. 2008;94:4837–4846. doi: 10.1529/biophysj.107.123265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma HR, Gruebele M. Kinetics are probe-dependent during downhill folding of an engineered lambda(6–85) protein. Proc Natl Acad Sci USA. 2005;102:2283–2287. doi: 10.1073/pnas.0409270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naganathan AN, Doshi U, Muñoz V. Protein folding kinetics: Barrier effects in chemical and thermal denaturation experiments. J Am Chem Soc. 2007;129:5673–5682. doi: 10.1021/ja0689740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cellmer T, Henry ER, Kubelka J, Hofrichter J, Eaton WA. Relaxation rate for an ultrafast folding protein is independent of chemical denaturant concentration. J Am Chem Soc. 2007;129:14564. doi: 10.1021/ja0761939. [DOI] [PubMed] [Google Scholar]
- 28.Godoy-Ruiz R, et al. Estimating free-energy barrier heights for an ultrafast folding protein from calorimetric and kinetic data. J Phys Chem B. 2008;112:5938–5949. doi: 10.1021/jp0757715. [DOI] [PubMed] [Google Scholar]
- 29.Naganathan AN, Perez-Jimenez R, Sanchez-Ruiz JM, Muñoz V. Robustness of downhill folding: Guidelines for the analysis of equilibrium folding experiments on small proteins. Biochemistry. 2005;44:7435–7449. doi: 10.1021/bi050118y. [DOI] [PubMed] [Google Scholar]
- 30.Sadqi M, Lapidus LJ, Muñoz V. How fast is protein hydrophobic collapse? Proc Natl Acad Sci USA. 2003;100:12117–12122. doi: 10.1073/pnas.2033863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferguson N, et al. Ultra-fast barrier-limited folding in the peripheral subunit-binding domain family. J Mol Biol. 2005;353:427–446. doi: 10.1016/j.jmb.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 32.Ikai A, Tanford C. Kinetics of unfolding and refolding of proteins. 1. Mathematical analysis. J Mol Biol. 1973;73:145–163. doi: 10.1016/0022-2836(73)90320-3. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, et al. Cooperative folding kinetics of BBL protein and peripheral subunit-binding domain homologues. Proc Natl Acad Sci USA. 2008;105:2397–2402. doi: 10.1073/pnas.0708480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubelka J, Eaton WA, Hofrichter J. Experimental tests of villin subdomain folding simulations. J Mol Biol. 2003;329:625–630. doi: 10.1016/s0022-2836(03)00519-9. [DOI] [PubMed] [Google Scholar]
- 35.Thompson PA, et al. The helix-coil kinetics of a heteropeptide. J Phys Chem B. 2000;104:378–389. [Google Scholar]
- 36.MacCallum JL, Moghaddam MS, Chan HS, Tieleman DP. Hydrophobic association of alpha-helices, steric dewetting, and enthalpic barriers to protein folding. Proc Natl Acad Sci USA. 2007;104:6206–6210. doi: 10.1073/pnas.0605859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwanzig R. Diffusion in a rough potential. Proc Natl Acad Sci USA. 1988;85:2029–2030. doi: 10.1073/pnas.85.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang CY, et al. Helix formation via conformation diffusion search. Proc Natl Acad Sci USA. 2002;99:2788–2793. doi: 10.1073/pnas.052700099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.