The availability of phosphorus and nitrogen are major limitations to plant growth, and as such our agricultural processes apply these nutrients at high concentrations to crop plants through fertilizer. Although fertilizer application has greatly enhanced food production it comes at a significant price: the chemical fixation of nitrogen depends on high levels of fossil fuels, making fertilizers a significant cost of food production and a major cause of greenhouse gas emissions from agriculture. A number of plants have entered beneficial interactions with microorganisms that facilitate the uptake of nitrogen and phosphorus from the soil. In this issue of PNAS Yano et al. (1) provide new insights into a novel genetic component in the plant that allows the establishment of these nutrient-capturing symbioses.
Plants enter a symbiosis with fungi of the taxonomic group Glomeromycota, which facilitates phosphate and mineral acquisition (2, 3). Fungal hyphae invade the plant root and form highly-branched intracellular intrusions into cortical cells, called arbuscules, where nutrient exchange occurs. A more limited set of plant species also forms interactions with rhizobial bacteria that facilitates nitrogen uptake. A novel organ, the nodule is formed on the roots of the host plants and bacteria are accommodated in plant membrane-bound compartments within the nodule cells where nitrogen fixation occurs (3). Legumes, peas and beans, are able to form both mycorrhizal and rhizobial symbioses. Genetic studies on nodulation in legumes revealed that several genes were required for both nodulation and mycorrhization, resulting in the identification of 7 distinct loci making up the so-called “common symbiosis signaling (Sym) pathway” (2, 4, 5). Thus far the Sym pathway consists of up to 2 putative cation channels (MtDMI1, LjCASTOR and LjPOLLUX), 1 leucine-rich repeat receptor-like kinase (MtDMI2, LjSymRK), 1 calcium- and calmodulin-dependent kinase (CCaMK), and 2 members of the nuclear pore complex (NUP85, NUP133). Today, we have reached a landmark in this pioneering age with the report by Yano et al. (1) of the cloning of the seventh gene in this pathway: CYCLOPS.
The Sym pathway is necessary for the recognition of the rhizobial signal Nod factor (6). Perception of Nod factor involves lysin motif-containing receptor-like kinases (LysM-RLK) that have a specific function in Nod factor recognition with no apparent role in the establishment of the mycorrhizal symbiosis (7–10). Downstream of these LysM-RLKs is the Sym pathway that is involved in the activation and perception of an oscillatory calcium signal: calcium spiking (reviewed in ref. 5). It is widely believed that CCaMK is responsible for decoding and transmitting the calcium signal. Specificity is possibly encoded in the frequency of the oscillation as calcium oscillations differ between Nod factor and mycorrhizal treatment (11). Downstream of CCaMK the pathway diverges with nodulation-specific transcription factors [NSP1, NSP2, and ERN1 (12–15)]. Analogous transcription factors in the mycorrhizal-specific branch have not yet been identified.
Mutations in CYCLOPS are less penetrant than mutants in other Sym pathway components.
Characterization of cyclops reveals that this gene functions at an equivalent position as CCaMK in the Sym pathway. CYCLOPS encodes a protein with a C-terminal coiled-coil motif and 2 nuclear localization signals that direct the protein to the nucleus, colocalizing with CCaMK. CYCLOPS is orthologous to the Medicago truncatula gene IPD3 that was identified in an interaction screen with M. truncatula CCaMK (16). CYCLOPS interacts with CCaMK in vivo, requiring a functional kinase domain, and CYCLOPS is phosphorylated in vitro by CCaMK. From this work it is difficult to predict the exact function of CYCLOPS: it may act downstream of CCaMK transmitting the calcium signal to the transcription factors, or it may act as a modulator of CCaMK that is itself modified by CCaMK.
CYCLOPS differs from other members of the Sym pathway in that cyclops mutants show more significant responses to mycorrhizal fungi and rhizobial bacteria: cyclops can initiate bacterial infection and nodule primordia and show greater levels of mycorrhizal infection than mutants in other Sym pathway components. These findings indicate that mutations in CYCLOPS are less penetrant than mutants in other Sym pathway components, although nup133 and nup85 also show some initiation of nodule primordia. A valuable assay has emerged for testing the relevance of nodulation genes for nodule organogenesis: a point mutation in the autophosphorylation site of CCaMK leads to a gain-of-function that induces the formation of nodules in the absence of rhizobia (17, 18). When cyclops was transformed with this construct spontaneous nodules were observed. From this Yano et al. (1) conclude that unlike other members of the Sym pathway CYCLOPS is not required for nodule organogenesis. However, while the CCaMK gain-of-function induced spontaneous nodules in cyclops, the numbers of nodulating plants and the number of nodules per plant was greatly reduced compared with wild type, which can be interpreted as a function for CYCLOPS during nodule organogenesis.
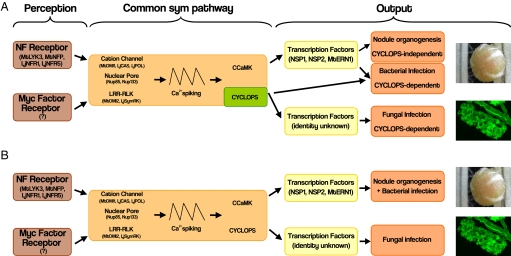
Yano et al. (1) suggest that CYCLOPS represents a branch point in the pathway: rhizobial infection is CYCLOPS-dependent, whereas nodule organogenesis is CYCLOPS-independent. CYCLOPS would then belong to an ancient pathway required for infection of fungi that was recruited later on for the infection of rhizobia (19). In this rationale, bacterial infection threads may have evolved from the prepenetration apparatus present in plant cells preparing for mycorrhizal invasion (19). An additional program encoding nodule organogenesis was then superimposed on this ancestral pathway and requires a pathway downstream of CCaMK but independent of CYCLOPS (Fig. 1A). However, this is an abrupt break from the current model as CYCLOPS would be the only Sym pathway component of seven not required for nodule primordium initiation. Also, it would imply a dual role for the downstream transcription factors NSP1, NSP2, and ERN1, all of which are involved in both nodule organogenesis and bacterial infection (Fig. 1A). A second interpretation of the spontaneous nodulation data are that the poor penetrance of cyclops mutants allows low levels of spontaneous nodulation, but that CYCLOPS is required for nodule organogenesis. The second model provides a much simpler model for the Sym pathway, where all components of the pathway are fulfilling equivalent functions in laying the developmental frameworks associated with bacterial infection, nodule organogenesis, and mycorrhizal invasion (Fig. 1B).
Fig. 1.
Two alternative models for the role of CYCLOPS in symbiosis signaling. (A) The model of the Sym pathway according to Yano et al. (1). Symbiotic signals are perceived by appropriate receptor complexes, activation of which will lead to a signaling cascade within the Sym pathway. Because cyclops mutants show spontaneous nodulation it is interpreted that CYCLOPS acts as a branching point in the pathway, where fungal and bacterial infection are regulated in a CYCLOPS-dependent manner and nodule organogenesis is regulated in a CYCLOPS-independent manner. This finding suggests that NSP1, NSP2, and ERN1 transcription factors have dual roles in both the bacterial infection branch and the nodule organogenesis branch as mutants in these genes are impaired in both. (B) An alternative model for the Sym pathway. Because cyclops mutants show greatly reduced levels of spontaneous nodulation it is interpreted that CYCLOPS is required for nodule organogenesis, but that the cyclops mutants have low penetrance. Low penetrance allows a more parsimonious Sym pathway in which CYCLOPS has analogous functions to all other members of the Sym pathway. The pathway bifurcates into nodulation-specific and mycorrhization-specific branches, each governed by their own set of transcription factors. NF, Nod factor. Pictures of a root nodule on L. japonicus and an arbuscule of Glomus versiforme in an M. truncatula root cortical cell stained with WGA Alexa Fluor 488 are shown at right.
This last decade has seen significant advances in the genetic dissection of the Sym pathway in legumes. The cloning of CYCLOPS represents the end of an era, being the final member of the Sym pathway to be defined in this early genetics work. This genetics has provided us with the anatomy of the Sym pathway. Our challenge now is to take this genetics into a coherent understanding of the mechanisms of symbiosis signaling.
Acknowledgments.
We thank Maria Harrison and Anne Heckmann for the photographs used in Fig. 1.
Footnotes
The authors declare no conflict of interest.
See companion article on page 20540.
References
- 1.Yano K, et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA. 2008;105:20540–20545. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parniske M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 3.Kistner C, Parniske M. Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci. 2002;7:511–518. doi: 10.1016/s1360-1385(02)02356-7. [DOI] [PubMed] [Google Scholar]
- 4.Kistner C, et al. Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell. 2005;17:2217–2229. doi: 10.1105/tpc.105.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oldroyd GE, Downie JA. Calcium, kinases, and nodulation signaling in legumes. Nat Rev Mol Cell Biol. 2004;5:566–576. doi: 10.1038/nrm1424. [DOI] [PubMed] [Google Scholar]
- 6.Lerouge P, et al. Symbiotic host specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 7.Limpens E, et al. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 8.Madsen EB, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 9.Radutoiu S, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 10.Arrighi JF, et al. The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosuta S, et al. Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc Natl Acad Sci USA. 2008;105:9823–9828. doi: 10.1073/pnas.0803499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heckmann AB, et al. Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a nonlegume. Plant Physiol. 2006;142:1739–1750. doi: 10.1104/pp.106.089508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalo P, et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- 14.Middleton PH, et al. An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell. 2007;19:1221–1234. doi: 10.1105/tpc.106.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smit P, et al. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- 16.Messinese E, et al. A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact. 2007;20:912–921. doi: 10.1094/MPMI-20-8-0912. [DOI] [PubMed] [Google Scholar]
- 17.Gleason C, et al. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature. 2006;441:1149–1152. doi: 10.1038/nature04812. [DOI] [PubMed] [Google Scholar]
- 18.Tirichine L, et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature. 2006;441:1153–1156. doi: 10.1038/nature04862. [DOI] [PubMed] [Google Scholar]
- 19.Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell. 2005;17:3489–3499. doi: 10.1105/tpc.105.035410. [DOI] [PMC free article] [PubMed] [Google Scholar]