Abstract
The α (immediate early) protein ICP0 of herpes simplex virus 1 enhances the expression of genes introduced by infection or transfection. Early in infection it performs two key functions: It blocks the silencing of the viral DNA by cellular proteins and it blocks the IFN stimulated host response to infection. Between 5 and 9 h after infection, ICP0 is translocated to the cytoplasm but remains dynamically associated with proteasomes. In this report we show that in permissive cells that are poor expressors of transfected genes (HEp-2, U2OS, etc.), ICP0 is retained in the nucleus if the cells had been transfected with DNA and then infected. The retention is DNA dose- and size-dependent but not DNA type-dependent. Retention of ICP0 is also a consequence of infection with wild-type virus concomitant with exposure of cells to sodium butyrate. ICP0 is not retained in transfected/infected cells that efficiently express transfected genes (HEK293, rabbit skin cells). The retention of ICP0 in the nucleus is concordant with failure to degrade PML and disperse ND10 structures, and delays in the transition to post α genes expression, translocation of components of the CoREST/REST/HDAC1 complex and histone relocation in the infected cell. The data suggest that (i) retention of ICP0 is linked to its function to remodel acetylated DNA but not DNA in heterochromatin. This function is independent of response elements embedded in the DNA and (ii) transfection-resistant cells do take up DNA but process it differently than cells that readily express transfected genes.
Keywords: DNA transfection, ND10 bodies, silencing
This report centers on ICP0, an α (immediate early) protein specified by herpes simplex virus 1 (HSV-1). ICP0 interacts with many diverse cellular and viral proteins and acts as a promiscuous transactivator of genes introduced into cells by transfection or infection (1–3). Its two most prominent functions are to render the infected cells resistant to antiviral activity of interferons and to block the silencing of viral DNA (4–7). At the molecular level, it acts as an E3 ubiquitin ligase to degrade PML and SP100. In consequence the components of ND10 nuclear bodies are dispersed (8, 9). In addition ICP0 blocks the silencing of viral DNA through dissociation of the HDAC1 or 2 from the CoREST/REST/LSD1 complex. The components of the complex are then translocated to the cytoplasm (5).
The genesis of current studies stemmed from two sets of observations. Briefly, ICP0 is translocated to the cytoplasm between 5 and 9 h after infection (10). The translocation of ICP0 to the cytoplasm involves three distinct events. Thus, ICP0 interacts with cyclin D3. Overexpression of cyclin D3 accelerates the translocation whereas substitution of D199 with alanine abolishes the interaction with cyclin D3 and results in the retention of ICP0 in the nucleus (11). The mutant carrying the D199A substitution replicates efficiently. Translocation of ICP0 also requires a late (γ1) gene expression inasmuch as inhibitors of viral DNA synthesis delay the translocation of ICP0. The ICP0 interacts dynamically with proteasomal components, inasmuch as exposure of infected cells to proteasomal inhibitor MG132 before 5 h of infection precludes the translocation to the cytoplasm, whereas exposure of infected cells to MG132 at 9 h after infection results in quantitative relocation of ICP0 from the cytoplasm to the nucleus (10). The second set of observations centers on complementation studies of HSV-1 mutants. In many instances it is convenient to transfect cells with a specific gene and then infect cells with a mutant virus to determine whether the transgene complements the mutant. Cells differ with respect to their ability to express the transfected genes and hence cell lines are frequently classified into “transfectable” (e.g., HEK293, rabbit skin cells) and non transfectable (human embryonic lung fibroblasts, HEp-2, U2OS, etc.). We report that in the “nontransfectable” cells transfected with DNA and then infected ICP0 was retained in nuclei in a manner dependent on DNA dose and size but not on the type of DNA. For the most part, ICP0 was initially trapped in ND10 structures and later filed the nucleus, but was not exported. The retention of ICP0 in ND10 structures was of particular interest in light of reports that DNA introduced into cells by transfection or infection colocalizes with or adjacent to ND10 structures (12, 13). The consequences of retention of ICP0 were failure or delay in degradation of PML, dispersal of ND10, dissociation and export of components of the HDAC1/CoREST/REST/LSD1 complex to the cytoplasm, impairment of post α genes expression and cell chromatin redistribution in the infected cells. ICP0 was not retained in nuclei of cells that effectively express transgenes (e.g., HEK293 or rabbit skin cells). Thus, consequences of retention of ICP0 in the nucleus by the mutant carrying the D199A substitution and those of cells transfected with DNA and then infected are distinctly different. We also noted that ICP0 was retained in cells exposed to sodium butyrate, an inhibitor of histone deacetylation.
The results presented here are consistent with three conclusions: (i) a key function of ICP0 is to remodel DNA introduced into cells by infection or transfection, (ii) the duration of its sojourn in the nucleus depends on the quantity of DNA suggesting that translocation occurs once its mission is accomplished, and (iii) the lack of expression of the transgenes in “nontransfectable cells” may be related to inhibition of processing of the DNA rather than failure to be taken up into the cells.
Results
Transfected DNA Causes Nuclear Retention of ICP0.
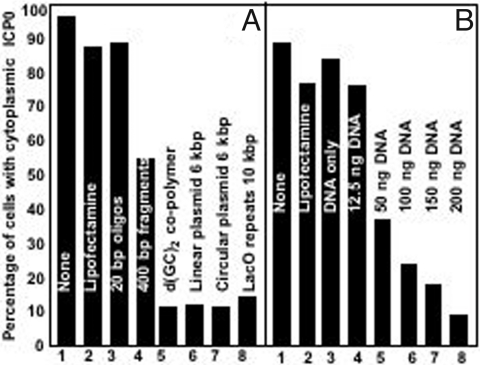
In the first series of experiments, HEp-2 cells were transfected with the aid of lipofectamine with 300 ng per well of 20-mer oligonuceotides, 400-bp fragments, 1-Kb copolymers, linear 6-Kb plasmids, or circular 6-Kb plasmids. The cells were fixed and examined 9h after infection. The results (Fig. 1A) indicate that ICP0 was translocated to the cytoplasm in untreated cells, in cells treated with lipofectamine only, and in cells transfected with 20-mer oligonucleotides with the aid of lipofectamine. ICP0 was also translocated to the cytoplasm in ≈50% of cells transfected with the 400-bp fragments. More than 85% of cells infected-transfected with larger size DNAs retained ICP0 in nuclei.
Fig. 1.
ICP0 is retained in the nuclei of infected cells that had been exposed to DNA and complete transfection mixture containing lipofectamine, and depends on the size, dose and not on the nature of the DNA. (A) HEp-2 cells were infected 24 h after either mock treatment (None), or exposure to Lipofectamine/Plus reagents (Lipofectamine), or transfection with lipofectamine and 300 ng of DNA per well. The DNAs varied with respect to size, presence of promoters or reading frames as detailed in Materials and Methods indicated above the bars in the figure. The cells were fixed at 9 h after infection, and reacted with rabbit polyclonal antibody to ICP0 as described in Materials and Methods. The images were captured with the same settings of a Zeiss confocal microscope. Approximately 300 cells from sequential fields were counted to determine the percentage of infected cells with cytoplasmic ICP0. (B) HEp-2 cells as above were transfected with increasing concentration of pcDNA3.1Zeo (+) (12.5, 50, 100, 150, or 200 ng per well). Controls were as in A. The cells were fixed at 9 h after infection and processed as in A. Cell counts were done as above.
Next, we transfected HEp-2 cells with DNA in amounts from 50 to 200 ng per well. As shown in Fig. 1B, ICP0 was translocated into the cytoplasm in untreated infected cells, or cells treated with lipofectamine only, DNA (300 ng per well) only, and in cells transfected with 12.5 ng DNA per well. The fraction of cells exhibiting ICP0 in the cytoplasm progressively decreased in cells transfected with increasing amount of DNA.
We conclude from these experiments that ICP0 is retained in the nucleus in HEp-2 cells transfected with DNA equal or >400 bp in size. Similar results were obtained when we applied different methods for transfection (e.g., electroporation or the calcium based protocols, data not shown).
Retention of the ICP0 in the Nucleus by Transfected DNA is Cell-Type-Dependent.
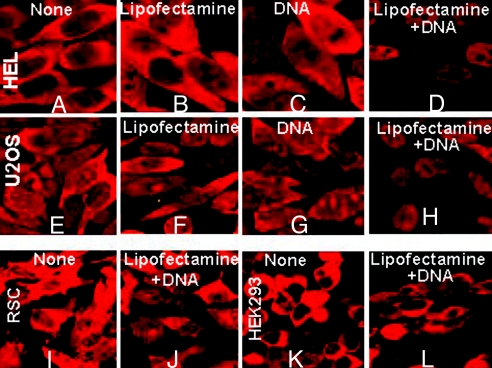
To determine whether the retention of ICP0 was cell-type-dependent, HEL, U2OS, RSC, or HEK-293 cells were untreated, exposed to Lipofectamine, DNA or transfected with 300 ng of DNA per well with the aid of Lipofectamine. The cells fixed, reacted with antibody and examined at 9h after infection. Fig. 2 A–D show that retention of ICP0 in HEL cells infected with wild-type virus is similar to that in Fig. 1. Thus, ICP0 was translocated in untreated cells (Fig. 2A), cells exposed to lipofectamine (Fig. 2B) or DNA only (Fig. 2C), but was retained in the nucleus in cells transfected with DNA and lipofectamine (Fig. 2D). Fig. 2 E–H show that in U2OS cells transfected with lipofectamine and DNA, ICP0 was retained in the nucleus. In contrast, in rabbit skin (Fig. 2 I and J) and HEK293 cells (Fig. 2 K and L), transfection of DNA with the aid of Lipofectamine did not impede the translocation of ICP0.
Fig. 2.
Retention of ICP0 in transfected-infected HEL or U2OS cells but not in rabbit skin (RSC) or HEK293 cells. HEL (A–D), U2OS (E–H), RSC (I and J) or HEK293 cells (K and L) were mock-treated (None), or treated with the Lipofectamine/Plus reagents (Lipofectamine), or only with 300 ng of pcDNA3.1Zeo (+) (DNA), or transfected with 300 ng per well pcDNA3.1Zeo + Lipofectamine + DNA, as detailed in Materials and Methods. At 24 h after transfection the cells were exposed to 10 PFU/cell. The cells were fixed at 9 h after infection, and stained with the rabbit polyclonal antibody to ICP0. The images were captured as above.
It is of interest to note that expression of transfected genes is efficient in rabbit skin and HEK-293 cells and relatively poor in HEp-2, HEL, or U2OS cells.
Effect of Transfected DNA on the Expression of ICP4, Transition from α to β Gene Expression, and Translocation of CoREST into the Cytoplasm.
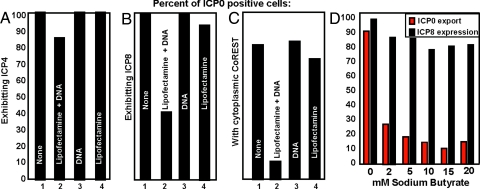
HEp-2 cells were treated as above. At 10 h after infection the cells were fixed and reacted with antibody to ICP0 and ICP4 (Fig. 3A), ICP0 and ICP8 (Fig. 3B), or ICP0 and CoREST (Fig. 3C). The results were as follows:
Fig. 3.
The consequences of transfection of DNA before infection with HSV-1 (F). (A) Transfected DNA has little or no effect on the expression of ICP4, a representative α protein. HEp-2 cells were either mock treated (None), or treated with the Lipofectamine/Plus reagents (Lipofectamine), or only with pcDNA3.1Zeo (+) (DNA), or transfected with 300 ng per well pcDNA3.1Zeo (+) (Lipofectamine + DNA). At 24 h after transfection the cells were exposed to 10 PFU/cell. The cells were fixed at 10 h after infection, and reacted with the rabbit polyclonal antibody to ICP0 and the mouse monoclonal antibody to ICP4. Cell counts were done as in Fig. 1. (B) Transfected DNA has a negative effect on post α genes expression. HEp-2 cells were transfected/infected and fixed as in A. After fixation the cells were reacted with rabbit polyclonal antibody to ICP0 and mouse monoclonal antibody to ICP8. (C) The nuclear translocation of CoRest is cells transfected with DNA and then infected with HSV-1 (F). In wild-type virus-infected cells, the CoREST/REST complex is translocated into the cytoplasm after infection (5). In independent studies, LSD-1, a protein tightly bound to CoREST, was also found to be translocated to the cytoplasm. In this experiment CoREST was selected as a surrogate marker for the translocation of all three proteins. The experimental procedures were as described in A except that the cells were fixed at 10 h after infection, and reacted with a rabbit polyclonal antibody to CoRest and the mouse monoclonal antibody to ICP0. (D) ICP0 is retained in the nuclei of cells treated with sodium butyrate and infected with HSV-1. Sodium butyrate was added to HEp-2 cells at the moment of infection (10 PFU/cell) at a final concentration of 0, 2, 5, 10, 15, or 20 mM. The cells were fixed at 9 h after infection and reacted with the rabbit polyclonal antibody to ICP0 and the mouse monoclonal antibody to ICP8. Approximately 300 cells from sequential fields were counted to determine the percentage of infected cells (ICP0 positive) that exhibit at the same time cytoplasmic ICP0 (red bars) and express ICP8 (black bars).
In control wells, 100% of cells exhibiting ICP0 also contained ICP4. In cells transfected with DNA with the aid of Lipofectamine, ICP4 was detected in 85% of the cells exhibiting ICP0. The results suggest that the DNA transfection did not significantly impede expression of ICP4.
In control wells nearly 100% of cells exhibiting ICP0 also accumulated ICP8. In contrast, in cells transfected with DNA and lipofectamine, <50% of the cells contained detectable amounts of ICP8 (Fig. 3B). In light of the time at which the cells were fixed (10 h after infection), the results indicate that the transition from α to β gene expression is impaired in transfected HEp-2 cells.
In controls, 70 to 80% of cells exhibiting ICP0 contained detectable CoREST protein in the cytoplasm. The number of ICP0 positive cells exhibiting detectable CoREST in the cytoplasm was slightly >10% in cultures transfected with DNA and lipofectamine (Fig. 3C). The results are consistent with the decrease in the number of ICP0 positive cells containing detectable amounts of ICP8.
Exposure of Cells to Sodium Butyrate Prevents the Translocation of ICP0 to the Cytoplasm.
Sodium butyrate inhibits histone deacetylases and renders DNA available for transcription. The posed question is whether the increase in the amount of template available for transcription by treatment with sodium butyrate would in effect, present a similar situation to that observed in cells transfected with DNA. HEp-2 cells exposed to the wild-type virus were treated with increasing concentrations of sodium butyrate and the distribution of ICP0 was monitored at 9 h after infection by immunofluorescence analysis. The results (Fig. 3D) were as follows: In untreated cultures ≈87% of the cells exhibited cytoplasmic localization of ICP0 by 9 h after infection (red bars). In contrast, only 27% of the infected cells exhibited translocation of ICP0 in the cytoplasm in cultures that had been treated with 2 mM sodium butyrate. Furthermore, there was a progressive decrease in the fraction of cells showing translocation of ICP0 in cells exposed to higher concentrations of sodium butyrate. To exclude the possibility that the nuclear retention of ICP0 was because of drug related toxicity we also monitored the post-α genes expression by costaining the ICP0-positive cells with the ICP8 antibody. The results shown on Fig. 3D (black bars) indicate that sodium butyrate had only a minor effect on the expression of ICP8 and therefore the nuclear retention of ICP0 is not due to a toxic effect of the drug.
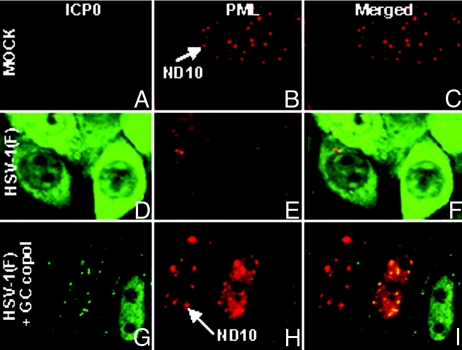
Degradation of PML and Dispersal of the Enlarged ND10 Structures Is Impaired in Infected Cells Transfected with DNA.
HEp-2 cells were infected 24 h after transfection with 300 ng of GC copolymer per well. The cells were fixed 10 h after infection and reacted with antibody to PML and ICP0. Representative images (Fig. 4) illustrate two key observations. Foremost, the ND10 structures were significantly larger in cells transfected with DNA and infected with wild-type virus (compare Fig. 4 H and B). Second, in infected cell that have not been transfected with DNA, ICP0 was largely in the cytoplasm and ND10 structures were dispersed (Fig. 4 D–F), whereas in cultures that have been transfected and infected some cells contain the intact, enlarged ND10 with relatively small amounts of ICP0 or reduced amounts of PML and large amount of ICP0 retained in the nucleus (Fig. 4 G–I). These results suggest that in transfected cells ICP0 accumulates at a slower rate than in infected untreated cells and that the degradation of PML and dispersal of ND10 are related to the accumulation of ICP0.
Fig. 4.
Delay in the degradation of PML in DNA transfected/infected cells. HEp-2 cells were either mock infected (A–C), exposed to 10 PFU/cell (D–F), or transfected for 24 h with 300 ng per well of the GC copolymer prior the exposure to virus (G–I). The cells were fixed at 10 h after infection and reacted with the mouse monoclonal antibody to ICP0 and the rabbit polyclonal antibody to PML. All of the images were captured with the same settings of a Zeiss confocal microscope. Arrows identify ND10 structure.
Distribution of Unmodified and Methylated Histones in Infected Cells.
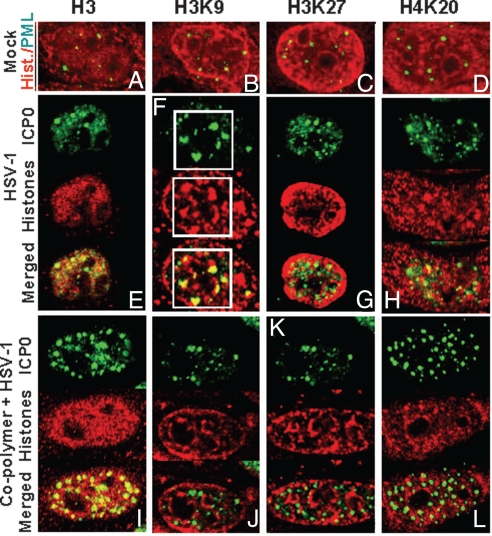
The objective of these studies was to determine whether transfected DNA alters the distribution of histones in infected cells. HEp-2 cells were either infected or transfected/infected, as described above, fixed at 4h after infection and reacted with antibodies to unmodified or modified histones H3 and H4 and either PML (mock) or ICP0 (infected cells). The results (Fig. 5) were as follows:
Examination of mock-infected cells failed to show obvious colocalization of ND10 structures with any of the histones tested (Fig. 5 A–D).
No obvious, colocalization of ICP0 and histones H3, H3K9, H3K27 or H4K20 in either cells that had been infected only or in cells that had been transfected with DNA and infected was observed.
Some infected cells reacted with ICP0 and H3K9 antibodies exhibited relatively large structures containing ICP0 juxtaposed to or partially overlapping structures containing H3K9 histone (Fig. 5F). These structures were significantly larger than ND10 bodies and may represent the initial formation of replication compartments in infected cells. These structures were not detected in transfected-infected cells (Fig. 5J).
Fig. 5.
The distribution of unmodified and methylated histones in infected and transfected/infected cells. HEp-2 cells were either mock infected, infected, or transfected/infected as in Fig. 4, except that the cells were fixed at 4h after infection. The mock infected cells were reacted with the mouse monoclonal antibody to PML and the rabbit polyclonal antibodies to histone H3, tri-methyl H3K9, tri-methyl H3K27, or tri-methyl H4K20. The infected or transfected/infected cells were reacted with the mouse monoclonal antibody to ICP0 and the histones antibodies, described above for mock. The images were captured as in Fig. 4.
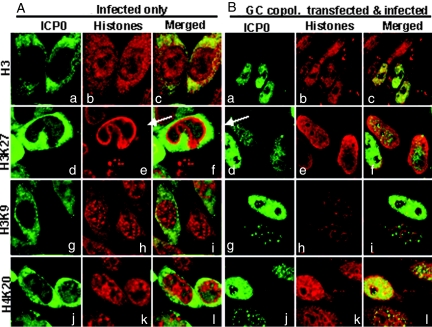
The second series of experiments was similar to those described above except that the cells were fixed at 9 h after infection. The results (Fig. 6) were as follows:
At 9 h after infection, ICP0 in untreated infected cells localized in the cytoplasm.
In untreated, infected cells all of the histones with the exception of H4K20 (Fig. 6Ak) were present in both nucleus and cytoplasm. The antibody against H3 illuminated numerous small structures in the cytoplasm, outlined the nuclear periphery, and reacted with granular structures in the nucleus similar in appearance to those present in the cytoplasm (Fig. 6Ab).
The pattern of proteins reacted with the anti-H4K20 in the nucleus (Fig. 6Ak) was similar to those of proteins reactive with the H3 antibody (Fig. 6Ab).
The antibody to H3K9 reacted with proteins that formed small granules, or were diffused in the cytoplasm and with much larger granules that were scattered throughout the nucleus and as well as outlined the nuclear membrane (Fig. 6Ah).
The antibody to H3K27 histone reacted very strongly with a dense mass that outlined the periphery of the nucleus (Fig. 6Ae). The confocal images of the cells reacted with both ICP0 and H3K27 suggests that H3K27 histone outline the nuclear side of the nuclear membrane (Fig. 6Af).
Fig. 6.
Transfected DNA blocks histones relocalization and chromatin margination induced upon virus infection. HEp-2 cells were either infected (A), or transfected with the GC copolymer and then infected with 10 PFU/cell (B), as in Fig. 5. The cells were fixed at 9 h after infection and reacted with the mouse monoclonal antibody to ICP0 and the rabbit polyclonal antibodies to histone H3 or to methylated forms of histone H3 and H4. The images were captured as in Fig. 4.
The distribution of histones in transfected-infected cells differed dramatically from that of infected, nontransfected cells. Specifically:
As expected, ICP0 was localized to the nucleus. The images examined did not reveal compelling evidence of colocalization of ICP0 and histones.
There was significantly less histone H3, H3K9, or H3K27 in the cytoplasm of infected-transfected cells than in infected cells only (compare Fig. 6Bb with Fig. 6Ab, Fig. 6Bh with Fig. 6Ah, and Fig. 6Be with Fig. 6Ae) (iii) The dense layer of H3K27 histones outlining the periphery of the infected cell nucleus was observed in transfected/infected cells in which ICP0 was present in abundant amounts (Fig. 6 Ae and Be, arrows). The H3K27 histone appeared to be more dispersed in cells in which ICP0 was less abundant.
Discussion
The salient feature of the results is that ICP0 is not translocated into the cytoplasm between 5 and 9 h after infection in cells transfected with DNA, as is the case in virtually all untreated cells infected with wild-type virus (10). The retention is transfected DNA size, dose and cell-type-dependent. Both, noncoding and coding DNAs cause retention of ICP0. Finally, the progression of viral replication can be impaired by the transfected DNA. Thus, for the same dose, 20-mers or 400-mers were less effective in retaining ICP0 than DNA 1 Kb in size. A significant finding that provides a clue to the events transpiring at early times in infection is that sodium butyrate, a HDAC inhibitor, also causes the retention of ICP0 in the nucleus even though at the same time it overcomes the silencing of HSV DNA in cells infected with a ΔICP0 mutant virus (15). The results reported here have significant implications regarding the function of ICP0.
ND10 structures appear to be centers of cellular transcriptional regulation. Reports of both positive and negative regulation have been attributed to PML and thus indirectly to ND10 (16). Several lines of evidence indicate that DNA introduced by infection or transfection aggregates at ND10 structures (12, 13). In this report we show that ND10 structures increase in size in cells transfected with DNA. The precise interaction of ND10 components with transfected DNA is unknown. Comparisons of the events in cells that readily express transfected genes with those that do not suggest that ND10 bodies act as keepers of repositories of DNA intended for repression. The inhibition of deacetylation caused by sodium butyrate and other inhibitors could be predicted to be potentially lethal to the cell. It is conceivable that the cell responds by repositioning newly acetylated DNAs to ND10 structures.
ND10 structures are also a destination of ICP0 immediately after its synthesis. Unambiguous evidence indicates that one function of ICP0 is to degrade PML and SP100. Degradation of PML results in the dispersal of ND10 structures (8, 17). A second function of ICP0 is to block silencing of viral DNA. This function includes dissociation of HDAC1 or 2 from CoREST/REST/LSD1 complex. In cells infected with a mutant virus in which ICP0 was replaced with a dominant negative CoREST, the silencing is blocked even though PML is not degraded and the ND10 structures remain intact. Hence the two functions are independent of each other although evidence supports the conclusion that they are executed in tandem (5).
The results presented in this report tie the retention of ICP0 in the nuclei to quantities of DNA transfected into cells. At times tested, 9 or 10 h after infection, in some cells ICP0 was still retained in small structures reminiscent of ND10 structures whereas in others ICP0 filled the nucleus but was nevertheless not exported to the cytoplasm. This result is identical with that observed in cells infected with a HSV-1 mutant in which the silencing function mapping close to the carboxyl terminus of ICP0 was impaired whereas the PML degradation function mapping more than 500 codons upstream was unaffected (5). The hypothesis we propose is that in the performance of its task to block silencing of DNA, ICP0 does not differentiate between the cellular and viral DNA accumulating at the ND10 bodies. The task confronting ICP0 in transfected/infected cells is far greater than that in cells that had been infected only. Because translocation of ICP0 to the cytoplasm requires a late viral gene function (10), the effective time of translocation is a surrogate measure of the time required to remodel viral DNA and enable initiation of viral replication. In transfected/infected cells, the delay translocation correlates with expression of ICP8 a protein essential for initiation of replication of viral DNA.
The key issue is what precisely is ICP0 remodeling. ICP0 interacts with CoREST and displaces HDAC1 or 2 from the CoREST/REST/LSD1 repressor complex. The role of ICP0 in this process is affirmed by the observation that a truncated CoREST incapable of binding the HDACs compensated the growth of a mutant virus lacking ICP0. HSV-1 contains a RE1 response element and hence it could be envisioned that displacement of HDAC1 and subsequent translocation of the repressor complex is sufficient to derepress viral DNA. This “cis” effect would not apply to transfected DNA because most DNAs tested lack REST binding sites. The hypothesis we propose is that the remodeling of DNA by ICP0 consists of two steps. The first is to block deacetylation of HSV-1 DNA by the displacement of HDAC1/2 from the CoRES/REST/LSD1 complex. The second step is to remodel or remove H3 inhibitory histones bound to the DNA to enable transcription. The effect of sodium butyrate is to augment the amount of acetylated DNA confronted by ICP0.
HSV DNA packaged in virions is linear; it contains nicks and gaps. On entry into nucleus the DNA must be repaired and circularize. ICP0 has been reported to affect the DNA repair machinery. The hypothesis that ICP0 is retained in the nucleus by the sheer amount of DNA that must be repaired and concatenated is inconsistent with the evidence that 300 ng of transfected 20-mer or 400-mer is less effective in retaining ICP0 in the nucleus than 300 ng of DNA consisting of 1 Kb copolymer of large size DNAs.
The studies on the localization of histone H3 and selected trimethylated histones were done for two reasons. Assuming that trimethylated histones bound to DNA were a target of ICP0, it was of interest to determine whether ICP0 colocalizes with any of the tested histones in cells transfected with DNA and then infected. In no instance was there unambiguous evidence that ICP0 colocalized with the histone H3 or trimethylated H3 or H4 histone in a defined structure. We did observe the juxtaposition of ICP0 and H3K9 trimethylated histone in cells that had been infected only. The structures formed by ICP0 resembled the initial stages of formation of replication compartments rather than the much smaller ND10 structures. It is conceivable that these H3K9 structures represented cellular DNA that had not yet been marginated.
The studies on localization of histones in infected cells yielded two significant observations. Foremost, the H3K27 histone is relocated relatively early in infection (4 h, compare Fig. 5 C and G). In mock infected cells the structures at the periphery of the nucleus were enriched in H3K27. At 4 h and especially at 9 h, the H3K27 histone was found almost exclusively in the periphery of the nucleus (Fig. 6Ae). One interpretation of the dense envelope of H3K27 histone at the periphery of the nucleus is that it represents cellular chromatin marginated during infection. The exclusive accumulation of H3K27 histones at the periphery of the nucleus is not a preeminent feature of cells that had been transfected and then infected (compare Fig. 6 Ae and Be). The other observation relevant to these studies is the accumulation of selected histones in the cytoplasm of cells that had been infected only, and to a much lesser degree in cells that had been transfected with DNA and then infected. Studies now in progress are designed to determine whether ICP0 plays a role in the relocation of histones to the cytoplasm after infection.
Transient gene expression of DNA introduced into cells by transfection is a powerful tool for studies of gene function and regulation. A fundamental problem of this tool is that the amounts of DNA required to elicit transgene expression are high and cell type dependent. Some cells appear to resist expression whereas others do not. The results presented in this report suggest that ND10 structures act as a filter to either block or enable transgene expression and that the limiting factor is the remodeling of the DNA rather than the take up of the DNA into cells.
Materials and Methods
Cells and Viruses.
The sources, properties, and propagation of HEp-2, rabbit skin cells, HEK293, U2OS and the telomerase transformed human embryonic lung (HEL) cells, and the properties of the limited passage HSV-1(F) strain were described elsewhere (14).
Transfections/Infections.
Cells grown in 4-well slides (Erie Scientific) were transfected when 60 to 70% confluent with quantities of DNA stated in results in mixtures with 1 μl of Lipofectamine and 1.5 μl of Plus reagents per well, as specified by the supplier (Invitrogen). At 3 h after transfection, the cells were rinsed extensively with DMEM supplemented with 10% FBS and further incubated for 24 h. The DNAs used in this study were as follows: The pcDNA 3.1Zeo (+) (Invitrogen) was purified with the Midi kit (Qiagen). The linear form of pcDNA 3.1Zeo (+) was obtained after digestion with EcoRV and purification from agarose. The 400-bp fragment from the neomycin ORF pEGFP-N3 (Clontech), but without initiation codon was digested with BspMI and extracted as above. The 20-base oligonucleotide corresponds to the sequence 5′-AAGCTTGATATCCTAGGGTA-3′ and was obtained from Integrated DNA technologies. The poly(deoxyguanylic-deoxycytidylic) acidic sodium salt, Poly(dG-dC)·Poly(dG-dC) double stranded copolymer of ≈1047 bp, was obtained from Sigma. To obtain the 10-Kb repeats of the Lac operator, the pLacO-SV2-neo plasmid was digested with the SalI/XhoI. In all experiments the cells were exposed to 10 PFU of HSV-1(F) per cell in medium 199 at 24 h after mock transfection or transfection with DNA (14).
Immunofluorescence.
The cells were fixed in 4% paraformaldehyde, at times indicated in the Results, permeabilized, blocked with 0.1% Triton X-100 in PBS in the presence of 10% horse serum and 1% BSA (PBS-TBH solution), and reacted with primary antibodies, diluted in PBS-TBH, as described earlier (14). The ICP4, ICP0, and ICP8 mouse monoclonal antibodies (Goodwin Institute for Cancer Research, Plantation, FL) were used at a 1:1,000 dilution. The ICP0 exon 2 rabbit polyclonal antibody was used at a 1:2,000 dilution (14). The CoREST rabbit polyclonal antibody (Upstate) was used at a 1:1,000 dilution. The rabbit polyclonal antibodies against histone H3, trimethyl-histone H3 (Lys-9), trimethyl-histone H4 (Lys-20) (Abcam), and trimethyl-histone H3 (Lys-27) (Upstate) were used at a 1:1,000 dilution. The rabbit polyclonal antibody and the mouse monoclonal antibody against PML (Santa Cruz) were used at a 1:500 dilution. The four-well cultures were then reacted with Alexa Fluor 594-conjugated goat anti-rabbit or Alexa Fluor 488-conjugated goat anti-mouse, diluted 1:1,000 in PBS-TBH. The cell images were captured with the aid of a Zeiss confocal microscope.
Acknowledgments.
These studies were aided by National Cancer Institute Grant R37 CA78766.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gelman IH, Silverstein S. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci USA. 1985;82:5265–5269. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Hare P, Hayward GS. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinlan MP, Knipe DM. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985;5:957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eidson KM, Hobbs WE, Manning BJ, Carlson P, DeLuca NA. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J Virol. 2002;76:2180–2191. doi: 10.1128/jvi.76.5.2180-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci USA. 2007;104:17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7- mediated activation of interferon-stimulated genes. J Virol. 2004;78:1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melroe GT, DeLuca NA, Knipe DM. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J Virol. 2004;78:8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee AV, Lopez P, Pandolfi PP, Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. 2003;77:7101–7105. doi: 10.1128/JVI.77.12.7101-7105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagglund R, Roizman B. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J Virol. 2004;78:2169–2178. doi: 10.1128/JVI.78.5.2169-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez P, Van SC, Roizman B. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J Virol. 2001;75:3832–3840. doi: 10.1128/JVI.75.8.3832-3840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van SC, Kawaguchi Y, Roizman B. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ching RW, Dellaire G, Eskiw CH, Bazett-Jones DP. PML bodies: A meeting place for genomic loci? J Cell Sci. 2005;118:847–854. doi: 10.1242/jcs.01700. [DOI] [PubMed] [Google Scholar]
- 13.Tsukamoto T, et al. Visualization of gene activity in living cells. Nat Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- 14.Kalamvoki M, Qu J, Roizman B. Translocation and colocalization of ICP4 and ICP0 in cells infected with herpes simplex virus 1 mutants lacking glycoprotein E, glycoprotein I, or the virion host shutoff product of the UL41 gene. J Virol. 2008;82:1701–1713. doi: 10.1128/JVI.02157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poon AP, Gu H, Roizman B. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc Natl Acad Sci USA. 2006;103:9993–9998. doi: 10.1073/pnas.0604142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- 17.Gu H, Roizman B. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci USA. 2003;100:8963–8968. doi: 10.1073/pnas.1533420100. [DOI] [PMC free article] [PubMed] [Google Scholar]