Abstract
Individual neuronal cell types are defined by the expression of unique batteries of terminal differentiation genes. The elucidation of the cis-regulatory architecture of several distinct, single neuron type-specific gene batteries in Caenorhabditis elegans has revealed a strikingly simple cis-regulatory logic, in which small cis-regulatory motifs are activated in postmitotic neurons by autoregulating transcription factors (TFs). Loss of the TFs results in the loss of the identity of the individual neuron type. I propose to term these TFs “terminal selector genes” and their cognate cis-regulatory target sites “terminal selector motifs.” Terminal selector genes assign individual neuronal identities by directly controlling the expression of downstream, terminal differentiation genes and act in specific regulatory network configurations. The simplicity of the cis-regulatory logic on which the terminal selector gene concept is based may contribute to the evolvability of neuronal diversity.
Keywords: Caenorhabditis elegans, neuronal differentiation, gene regulation, transcription factors, cis-regulatory motifs
Approximately a century ago, Ramon y Cajal (1) discovered and described in detail the anatomical complexity of cell types in nervous systems. His work framed the fundamental question of how such cellular complexity is generated during development. The molecular correlates to the anatomical diversity of neuron types in a mature nervous system are neuron-type specific gene batteries. The composition of neuron type-specific gene batteries is highly combinatorial. That is, the differentiated properties of individual neuron types are usually not defined by the unique expression of specific gene products, but rather by the unique combination of genes that may each be more broadly expressed. The question of how neuronal diversity is generated, that is, how individual neurons execute distinct and unique differentiation programs, can therefore be essentially boiled down to the question of how such combinatorial gene expression mechanisms are encoded in the genome, on both the level of cis-regulatory control elements and the trans-acting factors that read these control elements.
One strategy to tackle this problem is to use a bottom-up approach in which one first defines the nuts-and-bolts gene batteries that define the specific anatomical and functional properties of a neuron and then dissects the cis-regulatory control elements of these genes. Such an approach is particularly feasible in the experimentally easily amenable and neuroanatomically well characterized model organism Caenorhabditis elegans, which contains a nervous system of 302 neurons that fall into 118 anatomically precisely defined neuron classes (the term “neuron class” is used here interchangeably with the term “neuron type,” which is more frequently used in vertebrates) (2). Gene expression data exist for many individual neuron types, and cis-regulatory control regions can be easily dissected through mutational analysis of reporter genes expressed in transgenic animals.
I will first briefly describe several examples in which the regulatory program of individual neuron types has been elucidated in C. elegans in substantial detail (Table 1) (3) and will then discuss common principles that may apply across phylogeny.
Table 1.
Examples of neuronal terminal selector genes in C. elegans
| Terminal selector gene | Neuron class | Selector motif | Sufficiency of motif |
|---|---|---|---|
| CHE-1 zinc finger transcription factor | ASE sensory neurons | ASE motif | Yes |
 |
|||
| TTX-3/CEH-10 LIM/Prd homeodomain dimer | AIY interneurons | AIY motif | Yes |
 |
|||
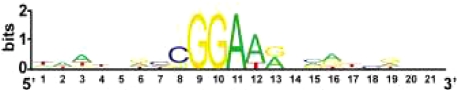
| AST-1 ETS-type transcription factor | All dopaminergic neurons | DA motif | Yes |
 |
|||
| MEC-3/UNC-86 LIM/POU homeodomain dimer | Mechanosensory neurons | 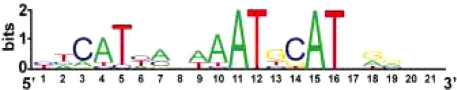 |
Yes |
| UNC-30 Prd-type homeodomain | GABAergic ventral cord motorneurons |  |
ND |
See text for references. ND, not determined.
ASE Gustatory Neurons
The ASE class of sensory neurons is composed of two bilaterally symmetric sensory neurons. Their transcriptome was determined by SAGE analysis (4). The analysis of the regulatory regions of ASE-expressed genes, performed by reporter gene analysis in transgenic worms, revealed a strikingly simple cis-regulatory logic. ASE-expressed genes contain in their proximity a small cis-regulatory motif of ≈12 bp, called the ASE motif (Table 1), which is absolutely required as well as sufficient by itself to drive gene expression in the ASE neurons (4). This motif is bound and activated by a zinc finger transcription factor (TF), CHE-1, that is exclusively expressed in the ASE sensory neurons. In the absence of the che-1 gene, ASE motif-containing genes fail to be activated, resulting in a complete loss of the identity of the mature ASE sensory neuron class (4–6). Panneuronal features are unaffected and no specific alternative fate appears to be executed upon loss of che-1, indicating that ASE is stuck in an indeterminate ground neuronal state. Moreover, CHE-1 is sufficient to induce ASE neuron fate if expressed in other sensory neurons (6).
AIY Interneurons
The identity of the AIY class of interneurons, which is composed of two bilaterally symmetric interneurons involved in processing sensory information and in behavioral plasticity (7, 8), is controlled by a similarly simple regulatory logic. A broad battery of genes that defines AIY identity shares a small cis-regulatory motif, called the AIY motif (Table 1), which, like the ASE motif, is required and sufficient to instruct gene expression in AIY (9). Like the ASE motif, the AIY motif commonly occurs in single copy in the vicinity of AIY-expressed genes. The AIY motif is synergistically activated by a dimer of two homeodomain proteins, the TTX-3 LIM homeodomain protein and the CEH-10 Paired-type homeodomain protein (9). Each of these factors is expressed in a few neurons, but their expression exclusively overlaps in the AIY neuron class, thereby making them, as is the case for CEH-1, a unique identifier for AIY identity (10). In either ttx-3 or ceh-10 null mutants, the identity of the mature AIY interneuron is completely lost. As in the case of che-1 and the ASE neuron class, the AIY interneurons are generated and still express pan-neuronal features in ttx-3 or ceh-10 mutants and have therefore “only” lost their neuron type-specific identity. Ectopic coexpression of ttx-3 and ceh-10 can transform neurons into an AIY-like fate, but, notably, only in specific cellular contexts (10).
Touch Sensory Neurons
In the most classic example of regulatory logic of neuronal specification in C. elegans, the identity of a group of six mechanosensory neurons is determined by the combinatorial activity of two homeobox genes mec-3 and unc-86 (11–13). A heterodimer of MEC-3 and UNC-86 acts synergistically to directly control the expression of mechanosensory-specific, terminal differentiation genes through a conserved cis-regulatory motif that is required and sufficient for touch neuron expression (Table 1) (14, 15). This cis-regulatory motif was subsequently found to be present in many other touch-neuron expressed genes as well (16).
Other Examples
The simple regulatory logic described above may apply broadly throughout the C. elegans nervous system. GABAergic motor neurons in the ventral nerve cord are specified by a Prd-type homeodomain protein, UNC-30, which also appears to directly control the expression of terminal differentiation markers through a small cis-regulatory motif (Table 1) (17, 18). The identity of all dopaminergic neurons in the worm are specified by an ETS domain TF that acts through a defined cis-regulatory motif, the DA motif, that is a required and sufficient determinant for gene expression in all DA neuron types (N. Flames and O.H., unpublished data). A more detailed phenotypic characterization of animals carrying mutations in other neuron type-specific TFs, e.g., ttx-1 (AFD neurons), odr-7 (AWA neurons), lim-4 (AWB neurons), and ceh-36 (AWC neurons) (3, 19), and the identification of their direct target gene batteries may reveal that those factors also broadly control neuron type-specific gene batteries through simple cis-regulatory motifs.
Terminal Selector Genes and Terminal Selector Motifs
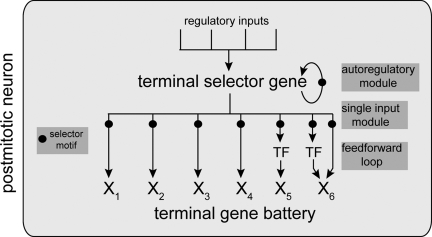
The examples described above and listed in Table 1 reveal a common underlying theme of neuronal specification in C. elegans. Neuron-type specific gene batteries contain a common “tag” in the form of a specific cis-regulatory motif, and this motif is controlled by a TF (either a single factor or a combinatorially acting TF complex) that specifies the identity of the neuron. I propose to term these TFs “terminal selector genes.” This term represents an extension of the selector gene concept introduced by Garcia-Bellido (20) >30 years ago, which describes genes that are required to determine the identity of a specific developmental field or organ. The original selector gene definition included genes that turned out to act relatively early in developmental pathways (21), and the definition included no specific mechanism of action. By terminal selector gene I mean to indicate that the gene is not only required to determine the identity of a specific neuron type but that it does so by directly regulating the expression of terminal differentiation genes [such terminal differentiation genes were coined “realizator” genes by Garcia-Bellido (20)]. A terminal differentiation gene battery defines the stable, unique properties of a specific, postmitotic neuron type, is maintained throughout the life of a neuron, and encompasses genes such as neurotransmitter receptors, ion channels, neurotransmitter synthesis pathway genes, structural protein, etc. Loss of the terminal selector gene results in a loss of the specific identity of the neuron type, without losing overall neuronal identity, though, as pan-neuronal features still remain expressed in terminal selector gene mutants. Gross morphology and position may also be unaffected by terminal selector genes (6, 22), underscoring their role in determining terminal identities rather than properties that are already predetermined in progenitor stages. Neuronal terminal selector genes act through what I propose to term “terminal selector motifs,” the simple cis-regulatory motifs described above (Table 1). Terminal selector genes are expressed throughout the life of the neuron and are continuously required for maintaining the differentiated state of a neuron. This maintenance function is mediated by autoregulation through terminal selector motifs that are present in the promoter of the terminal selector genes themselves (4, 9, 23) (Fig. 1).
Fig. 1.
Network configuration of terminal selector genes. Terminal selector TFs (acting either alone or in synergistic combination) activate downstream target genes directly via terminal selector motifs and also autoregulate their own expression via those motifs. Autoregulated, maintained expression of a terminal selector is critical to maintain the differentiated features of the cell. Downstream targets of terminal selectors (X) define differentiated properties of a neuron, such as neurotransmitter receptor, ion channels, adhesion proteins etc. Targets may also include TFs that regulate specific “subroutines.” The targets of these TFs may not harbor terminal selector motifs; therefore, not all genes expressed in a given neuron type must have a selector motif in their regulatory region (4). TFs that are induced by terminal selectors may also cooperate with terminal selector proteins in a feed-forward loop configuration to jointly control specific terminal genes (39, 44). Besides activating the expression of identity-defining genes it is also possible that terminal selectors may repress alternative fates through repressing the expression of selector genes for other cell types. This would explain neuronal identity switches observed upon removing putative terminal selector genes (26).
The terminal selector terminology only encompasses genes that directly bind to the regulatory regions of postmitotically expressed, terminal differentiation batteries, thereby setting it apart from the “neuron-type selector genes” coined by Jan and Jan (24) based on work in Drosophila or from the many TFs, in both invertebrates and vertebrates, that act at various upstream steps in neuronal commitment and determination and define neuronal progenitor identities (25).
The loss of a terminal selector gene results in a broad identity loss, as envisioned in the original Garcia-Bellido selector concept. Identity loss entails either the conversion into a nonrecognizable undifferentiated state, as appears to be the case for the examples listed in Table 1, or it may entail the switch to the identity of another neuron type. The mechanistic basis for the latter scenario may be that terminal selector genes may not only activate terminal differentiation genes, but also inhibit alternative fates by repressing the expression of other terminal selector genes. For example, vertebrate Tlx3 and Tlx1 genes may act in this manner (26).
The broad effect that terminal selector TFs have on the identity of a neuron sets them apart from the plethora of TFs that directly control specific subroutines of a neuron type and may themselves be targets of terminal selector TFs. For example, the ceh-23 homeodomain protein is a direct target of the TTX-3/CEH-10 terminal selector complex in the C. elegans AIY neuron class. Loss of ceh-23 affects the expression of a specific transmembrane receptor required for neuronal plasticity but does not affect any other known differentiated property of AIY (8, 10).
Do neuronal terminal selector gene exist in more complex nervous systems as well? The selector gene concept has already been applied to TFs acting in neuronal specification in flies and vertebrates (24, 26–29), yet it is not known whether in those cases the respective TF is truly a terminal selector gene that directly acts on terminal differentiation gene batteries. Terminal differentiation gene batteries that define individual neuron types are beginning to be identified in vertebrates by using transcriptome analysis from isolated cells (30). However, cis-regulatory control regions of such terminal differentiation gene batteries have not been analyzed on a scale that is required to draw conclusions about common regulatory features, such as the presence of terminal selector motifs. A striking exception is the vertebrate retina, which is composed of well characterized individual neuron types (31). Combinations of transcriptome analysis in wild-type and mutant backgrounds and bioinformatic analysis have revealed a terminal selector gene that appears to be very akin to the invertebrate examples discussed above, the Crx Paired-type homeodomain protein (32–34). Crx is a terminal selector gene that controls photoreceptor fate by directly regulating scores of terminal differentiation genes, acting also through a small cis-regulatory motif. Crx may cooperate in its terminal selector gene function with the closely related Prd-homeobox gene Otx2 (35, 36). The Crx/Otx2 terminal selector gene function appears to be conserved in flies as well (37, 38). The mouse homeobox genes Tlx3 and Tlx1, which determine postmitotic neuronal identity in the spinal cord (26), are among the many other known vertebrate TFs that are candidates to classify as terminal selector genes as well; identification of their direct target genes and a more extensive mutant phenotypic characterization will demonstrate whether these genes indeed fall into this category.
Integration of Terminal Selector Genes into Regulatory Networks
Gene regulatory networks have increasingly become recognized as an assembly of smaller network motifs, which endow these systems with specific functional properties (39, 40). Terminal selector gene function is characterized by the occurrence of three different network motifs (Fig. 1): (i) positive autoregulatory motifs, i.e., terminal selector genes regulate their own expression (4, 22, 23); (ii) single input modules (SIMs), which indicate the coregulation of the terminal gene battery; and (iii) feed-forward loop motifs in which a terminal selector gene controls the expression of another TF and then cooperates with this TF to regulate terminal differentiation genes. Examples include the above-mentioned vertebrate Crx TF that controls expression of the Nrl TF, which jointly control the expression of target genes in rod photoreceptors (32). Other examples are the C. elegans terminal selector gene complex TTX-3/CEH-10, which regulates the homeodomain protein CEH-23 to then jointly regulate the sra-11 downstream target gene (10) or the CHE-1 terminal selector that directly controls expression of the CEH-36 Otx-type homeodomain protein to then jointly regulate with CEH-36 the expression of the target gene gcy-7.
Feed-forward motifs and the implicit existence of additional layers of regulatory control downstream of terminal selector genes define another property of terminal selector genes. Neuron classes, whose identity is controlled by specific terminal selector genes, can often be subdivided into specific subclasses/subtypes. These subclasses essentially represent a variation of a theme, as illustrated by the example of vertebrate photoreceptors and the ASE gustatory neurons in C. elegans. Vertebrate photoreceptors, whose identity is controlled by the terminal selector Crx, can be subdivided into two subclasses, cone and rod cells. The identity of the rod subclass is controlled by a direct target of Crx, the Nrl basic leucine zipper protein (41). Crx and Nrl then cooperate in a feed-forward configuration to regulate rod-specific genes (32). Nrl also up-regulates NR2E2, a nuclear receptor TF that represses a subset of cone genes in rods (42, 43). Another example is the C. elegans ASE gustatory neuron class that can be subdivided into two distinct subclasses, the left ASE neuron (ASEL) and the right ASE (ASER) neuron, which are distinguishable by the expression of a different set of putative chemoreceptors (44). The C. elegans terminal selector gene CHE-1 controls overall ASE fate, but also directly controls the expression of a set of regulatory factors that distinguish ASEL from ASER and then cooperates with these factors to directly regulate downstream target genes specific for ASEL or ASER (44).
Taken together, terminal selector genes directly control the expression of the core set of identity-defining nuts-and-bolts terminal differentiation (realizator) genes, but they also control downstream regulatory events that further diversify individual neuron types.
Parallel Regulatory Programs
The genetic removal of neuronal terminal selector genes reveals the existence of regulatory programs that act in parallel to terminal selector genes and are therefore unaffected by the loss of the terminal selector. For example, neurons lacking terminal selector genes shown in Table 1 still express panneuronal features and, in the case of sensory neuron terminal selectors, may still express pan-sensory features (4, 9, 14). The latter features are controlled by the phylogenetically conserved RFX-type TF daf-19, which globally controls sensory neuronwide subroutines, such as the expression of genes that build sensory cilia (45). Pan-neuronal features may be controlled by a common trans-acting factor as suggested by the cis-regulatory analysis of some pan-neuronal genes (46), but others may be controlled in a more piecemeal manner by as-yet-unknown factors (47). The existence of parallel regulatory programs illustrates that terminal selector genes act by determining the specific identity of neuron types, rather than by determining broad and generic features of all or large groups of neurons.
The Importance of Context.
The activity of neuronal terminal selector genes is modulated by cellular context. Terminal selector genes, such as those shown in Table 1, can act only in specific cellular contexts and apparently only at specific developmental time points, as revealed, for example, by misexpression studies (9, 14). Moreover, genes may act as terminal selector genes in one cell type, but may have no such function in other neuron types in which they are expressed. For example, the ast-1 gene is a terminal selector gene for dopaminergic neuron fate, but in some nondopaminergic neurons it may have much more restricted functions in controlling axon outgrowth, but not overall cell fate (N. Flames and O.H., unpublished data). Likely context determinants are the presence of other regulatory factors with which terminal selector TFs may directly interact. For example, the ttx-3 homeobox selector gene is expressed in five neuron classes, but acts as a selector gene in only the one neuron class in which its interaction partner, the ceh-10 homeobox gene, is present (10). Context dependency is also evident from the perspective of selector motifs. In all cases tested selector motifs are alone sufficient to instruct neuron type-specific expression if placed into the context of a reporter gene vector (Table 1) (4, 9, 14) (N. Flames and O.H., unpublished data). However, the small size of terminal selector motifs and henceforth their abundance in genomic sequences (4) demands the existence of an additional layer of regulatory control, of which nucleosome-dependent binding accessibility is one possibility.
Upstream of Terminal Selectors
Many layers of transcriptional control mechanism funnel into the determination of a neuron type (25), and I propose here that terminal selector genes represent the regulatory endpoint of these complex gene regulatory cascades (Fig. 1). As revealed by mutant analysis and the dissection of their cis-regulatory control regions, upstream inputs into C. elegans terminal selectors are complex and composed of multiple different cis-regulatory elements that may serve to sample the developmental history of the neuron (48, 49) (unpublished data). The combination of multiple outputs and multiple inputs results in an hourglass-shaped regulatory topology, which can be considered to be one of the defining feature of terminal selector genes (Fig. 1).
Terminal selector genes can also have functions upstream of their terminal differentiation function and thereby act repeatedly in a given lineage. For example, the unc-86 terminal selector gene, a POU homeobox gene, acts early in specific neuronal lineages to determine the overall identity of the several neuroblasts (13, 14). Within a subset of those lineages, UNC-86 then acts several divisions later, now with another homeodomain protein, MEC-3, as a terminal selector complex to control the terminal specification of one specific neuron type, touch sensory neurons (13, 14). This example also illustrates again the context dependency of terminal selector genes; in the context of several neuroblasts, unc-86 acts to control neuroblast identity (likely in conjunction with as-yet-unknown TFs), whereas terminally differentiating touch neurons provide the context in the form of the MEC-3 homeodomain protein with which UNC-86 heterodimerizes to control terminal touch neuron differentiation.
Evolvability
Gene regulation by terminal selector genes provides a framework to think about the evolution of neuronal diversity. The simplicity of terminal selector motifs may mean that individual genes can be rapidly recruited into or lost from a neuron type-specific gene battery through loss or gain of terminal selector motifs. Such plasticity may provide a rich playground for evolution. The small size of terminal selector motifs and their consequent abundance in genomes suggests that many terminal selector motifs are not active; whatever regulates this dormancy, such as the above-mentioned possible nucleosome-mediated motif accessibility, provides another regulatory layer for evolution to play on. The addition of a functional selector motif into the control region of a TF may also add a specific “subroutine” to an existing neuronal class-specific gene expression program. Another way to view terminal selector genes in an evolutionary context is that the recruitment of terminal selector genes into a novel cellular context, e.g., by the acquisition of a novel cis-regulatory input into the terminal selector gene locus may generate a dramatically, rather than incrementally distinct, novel neuronal cell type, defined by the set of genes originally expressed in the cell type plus an entirely novel gene battery; such a “hybrid” cell is then subjected to Darwinian selection. Taken together, simplicity in regulatory architecture may provide evolutionary plasticity.
Outlook
The morphologically and lineally completely mapped out nervous system of C. elegans and its amenability to genetic analysis facilitated the identification of terminal selector genes in C. elegans. Most neuron classes studied in depth in C. elegans have revealed a common regulatory principle that can be subsumed into the terminal selector gene concept. More neuron types need to be examined as it is well conceivable that cases may surface in which neuronal identity is determined in a more “piecemeal” manner. In contrast to C. elegans, the grouping of neurons into specific classes is a much harder task in vertebrates. In the most extensively charted vertebrate neuronal domain, the retina, neuron types can be relatively clearly defined (31) and evidence for terminal selector genes indeed appears to exist, as discussed above. The identification and functional analysis of vertebrate TFs that fulfill the most fundamental criterion for a terminal selector gene, i.e., the maintained expression throughout the life of a neuron, may aid in defining neuron classes and cognate terminal selector genes in other regions of the vertebrate brain. As the terminal selector function of a TF may be obscured by earlier functions of the gene, temporally controlled gene knockouts are required to put the terminal selector gene concept to a rigorous test in vertebrates. Finally, even though I have discussed the terminal selector gene concept in the context only of the nervous system, it may apply to diversify other cellular fates as well. For example, the homeodomain TF Pax5 may serve as a terminal selector gene for B cell fate determination in the immune system (50).
Acknowledgments.
I thank R. Mann, P. Sengupta, C. Cepko, C. Desplan, I. Greenwald, and Y. Jin for comments on the manuscript. My work is supported by the Howard Hughes Medical Institute and National Institutes of Health Grants R01NS039996-05 and R01NS050266-03.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Gene Networks in Animal Development and Evolution,” held February 15–16, 2008, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS web site at http://www.nasonline.org/SACKLER_Gene_Networks.
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Cajal Ry. Histologie du Système Nerveux de L'Homme et des Vertébrés. Paris: Maloine; 1911. [Google Scholar]
- 2.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc London Ser B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 3.Hobert O The C. elegans Research Community, editor. Specification of the nervous system. WormBook. 2005 doi: 10.1895/wormbook.1.12.1. doi/ 10.1895/wormbook.1.12.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etchberger JF, et al. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang S, Johnston RJ, Jr, Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 2003;17:2123–2137. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130:1215–1224. doi: 10.1242/dev.00341. [DOI] [PubMed] [Google Scholar]
- 7.Tsalik EL, Hobert O. Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. J Neurobiol. 2003;56:178–197. doi: 10.1002/neu.10245. [DOI] [PubMed] [Google Scholar]
- 8.Remy JJ, Hobert O. An interneuronal chemoreceptor required for olfactory imprinting in C. elegans. Science. 2005;309:787–790. doi: 10.1126/science.1114209. [DOI] [PubMed] [Google Scholar]
- 9.Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Altun-Gultekin Z, et al. A regulatory cascade of three homeobox genes, ceh-10, ttx-3, and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- 11.Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- 12.Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24:59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- 13.Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 14.Duggan A, Ma C, Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–4119. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- 15.Xue D, Finney M, Ruvkun G, Chalfie M. Regulation of the mec-3 gene by the C. elegans homeoproteins UNC-86 and MEC-3. EMBO J. 1992;11:4969–4979. doi: 10.1002/j.1460-2075.1992.tb05604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- 17.Jin Y, Hoskins R, Horvitz HR. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature. 1994;372:780–783. doi: 10.1038/372780a0. [DOI] [PubMed] [Google Scholar]
- 18.Cinar H, Keles S, Jin Y. Expression profiling of GABAergic motor neurons in Caenorhabditis elegans. Curr Biol. 2005;15:340–346. doi: 10.1016/j.cub.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 19.Lanjuin A, Sengupta P. Specification of chemosensory neuron subtype identities in Caenorhabditis elegans. Curr Opin Neurobiol. 2004;14:22–30. doi: 10.1016/j.conb.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Bellido A. Genetic control of wing disc development in Drosophila. Ciba Found Symp. 1975;29:161–182. doi: 10.1002/9780470720110.ch8. [DOI] [PubMed] [Google Scholar]
- 21.Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12:592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- 22.Hobert O, et al. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 23.Way JC, Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 1989;3:1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- 24.Jan YN, Jan LY. Neuronal cell fate specification in Drosophila. Curr Opin Neurobiol. 1994;4:8–13. doi: 10.1016/0959-4388(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 25.Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 26.Cheng L, et al. Tlx3 and Tlx1 are postmitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- 27.Mangale VS, et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sieber MA, et al. Lbx1 acts as a selector gene in the fate determination of somatosensory and viscerosensory relay neurons in the hindbrain. J Neurosci. 2007;27:4902–4909. doi: 10.1523/JNEUROSCI.0717-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakatani T, Minaki Y, Kumai M, Ono Y. Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development. 2007;134:2783–2793. doi: 10.1242/dev.02870. [DOI] [PubMed] [Google Scholar]
- 30.Nelson SB, Hempel C, Sugino K. Probing the transcriptome of neuronal cell types. Curr Opin Neurobiol. 2006;16:571–576. doi: 10.1016/j.conb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Masland RH. Neuronal cell types. Curr Biol. 2004;14:R497–R500. doi: 10.1016/j.cub.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 32.Hsiau TH, et al. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS ONE. 2007;2:e643. doi: 10.1371/journal.pone.0000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, et al. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 34.Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 35.Nishida A, et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 36.Koike C, et al. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol. 2007;27:8318–8329. doi: 10.1128/MCB.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranade SS, et al. Analysis of the Otd-dependent transcriptome supports the evolutionary conservation of CRX/OTX/OTD functions in flies and vertebrates. Dev Biol. 2008;315:521–534. doi: 10.1016/j.ydbio.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tahayato A, et al. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev Cell. 2003;5:391–402. doi: 10.1016/s1534-5807(03)00239-9. [DOI] [PubMed] [Google Scholar]
- 39.Alon U. Network motifs: Theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 40.Davidson EH. Genomic Regulatory Systems. San Diego: Academic; 2001. [Google Scholar]
- 41.Mears AJ, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 42.Corbo JC, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005;1:e11. doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hobert O. Architecture of a microRNA-controlled gene regulatory network that diversifies neuronal cell fates. Cold Spring Harb Symp Quant Biol. 2006;71:181–188. doi: 10.1101/sqb.2006.71.006. [DOI] [PubMed] [Google Scholar]
- 45.Swoboda P, Adler HT, Thomas JH. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- 46.Ruvinsky I, Ohler U, Burge CB, Ruvkun G. Detection of broadly expressed neuronal genes in C. elegans. Dev Biol. 2007;302:617–626. doi: 10.1016/j.ydbio.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 47.Hwang SB, Lee J. Neuron cell type-specific SNAP-25 expression driven by multiple regulatory elements in the nematode Caenorhabditis elegans. J Mol Biol. 2003;333:237–247. doi: 10.1016/j.jmb.2003.08.055. [DOI] [PubMed] [Google Scholar]
- 48.Mitani S, Du H, Hall DH, Driscoll M, Chalfie M. Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans. Development. 1993;119:773–783. doi: 10.1242/dev.119.3.773. [DOI] [PubMed] [Google Scholar]
- 49.Baumeister R, Liu Y, Ruvkun G. Lineage-specific regulators couple cell lineage asymmetry to the transcription of the Caenorhabditis elegans POU gene unc-86 during neurogenesis. Genes Dev. 1996;10:1395–1410. doi: 10.1101/gad.10.11.1395. [DOI] [PubMed] [Google Scholar]
- 50.Hagman J, Lukin K. “Hands-on” regulation of B cell development by the transcription factor Pax5. Immunity. 2007;27:8–10. doi: 10.1016/j.immuni.2007.07.001. [DOI] [PubMed] [Google Scholar]