Abstract
Directed reprogramming of somatic cells by defined factors provides a novel method for the generation of patient-specific stem cells with the potential to bypass both the practical and ethical concerns associated with somatic cell nuclear transfer (SCNT) and human embryonic stem (hES) cells. Although the generation of induced pluripotent stem (iPS) cells has proven a robust technology in mouse and human, a major impediment to the use of iPS cells for therapeutic purposes has been the viral-based delivery of the reprogramming factors because multiple proviral integrations pose the danger of insertional mutagenesis. Here we report a novel approach to reduce the number of viruses necessary to reprogram somatic cells by delivering reprogramming factors in a single virus using 2A “self-cleaving” peptides, which support efficient polycistronic expression from a single promoter. We find that up to four reprogramming factors (Oct4, Sox2, Klf4, and c-Myc) can be expressed from a single virus to generate iPS cells in both embryonic and adult somatic mouse cells and we show that a single proviral copy is sufficient to generate iPS cells from mouse embryonic fibroblasts. In addition we have generated human induced pluripotent stem (hiPS) cell lines from human keratinocytes, demonstrating that a single polycistronic virus can reprogram human somatic cells.
Keywords: 2A peptide, four-factor reprogramming, iPS cell, polycistronic
Viral vector-mediated transduction of defined factors has been used to generate induced pluripotent stem (iPS) cells from embryonic or adult somatic cells in both mouse and human. By gene expression and developmental potential iPS cells are equivalent to embryonic stem (ES) cells maintaining a full capacity for differentiation with the ability to form teratomas, generate chimeras, and contribute to the germline (1–8). This technology can be readily applied to many cell types in addition to fibroblasts as numerous cell types have been shown to be amenable to direct reprogramming including pancreatic β cells, neural precursors, and terminally differentiated B cells (9–11). Without the ethical or practical concerns associated with human embryonic stem cells (hESC), iPS cell technology has emerged as the most promising method for cell-based therapies of regenerative medicine (12, 13). Yet the current protocols involve viral-based delivery of reprogramming factors leading to multiple proviral copies throughout the genome in addition to potentially harmful oncogenes, warranting serious concerns regarding insertional mutagenesis or potential reactivation of silenced viral transcripts in cells derived from iPS cell lines.
One improvement to the current strategy would be the delivery of reprogramming factors within the context of a single polycistronic vector. The use of internal ribosomal entry sites (IRES) in polycistronic vectors has been shown to express multiple genes from one promoter but this frequently leads to nonstoichiometric expression and significantly lower levels of the downstream cistron(s). One alternative to this method is autonomous “self-cleaving” 2A peptides originally identified and characterized in apthovirus foot-and-mouth disease virus (FMDV) (14, 15). Generally ≈18–22 aa long, 2A oligopeptides contain a highly conserved c-terminal D(V/I)EXNPGP motif that mediates “ribosomal skipping” at the terminal 2A proline and subsequent amino acid “2B” glycine (16). The most well-characterized 2A peptides are derived from FMDV (herein referred to as “F2A”), equine rhinitis A virus (ERAV, “E2A”), porcine teschovirus-1 (PTV-1, “P2A”), and insect Thosea asigna virus (TaV, “T2A”). Importantly the ability to express four proteins in vivo has been achieved using 2A peptides (17).
In this work we examined whether polycistronic vectors using 2A peptides can be applied to in vitro reprogramming with the goal of delivering the reprogramming factors in a single polycistronic virus. To test this we developed a four-factor 2A (4F2A) doxycycline (DOX)-inducible lentivirus encoding mouse cDNAs for Oct4, Sox2, Klf4, and c-Myc separated by three different 2A peptides (P2A, T2A, and E2A, respectively). Our findings are threefold: First, we demonstrate that up to three 2A peptides allow expression of the four reprogramming factors in a single vector being sufficient to reprogram both embryonic and adult murine somatic cells. Second, we find that a single proviral copy of the 4F2A is sufficient to reprogram mouse embryonic fibroblasts (MEFs), suggesting a single transgene could potentially reprogram somatic cells. Finally, we show that the 4F2A can be used in reprogramming human cells by generating several iPS lines from neonatal foreskin keratinocytes.
Results
Generation of Mouse Ips Cells Using a Single Polycistronic Virus.
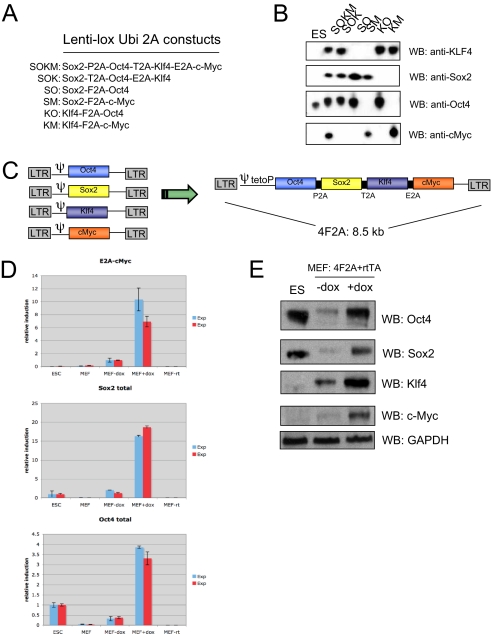
Our goal was to generate polycistronic viral vectors that would express multiple reprogramming genes from a single promoter using 2A peptides. For this to occur, one, two, or three 2A oligopeptides containing unique restriction sites were ligated into FUW lentivirus (18) backbones to allow efficient cloning of Oct4, Sox2, c-Myc, and Klf4 each separated by a different 2A sequence. Vectors carrying four, three, or two factors consecutively with different combinations of F2A, T2A, E2A, or P2A sequences (Fig. 1A) were tested for their ability to express individual factors by transient transfection in human 293 cells. Western blot analysis demonstrated that 2A peptides support efficient expression of two, three, or all four cistrons from a single polycistronic vector (Fig. 1B).
Fig. 1.
Generation of murine iPS cells using a single 4F2A polycistronic virus. (A) FUW lentivirus constructs tested by transient transfection. In total, four 2A peptides (F2A, T2A, E2A, and P2A) were used. (B) Transient transfection of 293 cells with FUW 2A lentiviruses. Cells were harvested after 48 h and analyzed by Western blot (WB). Efficient protein expression was observed in all constructs tested, indicating four unique 2A peptides support robust protein expression. Note: Sox2 protein is not detected in ES cells because only a short exposure was used. (C) Schematic of the 4F2A DOX-inducible lentivirus containing three types of 2A peptides (P2A, T2A, and E2A). Murine cDNAs for Oct4, Sox2, Klf4, and c-Myc. This particular sequence of factors and 2A peptides is subsequently referred to as “4F2A.” (D) RT-PCR anaylsis of mRNA induction in cells transduced with OSKM 4F2A + rtTA for 3 days. Total Oct4 or Sox2 induction was used to test levels of 4F2A induction relative to ES cells. E2A-cMyc primers were used to detect viral-specific transcripts. Error bars represent SD of the mean of triplicate reactions. (E) Western blot analysis of MEFs transduced with 4F2A + rtTA for 3 days. Cells infected with 4F2A DOX-inducible lentivirus + rtTA produce all four reprogramming factors upon addition of doxycycline (DOX).
A tetracycline inducible lentivirus vector was constructed where expression of the genes was controlled from the tetracycline operator minimal promoter (tetOP) (Fig. 1C). To test whether all four genes of a single four-factor (Oct4/Sox2/Klf4/c-Myc) virus could be expressed upon DOX addition, MEFs were infected with the polycistronic vector (referred to below as “4F2A”) and a constitutive FUW lentivirus carrying the tetracycline controllable transactivator (M2rtTA; abbreviated as rtTA). Two independent experiments were performed and drug inducible expression of the virus was tested 3 days postinfection by qRT-PCR. Using primers for viral specific transcripts (E2A-cMyc), robust induction was observed (7- to 10-fold) in cells cultured with DOX as compared to control medium (Fig. 1D). To test the relative induction compared to ES cells, Oct4 and Sox2 primers that cannot discriminate between viral or endogenous transcripts were used and in both experiments infected DOX-induced MEFs were significantly higher than in ES cells (≈3.5- and ≈17-fold over ES levels, respectively). Western blot analysis of cells isolated at 3 days after infection demonstrated that little or no protein was expressed when the cells were cultured without DOX whereas robust induction was seen in the presence of DOX with levels of Oct4 and Sox2 protein being similar to that in ES cells (Fig. 1E).
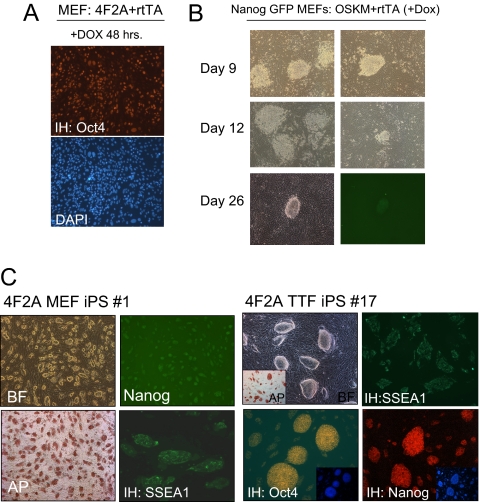
To test whether the 4F2A vector was able to reprogram somatic cells to a pluripotent state, MEFs containing a GFP reporter driven by the endogenous Nanog promoter were infected with virus (4F2A + rtTA). Eighty-five to 90% of the cells stained for Oct4 at 48 h after transduction indicated high titer infection (Fig. 2A). Morphological changes were observed a few days after addition of DOX (data not shown) with distinct colonies appearing after ?8 days and Nanog-GFP+ cells at ≈25 days after DOX induction (Fig. 2B). After mechanical isolation and subsequent passage, the cells had the typical morphology of ES cells and grew independently of DOX. Four independent 4F2A iPS cell lines were established that were positive for the pluripotency markers AP, SSEA1, and Nanog-GFP (Fig. 2C).
Fig. 2.
4F2A iPS cells express pluripotency markers. (A) Immunostaining of Oct4 protein indicates high titer infections can be achieved with the 4F2A. MEFs were cultured in DOX media for 2 days after transduction with 4F2A + rtTA. (B) Morphology changes in Nanog-GFP MEFs transduced with 4F2A + rtTA cultured in ES media + DOX. Colonies appeared at ≈8 days, similar to cells infected with single viruses. Nanog GFP+ colonies were observed by day 25 after DOX media removal at day 20. Two columns show typical colonies observed on the plate. (C) 4F2A iPS lines generated from Nanog-GFP MEFs or 14-week tail-tip fibroblasts (TTFs) that stain positive for pluripotency markers AP, SSEA1, Oct4 and have reactivated the endogenous Nanog locus (GFP+ for MEFs and by immunostaining for TTF).
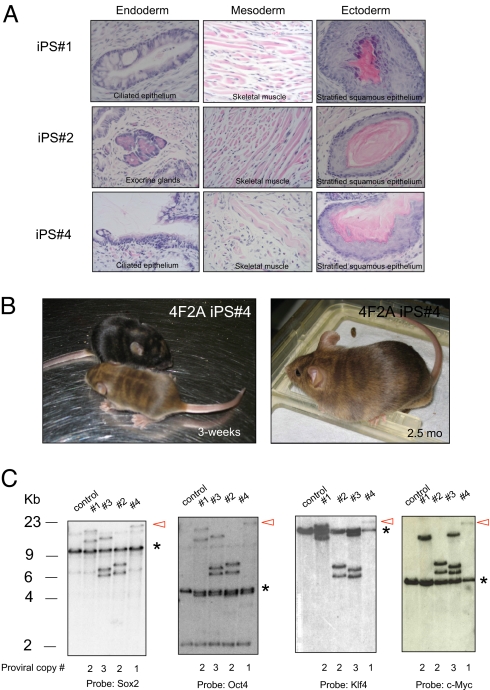
To investigate whether adult somatic cells could be reprogrammed using the 4F2A vector, we infected tail-tip fibroblasts (TTFs) from 14-week-old mice with the 4F2A + rtTA vectors. Similar to MEFs, typical morphological changes were observed a few days after addition of DOX media. Colonies appeared at ?8 days and continued to expand until they were picked (day 16) on the basis of morphology. After several passages, four stable iPS cell lines were established that stained positive for all pluripotency markers (Nanog, Oct4, SSEA1, and AP) (Fig. 2C). MEF iPS cell lines were injected s.c. into severe combined immunodeficiency (SCID) mice and were shown to induce teratomas that contained differentiated cells of all three germ layers (Fig. 3A). Finally, injection of MEF iPS cells (no. 4 in Fig. 3) into blastocysts generated postnatal chimeras (Fig. 3B), demonstrating that a single 4F2A polycistronic virus can reprogram MEFs to a pluripotent state.
Fig. 3.
4F2A iPS cells are pluripotent and contain between 1 and 3 proviral integrations. (A) In vivo differentiation of 4F2A MEF-iPS lines nos. 1, 2, and 4. Histological analysis of teratomas induced after s.c. injection into SCID mice indicates iPS lines contribute to all three germ layers. (B) Moderate-to-high contribution in postnatal chimeric mice as detected by agouti coat color from 4F2A iPS line no. 4. (C) Southern blot analysis of 4F2A proviral integrations in MEF-iPS cell line nos. 1–4. iPS cell DNA was digested with BamHI. Hybridization of the same molecular weight fragment using all four probes indicates presence of 4F2A provirus. Red arrow highlights iPS line no. 4, which contained one proviral copy of the 4F2A. * indicates endogenous allele.
To determine the number of proviruses carried in the 4F2A iPS cell lines, DNA was extracted and subjected to Southern blot analysis using an enzyme that does not cut in the vector sequences. Using Oct4, Sox2, c-Myc, and Klf4 probes for hybridization, we detected bands of identical molecular weight confirming that the factor sequences were carried in one provirus. The total number of proviruses was between one and three with iPS cell line no. 4 carrying a single viral insert (Fig. 3C). One of two integrations from iPS cell line no. 1 failed to produce a band after c-Myc hybridization, suggesting a 3′ deletion of the c-Myc sequences may have occurred. A second digest confirmed the proviral copy numbers (supporting information (SI) Fig. S1A).
To estimate reprogramming efficiency MEFs were infected with the 4F2A and rtTA vectors and plated at 0.25 × 106 per 10-cm-plate culture dish. About 70% of the MEFs were infected as estimated by immunostaining of Oct4 at 48 h after infection (Fig. S2A). Cells were cultured in ES media containing DOX for 20 days and subsequently transferred to ES cell medium until GFP+ colonies were counted on day 25. An average of ≈14.7 ± 4 colonies were detected in three independent dishes (10 + 10 + 17) indicating a relative efficiency of 0.0001%. This is two orders of magnitude lower than that of “secondary” fibroblasts or B cells carrying preselected DOX-inducible proviruses (19).
To test the kinetics of reprogramming using the 4F2A virus we performed DOX-withdrawl experiments where at specified days (i.e., 2, 4, 8, 12, etc.) DOX-containing media is replaced with ES media and the number of Nanog-GFP+ colonies are counted at day 25. Using separate drug-inducible viruses to deliver the four factors, it has been reported that ≈9–12 days is the minimum time required for the generation of stable iPS cells (20, 21). Cells are not passaged during this time to minimize duplication of reprogramming events. Two independent experiments were performed and in both cases single Nanog-GFP+ colonies were present on plates cultured in DOX media for 8 days, similar to the minimum time required using separate viruses (Fig. S1B).
These data demonstrate that a single polycistronic virus containing the four factors linked by three 2A peptides allows factor expression sufficient to generate iPS cells from embryonic or adult somatic cells. Importantly, our results also show that a single polycistronic proviral copy is sufficient to reprogram somatic cells to pluripotency.
Generation of Human Ips Cells Using a Single Polycistronic Virus.
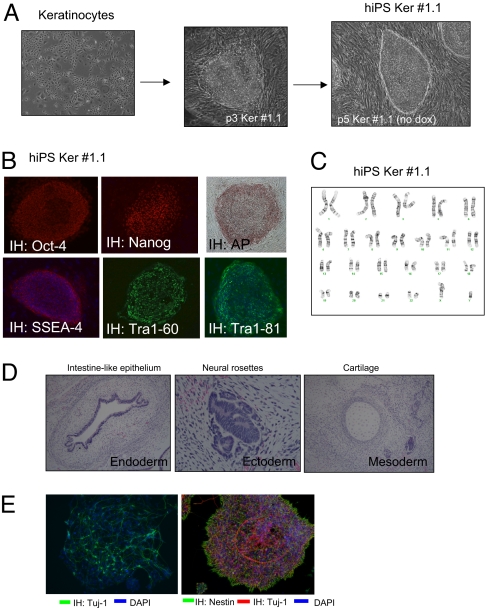
To investigate whether human cells could be reprogrammed with the polycistronic vector, neonatal human foreskin keratinocytes (NHFK) were transduced with both the constitutive rtTA and DOX-inducible 4F2A vectors. The fraction of infected cells was 10% as determined by staining for Oct4 at 48 h after transduction (Fig. S3A). Cells were incubated in keratinocyte medium + DOX and allowed to grow for 6 days until they were passaged and cultured in hESC media + DOX on gelatinized plates. Colonies were first detected at day 12 and most displayed transformed morphology with a few colonies exhibiting a distinct appearance that resembled hESC-like morphology. Two such colonies generated in independent infections were picked between 22 and 35 days after infection and found to expand as distinct colonies with morphology similar to hESC (Fig. 4A). These cells were expanded in the absence of DOX and gave rise to a homogenous population identical to hESC (Ker-iPS) after an additional two to five passages. The cells stained for the pluripotency markers AP, Oct4, Nanog, Sox2, SSEA4, Tra1–60, and Tra1–81 (Fig. 4B, Fig. S3B) and had a normal karyotype (Fig. 4C). DNA fingerprinting excluded that such Ker-iPS cell lines were contamination from previously established human iPS cells or hES lines from our lab (data not shown). To determine proviral copy number in Ker-iPS cell lines genomic DNA was extracted and subjected to Southern blot analysis using an enzyme that does not cut in the vector sequences. Probes for all four reprogramming factors show hybridization to similar molecular weight band(s) again indicating they were carried on a single virus. Two different digests (XbaI and BamHI) show the 4F2A proviral copy number is three (no. 1.1) and two (no. 3), respectively (Fig. S4 A and B).
Fig. 4.
Generation of human iPS lines using a single 4F2A polycistronic virus. (A) Neonatal human foreskin keritinocytes (NHFK) transduced with 4F2A (carrying mouse cDNAs) + rtTA. On day 22 a single colony was picked and expanded, giving rise to colonies resembling hES colonies. These colonies were picked and a stable hiPS line was established. (B) Ker-hiPS no. 1.1 immunostaining for pluripotency markers AP, Oct4, Nanog, SSEA-4, Tra1–60, and Tra1–81. DAPI stain is in lower panels. (C) Karyotype of Ker-hiPS no. 1.1 is normal 46 XY. (D) In vivo differentiation of Ker-hiPS no. 1.1. Hematoxylin and eosin staining of teratoma sections generated by Ker-hiPS no. 1.1. (E) In vitro differentiation of Ker-hiPS no. 1.1. (Left) Ker-iPS no. 1.1-derived neural precursors exposed to differentiation conditions for 6 days produce terminally differentiated neurons as detected by anti-Tuj1 immunostaining (green). (Right) Ker-iPS no. 1.1 neural precursors (NPs) undergo spontaneous differentiation. NPs were detected by anti-Nestin immunostaining and differentiated neurons by anti-Tuj1 (red). DAPI stain for DNA in both pictures is blue.
To test for pluripotency, one line, Ker-iPS no. 1.1, was injected s.c. into SCID mice. These cells induced teratomas and after histological examination differentiated into cells of all three germ layers (Fig. 4D). In addition, Ker-iPS no. 1.1 cells, when subjected to an in vitro neural differentiation protocol, produced nestin+ neural progenitor cell populations and Tuj1+ postmitotic neurons as detected by immunostaining. (Fig. 4E).
Discussion
The experiments described in this paper show that up to four different reprogramming factors inserted into a polycistronic vector separated by 2A sequences can be expressed at levels sufficient to achieve reprogramming. This is in contrast to IRES-based polycistronic vectors where the downstream genes are often expressed at substantially lower levels. Embryonic and adult murine fibroblasts and postnatal human keratinocytes were induced to form pluripotent iPS cells when infected with the FUW rtTA and 2A vector transducing Oct4, Sox2, Klf4, and c-Myc. Our finding that a single proviral copy of a polycistronic vector suggests that with sufficient expression it may be possible to reprogram using a stably expressed transgene. This is supported by the reports suggesting insertional mutagenesis is not necessary for reprogramming and that transient expression of reprogramming factors is sufficient to generate iPS cells (22, 23). Preliminary work by Okita et al. (2008) achieved reprogramming of murine embryonic fibroblasts using Moloney viruses expressing three factors separated by F2A peptides but with a significant reduction in efficiency. We extend these findings by demonstrating that even at a low efficiency, 2A vectors can reprogram adult murine somatic cells and human keratinocytes.
In addition, Okita et al. (2008) were able to reprogram murine embryonic fibroblasts using multiple transient transfections of two plasmids expressing Oct4-Klf4-Sox2 and c-Myc, respectively. Although iPS cells were generated with greatly reduced efficiency, iPS cell lines with no detectable vector integration were recovered. Interestingly, many of the iPS cell lines generated after transient transfection contained integrated plasmids, suggesting that stable integration of exogenous sequences can occur.
We observe a reprogramming efficiency significantly lower than previous experiments using single vectors to transduce each of the four factors (Fig. S2B and Table S1). It is possible that the lower reprogramming efficiency is because of the stochiometry of factor expression from the polycistronic vector, which may be suboptimal for inducing reprogramming. Transduction with separate vectors allows integration of different numbers of proviruses for each factor, therefore reprogramming may select for a specific set of proviral integrations that result in high expression or an optimal stochiometry between the different factors. This may not be possible using the 2A system, which has been reported to support near equimolar protein expression in vivo (17). Also, when separate vectors transducing each of the four factors were used for induction of iPS cells, Nanog-GFP+ cells were detected as early as 16 days after DOX induction in contrast to GFP+ cells observed 22–25 days after 4F2A vector transduction, consistent with less optimal reprogramming. Moreover, whereas iPS cells frequently carry multiple Oct4 or Klf4 proviruses, consistently fewer Sox2 proviruses were found, suggesting that a high level of Sox2 expression may be unfavorable for reprogramming (24).
Ultimately multiple technologies may converge to allow the generation of therapeutically acceptable iPS cells without viral integrations or lingering reprogramming factors within the genome. In a recent proof-of-principle work nonintegrating adenoviruses were used alone or in tandem with an inducible Oct4 transgene to reprogram mouse hepatocytes and fibroblasts, respectively, demonstrating insertional mutagenesis is not neccessary for reprogramming (23). iPS cells without detectable viral integrations were recovered; however, efficiencies were significantly lower than Moloney or lentivirus reprogramming of fibroblasts. It will be important to determine whether other more accessible cell types are amenable to adenovirus-mediated reprogramming as hepatocytes are harder to obtain and grow from human patients in comparison to keratinocytes or fibroblasts. Furthermore transient technologies such as adenovirus or DNA transfection of reprogramming factors may not have the ability to express reprogramming factors throughout the minimum time interval at levels necessary for acquisition of an iPS state, especially important for human reprogramming. Because the four factors are expressed from a defined location, the polycistronic vector system described in this paper should simplify the study of reprogramming mechanisms and may facilitate the excision of the vector resulting in iPS cells that carry no exogenous transgenes.
Materials and Methods
Viral Preparation and Infection.
Construction of 4F2A lentiviral vectors containing Oct4, Sox2, Klf4, and c-Myc under control of the tetracycline operator and a minimal CMV promoter was generated after EcoRI cloning from a FUW lentivirus backbone. All constructs were generated using unique restriction sites after amplification by PCR to place an individual factor between a respective 2A peptide (first, XbaI-NheI; second, SphI; third, XhoI; and fourth, AscI). Respective 2A sequences: P2A, GCCACGAAGCAAGCAGGAGATGTTGAAGAAAACCCCGG GCCT; T2A, GAGGGCAGAGGAAGTCTTCTAACATGCGGTGACGTGGAGGAGA ATCCCGGCCCT; and E2A, CAGTGTACTAATTATGCTCTCTTGAAATTGGCTGG AGATGTTGAGAGCAACCCAGGTCCC). Replication-incompetent lentiviral particles (4F2A and M2rtTA) were packaged in 293T cells with a VSV-G coat and used to infect MEFs containing a GFP allele targeted to the endogenous Nanog locus (7, 25). Fourteen-week-old tail-tip fibroblasts were derived from mice previously published (12). Human keratinocytes (NHFK) were obtained from Coriell Institute for Medical Research. Viral supernatants from cultures packaging each of the two viruses were pooled, filtered through a 0.45-μM filter and subjected to ultracentrifugation for concentration. Virus pellets were resuspended in ES cell medium (DMEM supplemented with 10% FBS (HyClone), leukemia inhibitory factor, β-mercaptoethanol (Sigma-Aldrich), penicillin/streptomycin, L-glutamine, and nonessential amino acids (all from Invitrogen) before being applied to cells for 24 h.
Western Blot.
One hundred microliters of lysis buffer containing 2% SDS, 10 mM DTT, 10% glycerol, 12% urea, 10 mM Tris-HCl (pH 7.5), 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture (Roche), 25 μM MG132 proteosome inhibitor, and boiled for 5 min. Proteins were then quantified using Bradford reagent (Pierce) and taking spectrophotometric readings at 590 nm. Concentrations were estimated against a standard curve generated using BSA. Total protein (5 μg) was subjected to electrophoreses in a denaturing 10% polyacrylamide gel containing 10% SDS. Proteins were then transferred onto Immobilon-P membranes (Millipore) using a semi-dry transfer apparatus. Membranes were blocked in PBS, 0.01% Tween 20 containing 2% nonfat powdered milk (Bio-Rad). Proteins were detected by incubating with antibodies at a concentration of 50 ng/mL in blocking solution. Antibodies used were Oct4 (h-134, Santa Cruz Biotechnology); Sox2 (mouse monoclonal, R&D Biosystems); c-Myc (06–340, Upstate); Klf4 (H-180, Santa Cruz Biotechnology); GAPDH (sc-25778, Santa Cruz Biotechnology).
Quantitative RT-PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen). Five micrograms of total RNA was treated with DNase I to remove potential contamination of genomic DNA using a DNA Free RNA kit (Zymo Research). One microgram of DNase I-treated RNA was reverse transcribed using a First Strand Synthesis kit (Invitrogen) and ultimately resuspended in 100 μL of water. Quantitative PCR analysis was performed in triplicate using 1/50 of the reverse transcription reaction in an ABI Prism 7000 (Applied Biosystems) with Platinum SYBR green qPCR SuperMix-UDG with ROX (Invitrogen). Equal loading was achieved by amplifying GAPDH mRNA and all reactions were performed in triplicate. Primers used for amplification were as follows:
Oct4 F, 5′-ACATCGCCAATCAGCTTGG-3′ and
R, 5′-AGAACCATACTCGAACCACATCC-3′
Sox2 F, 5′-ACAGATGCAACCGATGCACC-3′ and
R, 5′- TGGAGTTGTACTGCAGGGCG-3′
4F2A (E2A-cMyc) F, 5′-GGCTGGAGATGTTGAGAGCAA-3′ and
R, 5′-AAAGGAAATCCAGTGGCGC
GAPDH F, 5′-TTCACCACCATGGAGAAGGC-3′ and
R, 5′-CCCTTTTGGCTCCACCCT-3′.
Error bars represent SD of the mean of triplicate reactions.
Southern Blotting.
Ten micrograms of BamHI digested genomic DNA were separated on a 0.7% agarose gel, transferred to a nylon membrane (Amersham), and hybridized with 32P random primer (Stratagene) labeled probes for OCT4 (EcoRI-PstI fragment of pFUW-tetO-OCT4 plasmid), KLF4 (full-length KLF4 cDNA), c-MYC (full-length c-MYC cDNA), and SOX2 (full-length fragment of pFUW-tetO-SOX2 plasmid).
Immunofluorescent Staining.
Cells were fixed in 4% paraformaldehyde for 20 min at 25 °C, washed three times with PBS, and blocked for 15 min with 5% FBS in PBS containing 0.1% Triton-X. After incubation with primary antibodies against Oct4 (Santa Cruz, h-134); Sox2 (R&D Biosystems); Nanog (anti-ms and anti-h, R&D Biosystems); Tra-1–60 (mouse monoclonal, Chemicon International); hNANOG (goat polyclonal, R&D Bioystems); mNANOG (A300–398A, Bethyl Laboratories); Tra1–81 (mouse monoclonal, Chemicon International); and SSEA4 and SSEA1 (monoclonal mouse, Developmental Studies Hybridoma Bank) for 1 h in 1% FBS in PBS containing 0.1% Triton-X, cells were washed three times with PBS and incubated with fluorophore-labeled appropriate secondary antibodies purchased from Jackson Immunoresearch. Specimens were analyzed on an Olympus fluorescence microscope and images were acquired with a Zeiss Axiocam camera.
Mouse Chimera and Teratoma Formation.
Diploid blastocysts (94−98 h after hCG injection) were placed in a drop of Hepes-CZB medium under mineral oil. A flat tip microinjection pipette with an internal diameter of 16 μm was used for iPS cell injections. Each blastocyst received 8–10 iPS cells. After injection, blastocysts were cultured in potassium simplex optimization medium (KSOM) and placed at 37 °C until transferred to recipient females. About 10 injected blastocysts were transferred to each uterine horn of 2.5-day-postcoitum pseudopregnant B6D2F1 female. Pups were recovered at day 19.5 and fostered to lactating B6D2F1 mothers when necessary. Teratoma formation was performed by depositing 2 × 106 cells under the flanks of recipient SCID or Rag2−/− mice. Tumors were isolated 3–6 weeks later for histological analysis.
Human Teratoma Formation and Analysis.
hiPSCs were collected by collagenase treatment (1.5 mg/mL) and separated from feeder cells by subsequent washes with medium and sedimentation of iPSC colonies. iPSC aggregates were collected by centrifugation and resuspended in a ratio of 106 cells in 250 μL of iPSC culture media. iPSCs were injected s.c. by a 21-gauge needle in the back of SCID mice (Taconic). A tumor developed within 6 weeks and the animal was killed before tumor size exceeded 1.5 cm in diameter. Teratomas were isolated after killing the mice and fixed in formalin. After sectioning, teratomas were diagnosed on the basis of hematoxylin and eosin staining. Karyotype analysis was done with Cell Line Genetics.
In Vitro Differentiation of Human IPS Cells into Neuronal Progenitors.
Human keratinocyte iPS cells were allowed to outgrow in culture without pasaging for 2 weeks with daily medium change. At day 15 after passage, distinct neural rossets were observed and picked mechanically by pooled glass pipett (26). Rosettes were replated on dishes precoated with 15 μg/mL polyornithin/10 μg/mL of laminin (Po/Lam) in N2B27 medium supplemented with FGF2 (20 ng/mL) EGF (20 ng/mL) (all R&D Biosystems). After 5–7 days, cells were dissociated by scraping with cell lifter and pipetting to single cells in N2B27 medium and replated to Po/Lam culture dishes.
Differentiation and Immunocytochemistry.
Induction of differentiation of neural progenitors was performed by withdrawal of FGF2 and EGF from culture medium for 5 days. Cells were fixed in 4% paraformaldehyde for 20 min and stained for human nestin (Chemicon; 1:100) and Tuj-1 (1:100) and subsequently washed three times with PBS and incubated with fluorophore-labeled appropriate secondary antibodies purchased from Jackson Immunoresearch. Specimens were analyzed on an Olympus fluorescence microscope and images were acquired with a Zeiss Axiocam camera.
Supplementary Material
Acknowledgments.
We thank D. Hockemeyer and F. Soldner for the FUW M2rtTA (27) and M. Wernig, C. Lengner, M. Creyghton, R. Flannery, J. Dausman, D. Fu, and members of the Jaenisch lab for excellent assistance and helpful comments. R.J. is supported by grants from the National Institutes of Health: 5-RO1-HDO45022, 5-R37-CA084198, and 5-RO1-CA087869. J.H. is a Novartis Fellow by the Helen Hay Whitney Foundation.
Footnotes
Conflict of interest statement: R.J. is an advisor to Stemgent.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811426106/DCSupplemental.
References
- 1.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 4.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, et al. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 9.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JB, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 11.Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Curr Biol. 2008;18:890–894. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna J, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 13.Wernig M, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc Natl Acad Sci USA. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan MD, Drew J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994;13:928–933. doi: 10.1002/j.1460-2075.1994.tb06337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan MD, King AM, Thomas GP. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J Gen Virol. 1991;72(Pt 11):2727–2732. doi: 10.1099/0022-1317-72-11-2727. [DOI] [PubMed] [Google Scholar]
- 16.Doronina VA, et al. Site-specific release of nascent chains from ribosomes at a sense codon. Mol Cell Biol. 2008;28:4227–4239. doi: 10.1128/MCB.00421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 18.Lois C, et al. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 19.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stadtfeld M, et al. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okita K, et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 23.Stadtfeld M, et al. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eminli S, Utikal JS, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into iPS cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467–2474. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- 25.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 26.Zhang SC, et al. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 27.Hockemeyer D, et al. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.