Abstract
Leukotrienes (LTs) are signaling molecules derived from arachidonic acid that initiate and amplify innate and adaptive immunity. In turn, how their synthesis is organized on the nuclear envelope of myeloid cells in response to extracellular signals is not understood. We define the supramolecular architecture of LT synthesis by identifying the activation-dependent assembly of novel multiprotein complexes on the outer and inner nuclear membranes of mast cells. These complexes are centered on the integral membrane protein 5-Lipoxygenase-Activating Protein, which we identify as a scaffold protein for 5-Lipoxygenase, the initial enzyme of LT synthesis. We also identify these complexes in mouse neutrophils isolated from inflamed joints. Our studies reveal the macromolecular organization of LT synthesis.
Keywords: inflammation, multiprotein complex, 5-lipoxygenase, 5-lipoxygenase-activating protein
Leukotrienes (LTs) are lipid signaling molecules derived from arachidonic acid (AA) that initiate and amplify innate and adaptive immune responses by regulating the recruitment and activation of leukocytes in inflamed tissues (1–3). Cells employ multiple mechanisms to prevent the inappropriate onset of pro-inflammatory signaling while requiring tightly coupled processes to trigger the generation of pro-inflammatory signaling molecules. The interplay of these processes is epitomized by the synthesis of LTB4 and LTC4. In unstimulated myeloid cells, including mast cells, LT formation is held in abeyance by the cytosolic compartmentalization of cytosolic phospholipase A2 (cPLA2) (4, 5) and the cytosolic/nucleoplasmic localization of 5-lipoxygenase (5-LO) (6). LT formation is initiated by translocation of cPLA2 to the Golgi and ER/nuclear envelope to release AA (4, 5). In parallel, 5-LO targets to the inner and outer nuclear membranes to initiate LT synthesis by converting AA to 5-HPETE and LTA4, a process that requires the integral nuclear envelope protein 5-lipoxygenase-activating protein (FLAP) (7–9). Although we have shown that LTC4 synthase and FLAP constitutively interact on the nuclear envelope (10), how or whether 5-LO interacts with these proteins on the nuclear envelope in response to cell signaling is unknown.
Understanding how cells couple the reorganization of the LT biosynthetic enzymes to efficient AA utilization is central to understanding the signal transduction pathways that control the synthesis of bioactive lipids derived from AA. Scaffold/docking proteins can localize components of biochemical reactions at membrane interfaces; examples include protein kinase A anchoring proteins (11) and the integral membrane proteins caveolins 1–3 (12). The dependence of cellular LT synthesis on FLAP (7, 8) and its membrane localization (9) make it a conceptually appealing candidate for a 5-LO scaffold. However, no interaction of 5-LO with any membrane protein has been detected.
A closely related question is: how do different combinations of extracellular signals lead to LT generation? For example, in mast cells, the engagement of FcεR1 by IgE/antigen triggers LT synthesis, whereas in eosinophils and polymorphonuclear leukocytes (PMN), a combination of cytokines, G protein-coupled receptor ligands, or bacterial lipopolysaccaharide perform this function (13–15). An emerging theme in cell biology and immunology is that assembly of multiprotein complexes transduces apparently disparate signals into a common read-out. We therefore sought to identify multiprotein complexes that include 5-LO associated with FLAP and are assembled in response to inflammatory signaling. We report the identification of LT synthetic complexes, structures regulating inflammation, which are assembled on inner and outer nuclear membranes in response to cell activation. These structures are found in both RBL-2H3 cells stimulated via FcεR1 and in mouse PMN isolated from inflamed joints in an IgE-independent model of arthritis. These complexes are centered on FLAP, which we identify as a nuclear envelope scaffold protein for 5-LO, and also contains additional proteins. Our studies reveal the macromolecular organization of the synthetic machinery for LTs. Furthermore, by defining a supramolecular organization to LT synthesis, we establish a conceptual model that allows the understanding of how diverse signals are integrated on nuclear membranes to initiate LT synthesis by assembling a common molecular platform.
Results
5-LO and FLAP Interact on the Nuclear Envelope.
Our approach to establishing the membrane organization of LT synthesis was to identify FLAP as a scaffold protein for 5-LO and use this interaction to locate LT synthetic complexes in vivo and then as a focal point to probe for other complex components. We initially used RBL-2H3 cells stimulated via FcεR1 because they are an established system for analyzing the synthesis of LTs. Several considerations made antibody (Ab)-based Fluorescence Lifetime Imaging Microscopy (FLIM) the approach of choice to image the interaction between 5-LO and FLAP. First, based on its crystal structure (16) the N- and C- termini of FLAP are on the opposite side of the membrane from 5-LO. Second, the placement of fusion proteins on 5-LO must be on the N-terminal of the enzyme to preserve catalytic function. Because FLIM experiments in cells using fusion proteins would require energy transfer across membranes, they were projected and found to be unsuccessful. Finally, although YFP, CFP, and GFP fusion proteins of FLAP localize correctly within cells, they could not reconstitute catalytic activity when paired with fusion proteins of 5-LO, presumably due to steric constraints.
RBL-2H3 cells stimulated via FcεR1 have been reported to generate LTs for 10–15 min (17) or up to 20–30 min (18). We therefore performed our analysis within this time interval. Initially, we examined the interaction of 5-LO with FLAP at 10 min poststimulation, where cells generated 32 ng LTC4/106 cells (n = 2) and then at times up to 30 min. RBL-2H3 cells cultured on slides were primed with anti-DNP IgE and activated with DNP-conjugated BSA. The cells were fixed and 5-LO detected with Alexa Fluor 488-conjugated secondary Ab (donor fluorophore) and FLAP with Alexa Fluor 594-conjugated secondary Ab (acceptor fluorophore). Analysis by immunofluorescence microscopy was followed by FLIM (Fig. 1A). Significant amounts of 5-LO are localized in the nucleus in IgE primed cells (panel 1), and a slight redistribution to the nuclear envelope occurred (panel 2) 10 min after antigen addition. Panel 3 shows the distribution of FLAP in the same cells.
Fig. 1.
5-LO and FLAP interact. (A) Imaging 5-LO/ FLAP interactions. The localization of 5-LO in unstimulated cells (1), and 5-LO (2) and FLAP (3) in IgE-primed cells. Pseudocolor FLIM image of primed cells probed for 5-LO (4). Pseudocolor image of a representative cell probed for 5-LO 10 min after stimulation with DNP-BSA (5) and of an IgE-primed cell probed for 5-LO and FLAP (6). Pseudocolor images of cells 10 (7 and 8) and 15 min (9) after stimulation. FLAP was detected with Ab(N) (7 and 9) or Ab(C) (8). n, nucleus; ne, nuclear envelope; IgE, cells primed with DNP-specific IgE; IgE + DNP, primed cells stimulated by the addition of DNP-BSA. (B) Biochemical analysis of 5-LO interactions. Western blots of cell extracts (Left) at times after antigen addition. Proteins immunoprecipitated with anti-5-LO Ab were resolved by SDS/PAGE and analyzed by Western blotting for 5-LO or FLAP (Right). The control lane “C” has been consolidated with the other lanes. (C) Biochemical interactions of FLAP. Proteins were cross-linked with DMP and cell extracts analyzed by Western blotting for FLAP (Left). Noncross-linked (1) or cross-linked (2) controls before activation; 3: IgE-primed cells; 4: 30 min postactivation. The time course for FLAP/AP-10 dissolution is shown in the right panel.
As an initial control for FLIM, unprimed cells were probed solely for 5-LO (not shown). After stimulation at 800 nm, the lifetime of the donor fluorophore was 2059+/−30 picoseconds (ps, n = 5). After IgE priming, the results were essentially the same (as depicted in the pseudocolor image in Fig. 1A panel 4, and in the bar graph in Fig. S1. Almost identical results were also seen in cells stimulated for 10 min by the addition of antigen but probed only for 5-LO (panel 5). When IgE-primed cells were probed for both 5-LO and FLAP (panel 6), no decrease in the lifetime of the donor fluorophore was observed, indicating no interaction between 5-LO and FLAP.
Ten minutes after the addition of antigen the donor lifetime decreased to 608 ± 74 ps (using antibody to the N-terminal of FLAP, AbN, n = 6, panel 7) and 606+/−53 ps (using antibody to the C-terminal, AbC, n = 6, panel 8) respectively, as represented by the yellow-orange nuclear envelope, indicating the interaction of 5-LO and FLAP. No interaction was observed within the nucleus (blue) or cytosol. The interaction of 5-LO and FLAP was also seen at 15 min (panel 9).
Biochemical Analysis of the Interactions of 5-LO.
Biochemical analysis confirmed the imaging data (Fig. 1B). RBL-2H3 cells were stimulated via FcεR1 for times between 5 and 30 min. At each time of analysis the buffer was removed and analyzed for LTC4. Cellular proteins were cross-linked with the membrane-permeant cross-linker dimethyl pimelimidate dihydrochloride (DMP). Cells were extracted, and the proteins resolved directly by SDS gels and analyzed by Western blotting for 5-LO. Alternatively, proteins were immunoprecipitated with anti-5-LO Ab before analysis for either 5-LO or FLAP. The association of 5-LO with FLAP was seen as a faint 100 kDa band in cell extracts probed with anti-5-LO Ab 5 min after activation. This corresponded to approximately 5% of cellular 5-LO (not shown). By 15 and 20 min this was approximately 12% of total 5-LO (Fig. 1B Left), and remained unchanged up to 45 min. LTC4 synthesis was complete by 10 min. Western blots of immunoprecipitates probed with anti-5-LO Ab identified monomeric 5-LO at approximately 78 kDa and a second 100 kDa protein species (Fig. 1B Upper Right). Using anti-FLAP Ab, only a 100 kDa band was detected (Fig. 1B Bottom Right), identifying it as a FLAP/5-LO dimer. Without cross-linking, only the 78 kDa 5-LO species was detected. There was some discordance between the percentage of 5-LO complexed to FLAP in immunoprecipitates and total extracts. At least half the immunoprecipitated 5-LO was dimerized to FLAP by 15 min, and by 30–40 min almost all immunoprecipitated 5-LO was FLAP-associated. Because the percentage of 5-LO complexed to FLAP in direct analysis of cell extracts remained constant after 15 min, the results in immunoprecipitates is likely due to the preferential immunoprecipitation of cross-linked 5-LO. Because all LTC4 synthase is associated with FLAP (19), we probed whether an interaction between LTC4 synthase and 5-LO could be detected either by FLIM, or as a 100 kDa heterodimer by Western blotting; no interaction was found (data not shown).
Biochemical Analysis of the Interactions of FLAP.
The interactions of FLAP were more complicated than those of 5-LO (Fig. 1C Left). In extracts of non-activated cells only FLAP monomers were identified without cross-linking. When proteins were cross-linked, multiple previously identified protein species were detected (19): monomeric FLAP (18 kDa); a 37 kDa species corresponding to either homodimers of FLAP or heterodimers of FLAP and LTC4 synthase, and a FLAP trimer (approximately 55 kDa). In addition, a 28 kDa species was observed. This was felt to most likely represent the association between FLAP and a 10kDa protein (Associated Protein 10kDa, AP-10). After cell activation, several clear changes were observed that are accentuated by 30 min. First, the 5-LO/FLAP dimer was observed at 100kDa (see Fig. 1B). In addition, the 28 kDa species was no longer detected. A minor species at approximately 200 kDa was also observed but not further characterized. Because the disappearance of the FLAP/AP-10 dimer was a postactivation change, this was examined over a 30 min time period. The loss of the FLAP/AP-10 dimer clearly progressed over time (Fig. 1C Right); whether this was due to dissociation or metabolic degradation/processing, is unknown. We also explored whether cPLA2 or LTA4 hydrolase were associated with either FLAP or 5-LO after cell activation. Neither enzyme was incorporated into LT membrane synthetic complexes (Fig. S2). In the context of the known trimeric structure of FLAP the ratio of monomers: dimers: trimers of FLAP seen is best explained by the efficiency of the cross-linking reaction which is not 100% and decreases after the first reaction.
LT Membrane Synthetic Complexes Are Assembled in Vivo.
To determine whether LT membrane synthetic complexes are assembled in vivo, we used the K/BxN serum transfer model of arthritis. Joint inflammation is dependent on LTB4 generation by synovial PMN (20) and independent of FcεR1 stimulation, with no differences seen in FcεR1−/− and WT mice (21). If the formation of LT membrane synthetic complexes is a general property of LT formation rather than restricted to cells stimulated via FcεR1, synovial PMN should have LT membrane synthetic complexes which are not present in quiescent mature bone marrow PMN. To test this, proteins were extracted from bone marrow and synovial PMN before or after cross-linking with DMP (Fig. 2 A and B) and analyzed for the presence of membrane complexes. No complexes were observed in bone marrow cells, whereas 5-LO/FLAP dimers were present in synovial cells probed for 5-LO (Fig. 2A). FLAP homodimers, dimers, and trimers were also identified in synovial PMN (Fig. 2B). In addition, in cellular extracts probed for FLAP, the 28 kDa species seen in vitro was also observed. These results confirm the ubiquitous nature of the interactions seen in RBL-2H3 cells.
Fig. 2.
LT membrane synthetic complexes are assembled in vivo. (A) Biochemical analysis of mature bone marrow PMN and synovial for 5-LO. PMN proteins were extracted and analyzed before (−) or after (+) cross-linking with DMP. Extracts (20 μg per lane) were resolved by SDS PAGE and then analyzed by Western blotting for 5-LO. (B) Biochemical analysis of mature bone marrow PMN and synovial for FLAP. PMN proteins were extracted and analyzed before (−) or after (+) cross-linking with DMP. Extracts (20 μg per lane) were resolved by SDS PAGE and then analyzed by Western blotting for FLAP. The asterisk (*) identifies the 5-LO/FLAP heterodimer. The MW of each protein species in (A) and (B) is indicated on the right.
Defining Inner and Outer Nuclear Membrane Complexes.
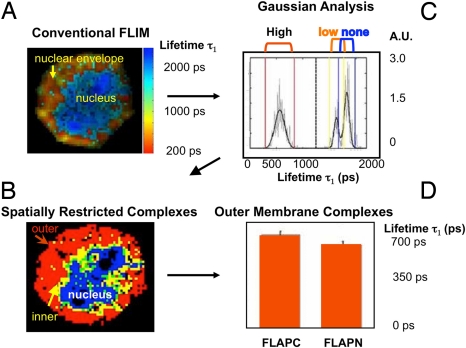
FLIM identifies pixels of interactions, and lifetimes are quantified by taking the mean over all pixels in the area of analysis and includes pixels of strong, medium, weak, and no interaction. This limits the resolution to a relatively broad area such as the entire nuclear envelope (Fig. 3A). The calculated lifetime, therefore, may not directly represent the interaction being studied and can be significantly different. This is particularly problematic in a setting where an area of no interaction (the nucleus) is analyzed along with an area of high affinity (the nuclear envelope). In addition, areas of intermediate affinities are not identified. To overcome these constraints, we used an approach based on the assumption that lifetimes in a FLIM experiment conform to a series of Gaussian distributions around mean values and that each mean qualitatively defines a population of interacting proteins. Fitting an intensity-weighted histogram of lifetimes from the region of interest with a multiple Gaussian function allows the recovery of multiple lifetimes, even when their distributions overlap (SI Text). These lifetimes can then be assigned to pixels in the original image. This approach was named “Multiple Gaussian for Lifetime Evaluation” (MUGLE, 22).
Fig. 3.
Identification of spatially restricted populations of LT synthetic complexes on the nuclear envelope. (A) Pseudocolor image of the cell nucleus. (B) Intensity weighted histogram (gray) of the lifetimes depicted in (A). A multi Gaussian fit to the histogram is shown (heavy black) (R2 = 0.8). Vertical bars show the centroid ± 3 times the standard deviation for the short (red) and long (yellow) lifetime distributions as well as a third distribution (bleed-through). The crude mean of the lifetimes (dashed line) does not represent the mean of either distribution. The data are transformed to a pseudocolor scale of absorbance units for spatial plotting. (C) Spatial extent of the three distributions is established by plotting the pixels whose lifetimes fall within the colored vertical lines in B. Red = strong interactions; yellow = weak interactions; blue = no FRET. Green depicts interactions that could be attributed to yellow or blue groups. (D) 5-LO is close to the N-terminal of FLAP in the outer nuclear membrane. The characteristic decay constants of the shorter (τ1) component of the bi-exponential fit to the donor decay profile. The average lifetime for the shortest component was recovered using MUGLE and averaged over five cells. A students t test shows significance (P ≈ 0.002).
As described above, a conventional FLIM pseudocolor image demonstrates fluorescence resonance energy transfer (FRET) between 5-LO and FLAP on the nuclear envelope 10 min after activation of RBL-2H3 cells (Fig. 3A). The mean lifetime was 1255 ps. This data were then plotted (Fig. 3B) as an intensity-weighted histogram (gray) of the lifetimes, and analyzed as series of Gaussian distributions around mean values (heavy black line). Three clear lifetime means (τn) and standard deviations (σn) of the distributions were detected: 569 ± 92 ps, 1634.42 ± 45.06 ps and 1839.38 ± 50.05 ps. The boundaries of the distributions are shown as vertical colored lines and bound lifetimes of τn ± 3σn, which include >99% of the pixels contained in each distribution. Each pixel was then assigned a distribution and mapped to the original image, giving a map of the spatial distributions of the three lifetimes (Fig. 3C). Red pixels correspond to the shortest lived distribution (strongest interaction), yellow the middle (weaker interaction), and blue the longest (no interaction). Pixels potentially belonging to either middle or long lifetimes are green. Weaker interactions between 5-LO and FLAP occur solely on the inner nuclear membrane, where 5-LO molecules are approximately 50% further from FLAP than on the outer membrane. The blue pixels depict non-interacting nucleoplasmic 5-LO. The spatial distribution of the complexes and the apparent relative proportion of the inner and outer nuclear membrane populations can be explained by recognizing that the figure is an image of an optical slice taken above the center of the cell. By visualizing a carved pumpkin with the top off, the inside of the shell (yellow pixels) appears as a thin rim, while the outside skin (red pixels) expands toward the midline. The blue area represents the empty cavity of the pumpkin. The yellow ring, which represents the middle lifetime, is not due to a partial volume effect from spatial binning. This was established by the observation that in some images of the nuclei the yellow distributions occupy a large region, several pixels across, in both lateral dimensions (data not shown). A distinct inner membrane population of complexes has been seen in all activated cells analyzed to date.
To define the relationship between 5-LO and FLAP in outer and inner nuclear membranes we probed cells with anti-5-LO Ab in combination with FLAPN Ab or FLAPC Ab. MUGLE (Fig. 3D) revealed no difference in lifetimes between the N- and C-terminal of FLAP for inner membrane complexes. However, in the outer nuclear membrane, the average lifetimes were 541± 26 ps (mean +/− SD, n = 5) using FLAPN Ab and 607± 21 ps (n = 5) using FLAPC Ab (p ≈ 0.002), indicating that 5-LO is closer to the N terminus of FLAP.
The 5-LO Interaction Site of FLAP Is Distinct from the Inhibitor and AA Binding Domain.
Based on photoaffinity and mutagenesis studies the binding sites for AA and for the indole-based FLAP inhibitor MK-886 overlap (23). The 3D structure of FLAP (16) also indicates that the binding site of the second generation indole FLAP inhibitor MK-591 overlaps with that of AA. We reasoned that if 5-LO and FLAP interact at the AA binding site, then LTC4 synthesis and the interaction of 5-LO and FLAP should be inhibited by the same concentration range of MK-886. RBL-2H3 cells were activated by IgE/antigen in the presence of 200 nM to 1.0 μM MK-886 for 10 or 15 min, and the interaction of 5-LO and FLAP analyzed by MUGLE. In parallel, cells were plated in 90 mm dishes and activated by IgE/antigen in the presence of the same concentrations of MK-886 and supernatants analyzed for LTC4 generation. Concentrations of MK-886 of 200 nM or greater completely inhibited LTC4 generation (Fig. 4), whereas no effect on the interaction of 5-LO and FLAP was observed. In the outer nuclear membrane, the lifetime after cell activation was 544 ± 25 ps (mean +/− SEM) and 546 ± 51 ps in the presence of 400 nM MK-886 indicating no effect of MK-886 (Fig. 4B). This was true for outer membrane complexes using AbC and for all inner membrane complexes. Only at a concentrations of 1 μM was a non-specific disruption of all FLAP interactions seen (Fig. S3). These results imply that the site(s) of FLAP-5-LO interaction are distinct from that presenting AA.
Fig. 4.
5-LO and FLAP do not interact at the AA/Inhibitor binding site. RBL-2H3 cells in 90 mM dishes were primed with DNP-specific IgE. They were then stimulated for 15 min by the addition of DNP-BSA in the presence or absence of 400 nM MK-886. LTC4 generation in the supernatants was measured. In parallel, the interaction of 5-LO and FLAP was analyzed by MUGLE and the effect of 400 nM MK-886 on the lifetime of the donor fluorophore (Alexa Fluor 488) measured in both outer (O) and inner (I) membrane complexes. O/I indicates the lack of interaction in non-stimulated cells. The data represent the mean ± S.E.M from five cells transformed by MUGLE in a representative experiment.
Discussion
We have combined molecular imaging analysis, biochemical approaches, and in vivo studies to identify the activation-dependent reorganization of the LT synthetic enzymes into spatially distinct multiprotein complexes on inner and outer nuclear membranes. These structures link cell activation with the initiation of inflammation by LTs. The identification of the interaction of 5-LO and FLAP in synovial PMN (Fig. 2) confirms the in vivo relevance of our observations. PMN and eosinophils employ a combination of signaling by cytokine or toll-like receptors and G protein-coupled receptors to initiate LT synthesis (13–15). The identification of the same FLAP-containing protein species in both synovial PMN and in IgE-stimulated RBL-2H3 cells support a ubiquitous role for these structures in LT synthesis in IgE dependent and independent systems.
Scaffold proteins bring components of biochemical reactions in close apposition to facilitate their interaction and amplify downstream signaling, they also compartmentalize reactions within cells. FLAP fulfills each of these requirements. First, it localizes 5-LO to the inner and outer nuclear membranes. Secondly, the combination of mutagenesis and structure determination shows a partial overlap between the AA and inhibitor binding sites of FLAP (16, 23). These sites are positioned within the membrane, ideally situated to capture laterally diffusing AA generated exogenously to the complex; FLAP-associated AA could be made available to 5-LO (16, 23). The structural basis of how FLAP brings 5-LO into apposition with AA remains a key question, as does the site(s) at which 5-LO interacts with FLAP. The identity and role of AP-10 remains to be determined. How or whether the progressive loss of the FLAP/AP-10 dimer (Fig. 1C) is coupled with the interaction of 5-LO to FLAP is not known. However, the common time course of the disappearance suggests that these processes are likely linked.
Fig. 5 depicts a basic synthetic unit in which AA diffusing within the membrane is “trapped” by FLAP and presented to 5-LO. AP-10 is dissociated from FLAP concomitant with 5-LO targeting. Whether additional protein species are present in synthetic complexes or interact with FLAP remain to be determined. We have taken two additional observations into consideration. First, we have shown that all cellular LTC4 synthase is associated with FLAP (19) and that LTC4 synthase is likely to be enriched in the outer nuclear membrane (10). Second, structural studies have indicated that both FLAP and LTC4 synthase must exist as independent homotrimers (16, 24, 25). When combined with our results, this indicates that LTC4 synthase and FLAP interact at a site distinct from the site where 5-LO and FLAP interact. Recent studies of the integral membrane protein syntaxin suggests a testable model that can accommodate each of these observations. Syntaxin self-associates into dense supramolecular clusters of approximately 75 molecules with a diameter of 50–60 nm, with the ultimate size and composition of the clusters determined by a balance between self-association and steric repulsions (26). The association of FLAP with LTC4 synthase depicted in Fig. 5 is likely mediated by the “almost self” association of these two highly identical proteins within the membrane. Our model allows LTA4 generated in a FLAP molecule to be efficiently transferred to a tightly associated LTC4 synthase molecule. It also explains why a close interaction between 5-LO and LTC4 synthase was not be detected. These units would be assembled into larger supramolecular clusters whose size and composition remains to be determined.
Fig. 5.
The membrane organization of leukotriene synthesis. The localization and interactions of the synthetic enzymes in IgE-primed RBL-2H3 cells (Upper) and in cells subsequently activated with antigen (blue triangle, Lower) are shown. FLAP is depicted on the outer and inner nuclear membranes LTC4 synthase (LTC4S) is depicted in the outer nuclear membrane associated with FLAP. In the primed cell, AP-10 is associated with FLAP, 5-LO is in the cytosol and nucleoplasm, and cPLA2 is in the cytosol. In the activated cell AP-10 dissociates from FLAP, cPLA2 and 5-LO are translocated to cell membrane, and 5-LO becomes associated with FLAP. LTA4 hydrolase (LTA4H) is not membrane or complex-associated. The possibility that inner membrane complexes contribute LTA4 to LTB4 synthesis and outer membrane complexes contribute the majority of LTA4 to LTC4 generation is shown.
MUGLE (Fig. 3) identified clear qualitative differences between inner and outer nuclear membrane complexes. This observation is intriguing in the context of previous studies. First, although no EM studies localizing 5-LO and FLAP in RBL-2H3 nuclear membranes have been reported, we have previously shown that LTC4 synthase is preferentially localized to the outer nuclear membrane in RBL-2H3 cells (10). Secondly, in human PMN activated with calcium ionophore, the ratio of inner nuclear membrane 5-LO or FLAP to outer nuclear membrane 5-LO or FLAP respectively is at least 4:1 (9). In addition, functional studies have suggested that nuclear 5-LO may be associated with the production of LTB4 (e.g., ref. 27). Whether inner membrane complexes preferentially generate LTA4 associated with LTB4 synthesis suggested in Fig. 5, or how or whether inner and outer membrane complexes differ structurally remain to be determined. Our findings provide a framework to probe this hypothesis. One possibility is that the different relationship between 5-LO and FLAP seen in the inner and outer membranes (Fig. 3D) could be explained by the enrichment of LTC4 synthase in outer membrane supramolecular clusters. Furthermore, detecting spatially restricted, qualitatively different compartmentalized interactions between two molecules within a cell has broad application in immunology and cell biology. In our studies, the synthesis of LTs terminated between 5 and 10 min in response to stimulation of FcεR1. A consistent amount of 5-LO is incorporated into the LT synthetic complex well past this time frame, suggesting that multiple processes can contribute to the termination of LT formation. These include re-esterification of AA, oxidative inactivation of 5-LO, and the metabolism of the assembled membrane synthetic complex. The contribution of each of these mechanisms remains to be determined.
Traditionally, the role of LTs, and prostanoids has been investigated using knockout mice for a specific biosynthetic enzyme (e.g., 5-LO) or receptor (e.g., CysLT1R). This approach assumes a linear aspect in the signal transduction pathways. The inflammasome is the multiprotein complex, which comprises NALPs, ASC, caspase-1, and caspase-5 and is involved in the proteolytic cleavage of pro-IL-1β to IL-1β to activate the innate immune system (28). It is assembled in response to diverse stimuli including bacterial pathogens and uric acid. The LT membrane synthetic complex plays an analogous role for the generation of LTs by providing a common macromolecular platform that transduces diverse extracellular signals to initiate LT signaling in inflammation.
Materials and Methods
Cell Culture, Activation, and Immunofluorescence Microscopy of RBL-2H3 Cells.
RBL-2H3 cells (transformed mast cell line) were grown on two chambered slides and analyzed as previously described (10, 19). For activation (also see SI Text), they were washed twice with HBSS containing 1 mM CaCl2, 1 mM MgCl2, and 0.1% BSA (HBSA2+) and primed with 2 μg/ml of mouse monoclonal anti-DNP-specific IgE (Sigma) for 1 h at room temperature in HBSA2+. They were then washed twice with HBSA2+ and activated by the addition of 50 ng/ml of DNP-BSA (Calbiochem) added in 500 μl of HBSA2+. Controls were cells that were neither activated nor primed or cells that were only primed. The primary Abs were rabbit polyclonal anti-5-LO Ab and goat polyclonal anti-FLAP Ab (1:50 dilution). The secondary antibodies were Alexa Fluor 488 donkey anti-rabbit IgG, 10 μg/ml, or Alexa Fluor 594 donkey anti-goat IgG, 10 μg/ml). For experiments in which biochemical analysis was performed, cells were seeded in six well culture dishes and then activated as above. In these experiments, the supernatants were removed and assayed at various time intervals for the generation of LTC4.
in Situ Cross-linking of Proteins and Western Blot Analysis.
Cross-linking and extraction of RBL cells was performed as previously described (19) with the modification that 4 mM DMP was used as the cross-linker and extraction was performed using M-PER lysis buffer (Pierce, SI Text). For quantification of band intensity, blots were scanned and then imported in JPEG format for analysis by IMAGEJ software (National Institutes of Health).
FLIM Analysis.
Ab-based FLIM was performed as previously described (29, 30, SI Text).
MUGLE Analysis. See SI Text.
Assay for LTC4.
The formation of LTC4 from activated RBL-2H3 cells was assayed by an internal Fluorescence-Linked Immunoabsorbent assay at Merck Research Labs modified for the detection of LTC4 (31).
K/BxN Model of Arthritis and Isolation of Synovial PMN from Mice.
Arthritis was induced by the i.p. injection of arthritogenic serum on days 0 and 2 of the experiment and evaluated as previously described (20, SI Text). All animal studies were approved by the Partners Institutional Animal Care and Use Committee.
Isolation of PMN from mouse bone marrow. Mature PMN (>90%) were isolated from bone marrow at the 65%/75% interface of discontinuous Percoll gradients as previously described (20, 32).
Supplementary Material
Acknowledgments.
The authors would like to thank Drs. Marty Springer (Merck Research Labs) and Bob Young, Merck Frosst, Canada, for their support. The work was supported by R56 AI068771 and R01 AI068771, 5R01 GM61823 (AM, AMB, BN, PC, and RJS); 5P50 NS010828 (PJ); 5R01 AG08487 (BH); 5R01 EB000768 (BB); and R01 AI 059746 and P01 AI065858 (DML and MC) T32 DK001540 (A.M.B.) from the National Institutes of Health. Grants from Merck Research Labs, Rahway NJ (PC) and a charitable gift from Merck and Merck Frosst (RJS); and a grant form the Fidelity Foundation (BH), and an Arthritis Foundation Fellowship (MC).
Footnotes
Conflict of interest statement: All financial and material support for this research and work are clearly identified in the manuscript. In particular, Dr. Peter Christmas was supported by a grant from Merck Research Labs, Rahway, NJ, and Dr. Roy Soberman received unrestricted charitable gifts from Merck Research Labs and from Merck Frosst, Canada.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808211106/DCSupplemental.
References
- 1.Ford-Hutchinson AW, Bray MA, Doig MV, Shipley NE, Smith MJH. Leukotriene B, a potent chemokinetic and aggregating substance released from polymorphonuclear leukocytes. Nature. 1980;286:264–265. doi: 10.1038/286264a0. [DOI] [PubMed] [Google Scholar]
- 2.Ott VL, Cambier JC, Kappler J, Marrack P, Swanson BJ. Mast cell-dependent migration of effector CD8+ T cells through production of leukotriene B4. Nat Immunol. 2003;4:974–981. doi: 10.1038/ni971. [DOI] [PubMed] [Google Scholar]
- 3.Tager AM, et al. Leukotriene B4 receptor BLT1 mediates early effector T cell recruitment. Nat Immunol. 2003;4:982–990. doi: 10.1038/ni970. [DOI] [PubMed] [Google Scholar]
- 4.Glover S, et al. Translocation of the 85-kDa phospholipase A2 from cytosol to the nuclear envelope in rat basophilic leukemia cells activated with calcium ionophore or IgE/antigen. J Biol Chem. 1995;270:15399–153407. doi: 10.1074/jbc.270.25.15359. [DOI] [PubMed] [Google Scholar]
- 5.Evans JH, Spencer DM, Zweifach A, Leslie CC. Intracellular calcium signals regulating cytosolic phospholipase A2 translocation to internal membranes. J Biol Chem. 2001;276:30150–30160. doi: 10.1074/jbc.M100943200. [DOI] [PubMed] [Google Scholar]
- 6.Brock TG, McNish RW, Bailie MW, Peters-Golden M. Rapid import of cytosolic 5-lipoxygenase into the nucleus of PMNs after in vivo recruitment and in vitro adherence. J Biol Chem. 1997;272:8276–8280. doi: 10.1074/jbc.272.13.8276. [DOI] [PubMed] [Google Scholar]
- 7.Miller DK, et al. Identification and isolation of a membrane protein necessary for leukotriene production. Nature. 1990;343:278–281. doi: 10.1038/343278a0. [DOI] [PubMed] [Google Scholar]
- 8.Dixon RAF, et al. Requirement of a 5-lipoxygenase-activating protein for leukotriene synthesis. Nature. 1990;343:282–284. doi: 10.1038/343282a0. [DOI] [PubMed] [Google Scholar]
- 9.Woods J, et al. 5-lipoxygenase and 5-lipoxygenase-activating protein are localized in the nuclear envelope of activated human leukocytes. J Exp Med. 1993;178:1935–1946. doi: 10.1084/jem.178.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christmas P, Webber BM, McKee M, Brown D, Soberman RJ. Membrane localization and topology of leukotriene C4 synthase. J Biol Chem. 2002;277:28902–28908. doi: 10.1074/jbc.M203074200. [DOI] [PubMed] [Google Scholar]
- 11.Wong W, Scott JD. AKAP signalling complexes: Focal points in space and time. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto T, Amnon S, Scherer PE, Lisanti MP. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- 13.DiPersio JF, Billing P, Williams R, Gasson JC. Human granulocyte-macrophage colony-stimulating factor and other cytokines prime human neutrophils for enhanced arachidonic acid release and leukotriene B4 synthesis. J Immunol. 1988;140:4315–4322. [PubMed] [Google Scholar]
- 14.Takafuji S, Bischoff SC, De Weck AL, Dahinden CA. IL-3 and IL-5 prime normal human eosinophils to produce leukotriene C4 in response to soluble agonists. J Immunol. 1991;147:3855–3861. [PubMed] [Google Scholar]
- 15.Surette ME, Dallaire N, Jean N, Picard S, Borgeat P. Mechanisms of the priming effect of lipopolysaccharides on the biosynthesis of leukotriene B4 in chemotactic peptide-stimulated human neutrophils. FASEB J. 1998;12:1521–1531. doi: 10.1096/fasebj.12.14.1521. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson AD, et al. Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science. 2007;317:510–512. doi: 10.1126/science.1144346. [DOI] [PubMed] [Google Scholar]
- 17.Yamashita M, et al. Inhibition by troglitazone of the antigen-induced production of leukotrienes in immunoglobulin E-sensitized RBL-2H3 cells. Br J Pharmacol. 2000;129:367–373. doi: 10.1038/sj.bjp.0703044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming TJ, et al. Negative regulation of FcεR1-mediated degranualtion by CD81. J Exp Med. 1997;186:1307–1314. doi: 10.1084/jem.186.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandal AK, et al. The membrane organization of leukotriene synthesis. Proc Natl Acad Sci USA. 2004;101:6587–6592. doi: 10.1073/pnas.0308523101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M, et al. Neutrophil-derived leukotriene B4 is required for inflammatory arthritis. J Exp Med. 2006;203:837–842. doi: 10.1084/jem.20052371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nigrovic PA, et al. Mast cells contribute to initiation of auto antibody-mediated arthritis via IL-1. Proc Natl Acad Sci USA. 104:2325–2330. doi: 10.1073/pnas.0610852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones PB, et al. Two post-processing techniques for the elimination of background autofluorescence for FLIM. J. Biomed Optics. 2008;13:-14008–8. doi: 10.1117/1.2837169. [DOI] [PubMed] [Google Scholar]
- 23.Mancini JA, et al. 5-lipoxygenase-activating protein is an arachidonic acid binding protein. FEBS Lett. 1993;318:277–281. doi: 10.1016/0014-5793(93)80528-3. [DOI] [PubMed] [Google Scholar]
- 24.Ago H, et al. Crystal structure of a human membrane protein involved in cysteinyl leukotriene biosynthesis. Nature. 2007;448:609–612. doi: 10.1038/nature05936. [DOI] [PubMed] [Google Scholar]
- 25.Molina DM, et al. Structural basis for synthesis of inflammatory mediators by human leukotriene C4 synthase. Nature. 2007;448:613–616. doi: 10.1038/nature06009. [DOI] [PubMed] [Google Scholar]
- 26.Sieber JJ, et al. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–1076. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- 27.Luo M, Jones SM, Peters-Golden M, Brock TG. Nuclear localization of 5-lipoxygenase as a determinant of leukotriene B4 synthetic capacity. Proc Natl Acad Sci USA. 2003;100:12165–12170. doi: 10.1073/pnas.2133253100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tschopp J, Martinon F, Burn K. Nalp3's: A novel protein family involved in inflammation. Nat Rev Mol Cell Biol. 2003;4:95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- 29.Kinoshita A, et al. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J Cell Sci. 2003;116:3339–3346. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- 30.Lennon NJ, et al. Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J Biol Chem. 2001;278:50466–50473. doi: 10.1074/jbc.M307247200. [DOI] [PubMed] [Google Scholar]
- 31.Miller DK, et al. Development of a high-capacity homogeneous fluorescent assay for the measurement of leukotriene B4. Anal Biochem. 2006;349:129–135. doi: 10.1016/j.ab.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 32.Christmas P, et al. Cytochrome P-450 4F18 is the leukotriene B4 ω-1/ω-2 hydroxylase in mouse polymorphonuclear leukocytes: Identification as the functional orthologue of human polymorphonuclear leukocyte cyp4f3a in the down-regulation of responses to LTB4. J Biol Chem. 2006;281:7189–7196. doi: 10.1074/jbc.M513101200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.