Abstract
The neural crest, a multipotent embryonic cell type, originates at the border between neural and nonneural ectoderm. After neural tube closure, these cells undergo an epithelial–mesenchymal transition, migrate to precise, often distant locations, and differentiate into diverse derivatives. Analyses of expression and function of signaling and transcription factors in higher vertebrates has led to the proposal that a neural crest gene regulatory network (NC-GRN) orchestrates neural crest formation. Here, we interrogate the NC-GRN in the lamprey, taking advantage of its slow development and basal phylogenetic position to resolve early inductive events, 1 regulatory step at the time. To establish regulatory relationships at the neural plate border, we assess relative expression of 6 neural crest network genes and effects of individually perturbing each on the remaining 5. The results refine an upstream portion of the NC-GRN and reveal unexpected order and linkages therein; e.g., lamprey AP-2 appears to function early as a neural plate border rather than a neural crest specifier and in a pathway linked to MsxA but independent of ZicA. These findings provide an ancestral framework for performing comparative tests in higher vertebrates in which network linkages may be more difficult to resolve because of their rapid development.
Keywords: neural plate border, transcription factor, agnathan
A vertebrate innovation, the neural crest is a multipotent embryonic cell type characterized by its site of origin at the border between neural and nonneural ectoderm, extensive migratory ability, and capacity to differentiate into multiple, diverse derivatives (1, 2). Neural crest cells are first morphologically recognizable when they begin migration, which occurs in most species at the time of neural tube closure. As a consequence, neural crest induction was originally thought to occur as the neural folds elevate just before the onset of migration. However, recent data suggest that this process initiates much earlier, during gastrulation (3–5).
Several signaling molecules (e.g., Wnts, FGFs, BMPs), and transcription factors (e.g., Msx, Pax3/7, Snail, SoxE genes), have been implicated in neural crest formation. Cumulative analysis of their expression patterns and functions, assembled from several model jawed vertebrates, has led to formulation of a neural crest gene regulatory network (NC-GRN) proposed to underlie neural crest formation (6).
The function of NC-GRN genes typically has been examined by looking at 1 or 2 genes at a time in studies in Xenopus, zebrafish, or chick. Recently, a more comprehensive functional analysis of multiple components with the network was systematically tested in a single species (7). The results suggested that much of the NC-GRN is highly conserved across vertebrates (7), contrasting with nonvertebrate chordates (6, 8–15), which appear to lack a module involved in neural crest specification (16). However, proximal portions of this network appear conserved to the base of the chordate lineage (17).
The precise time of action, order, and interrelationships between network genes remain poorly understood. This is partially because early interactions are difficult to ascribe in rapidly developing higher vertebrates like Xenopus and zebrafish, because of the narrow time window separating neural plate border and neural crest specification and the multiple roles for some of inducing factors. To address the connections between network genes involved in specification of neural crest progenitors at the border, we turned to the basal vertebrate, lamprey, whose slow development makes it possible to resolve early events in neural crest induction, 1 regulatory step at a time.
The basal-most extant vertebrates, lamprey and hagfish, are both agnathans (jawless vertebrates) that have migrating neural crest cells and most of the neural crest derivatives (7, 17–20). Only lampreys reliably produce embryos that are accessible to experimental manipulation in the laboratory. Because of its parasitic lifestyle, the lamprey body plan may not reflect that of the early chordate ancestor. However, it is interesting to note that modern lampreys closely resemble lamprey fossils >360 million years old (21), suggesting that their body plan has remained fixed. This raises the intriguing possibility that the core modules of the NC-GRN of a modern lamprey may reflect the ancestral vertebrate state. As the neural crest is unique to vertebrates, analysis of this network in a basal vertebrate holds the promise of informing on the generic architecture of a prototypic network.
In a previous study, we identified lamprey homologues of numerous neural crest network genes and examined their effects on the appearance of neural crest cells (7). However, the interactions among neural plate border specifiers were not clear because of the relatively late stage of analysis. Here, we interrogate the lamprey NC-GRN by focusing on early stages during the most proximal events in neural crest formation. Specifically, we test the effects of perturbing members of the neural plate border specifier module, to establish their order and interrelationships, and their connection to an early acting subset of neural crest specifiers that are also present at the neural plate border. Our results allow refinement of the NC-GRN and reveal some unexpected order and linkages therein. Moreover, this putative network in lamprey provides an experimental scaffold that can be compared, translated, and tested in higher, more rapidly developing vertebrates.
Results and Discussion
Temporal Sequence of Neural Plate Border and Early Neural Crest Specifier Genes Expression from Early Gastrula to Early Neurula [Embryonic Day (E) 3.5 to E4.5].
To resolve events surrounding the time of neural plate formation and their temporal and regulatory sequence, we first examined the dynamic spatial and temporal expression pattern and function of selected lamprey neural plate border and neural crest specifier genes, focusing on the time points between E3 and E4.5. At E3, the embryos have just initiated gastrulation, whereas at E3.5, they are actively gastrulating and resemble round balls of cells, comparable to stage 10.5 Xenopus embryos, but with a smaller blastopore. By E4, the prospective neural plate develops anteriorly by thickening and flattening of the ectoderm, although it is difficult to distinguish in the absence of molecular markers. However, by E4.5, the neural plate is morphologically visible but has yet to condense into a neural rod.
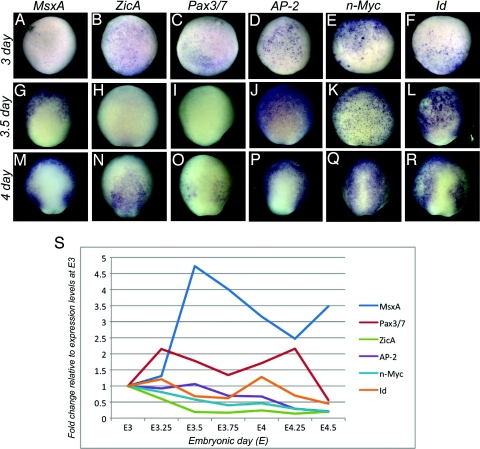
We analyzed expression of 3 neural plate border specifier genes (MsxA, Pax3/7, ZicA) and 3 early neural crest specifier genes (n-Myc, Id, AP-2) by in situ hybridization and quantitative PCR (QPCR) (Fig. 1). All transcripts are present at E3, although MsxA is only expressed in a few cells. MsxA levels progressively increase to peak approximately E3.5, when transcripts are in anterior-half ectoderm, but most prominent in newly ingressed lateral mesoderm (Fig. 1 A and G and data not shown). Interestingly, expression levels rapidly decline at approximately E3.75 to peak again between E4 and E4.5, predominantly within the neural plate border and nonneural ectoderm (Fig. 1 M and data not shown). ZicA and Pax3/7 hybridized embryos show early expression at E3 in the embryonic ectoderm (ZicA) and future mesoderm (Pax3/7), but their levels decrease by E3.5 (Fig. 1 B, C, H, and I) to reappear by E4 in the future neural plate border, neural (ZicA; Fig. 1N) and nonneural ectoderm (Pax3/7; Fig. 1O). In contrast, all 3 early neural crest specifiers show high levels of expression during gastrulation, with AP-2 and n-Myc transcripts present in the embryonic ectoderm (Fig. 1 D and E) and Id transcripts found mainly in the future posterior areas surrounding the blastopore (Fig. 1F; data not shown). By E3.5 n-Myc transcripts are present quasi-ubiquitously in the ectoderm, whereas AP-2 and Id are expressed in the anterior portion of the embryo but transcript-negative posteriorly, close to the site of invagination (Fig. 1 J–L). As the neural plate thickens at E4, Pax3/7 transcripts appear, mostly confined to the broad medioposterior border territory between the neural plate, labeled by ZicA, and nonneural ectoderm, labeled by AP-2 and MsxA (Fig. 1 M–O). Whereas MsxA, ZicA, Pax3/7, and Id transcripts encompass the more posterior border territory, n-Myc and AP-2 are evenly distributed throughout the anterior–posterior extent of the neural plate border (Fig. 1 M–R).
Fig. 1.
Expression of neural crest network genes in the midgastrula to early neurula. (A–R) Dorsal view of in situ hybridization patterns showing expression of neural plate border (MsxA, Pax3/7, and ZicA) and early neural crest specifiers (AP-2, n-Myc, and Id) in lamprey at early gastrula (E3; A–F), midgastrula (E3.5; G–L), and early neurula stages (E4; M–R). Anterior is to the top. (A–F) All of the transcripts, except MsxA, are clearly present at the onset of gastrulation (E3). (G and J–L) At midgastrula, MsxA is expressed in the ventral ectoderm, but not in the prospective neural plate (G), whereas AP-2, n-Myc and Id are expressed ubiquitously throughout the ectoderm (J–L). (H and I) Expression of Pax3/7 and ZicA declines by E3.5. (N and O) At E4 ZicA is seen in the neural plate (N), whereas Pax3/7 is confined to the neural plate border (O). (M–R) By E4, MsxA, AP-2, and Id are expressed in the ectoderm and neural plate border, whereas n-Myc is present throughout the ectoderm including the neural plate. (S) Quantitative progression of border/early specifier expression between E3 and E4.5 by QPCR. The dynamic changes in transcription level of individual genes are depicted as fold-changes relative to their levels at E3. (Magnification: A–R, 20×.)
QPCR was used to examine dynamic changes in gene expression between E3 and E4.5, using 7 different time points, each 6 h apart (Fig. 1S). Changes in transcript levels of each gene are presented in the same graph, as fold-change relative to its level at E3 (≈12 h after onset of zygotic transcription), set arbitrarily at 1 for each gene. Conversely, the detailed in situ hybridization analysis (Fig. 1 A–R and data not shown) allows comparison of transcript levels among different genes. Interestingly the in situ patterns closely follow the changes quantified by QPCR. In the early embryo, all of the transcription factors examined are initially present as maternal transcripts. QPCR data reveal that AP-2, ZicA, Id, and Pax3/7 exhibit 2 peaks of expression during early embryonic stages (Fig. 1S). However, QPCR analysis alone fails to demonstrate the timely appearance of 2 peaks of expression in the case of MsxA transcripts, which are evident from our in situ data. The initially very high concentration of MsxA transcripts accumulated in the mesoderm and the embryonic ectoderm during gastrulation likely masks the increase in transcript levels in the newly forming neural plate border. The second peak only becomes evident after E4.25. The first peak in expression of MsxA, AP-2, ZicA, Id, and Pax3/7 most likely corresponds to different roles these genes play during gastrulation (i.e., prominent presence of MsxA activity in the lateral mesoderm, as shown by in situ analysis). The second peak of expression at E4–4.5 of the above-mentioned genes corresponds to their function in the neural plate border. n-Myc appears to be present at relatively high concentrations in dorsal ectoderm and its levels remain approximately constant during this period with a slight decline at approximately E3.75, as it becomes confined to the neural plate border. This finding suggests that with respect to border function, E4 is the time of maximal developmental activity of their gene products, consistent with the possibility that this time point corresponds to ongoing inductive events (Fig. 1S).
The data suggest that AP-2 and n-Myc act earlier that anticipated, based on their previous classification as “neural crest specifiers.” In fact, our more detailed analysis suggests that they may act concomitant with neural plate border genes. Interestingly, other neural plate border specifiers, ZicA and Pax3/7, are induced slightly later, suggesting they may be either downstream of AP-2 and MsxA or in a parallel pathway but up-regulated slightly later. The transcriptional regulators, Id and n-Myc, appear to be in a separate regulatory module that acts early in gastrulation, likely involved in cell cycle control, and later also in neural plate border progenitors.
Although the precise temporal sequence of neural plate border specifiers has yet to be precisely resolved in frog, several reports suggest similarities to our observations in lamprey. For example, it is clear that induction of Msx1 and Zic1/Pax3/7 represent independent events in the frog (3, 4, 22). Although classified as neural crest specifier genes, c-Myc, Id, and AP-2 are first deployed at the neural plate border of the early neurula, rather than in nascent neural crest cells (23–29). Because recent studies suggest that onset of neural crest formation and specifier expression in both frog and chick may occur in the gastrula, these genes may act earlier in neural crest specification than formerly assumed. The coregulation of neural plate border and neural crest specifiers at the border may reflect a previously unrecognized pan-vertebrate characteristic of the NC-GRN, illustrating how information from lamprey may translate because of its tight conservation and ancient origin.
Effects of Knockdown of Neural Crest Genes with Antisense Morpholinos (MOs).
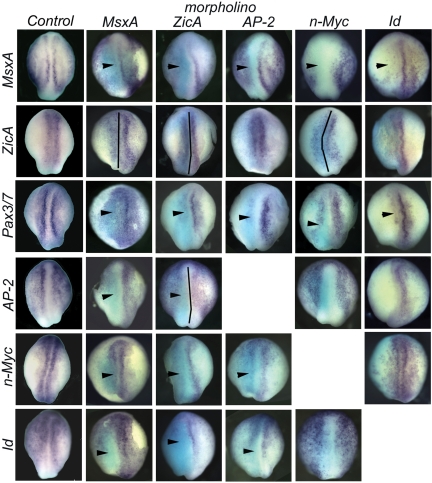
To unravel regulatory relationships among early neural crest transcription factors MsxA, Pax3/7, ZicA, n-Myc, Id, and AP-2, we tested the effects of loss of each gene on expression of the other 5 (Fig. 2 and supporting information (SI)Fig. S1). To this end, 1 blastomere of 2-cell stage embryos was injected with MO antisense oligonucleotides, and embryos were collected for analysis at E4.5. This time point was chosen to address interactions between these early regulatory factors at the neural plate border itself rather than at the time by which the neural crest proper has formed. This represents the earliest analysis in any model organism, largely, aimed to separate events at the neural plate border from neural crest specification.
Fig. 2.
Effect of MO-mediated knockdown of 3 neural plate border specifiers and 3 neural crest specifiers on neural plate border formation and gene expression. Dorsal view of stage E4.5 lamprey neurula, anterior is to the top. MO-injected side, marked with the light blue stain after anti-FITC antibody staining, is to the left (MsxA, ZicA, Ap-2, and n-Myc columns). Id MO was coinjected with rhodamine-dextran as a tracer (Id column). MsxA MO injection caused loss of MsxA, Pax3/7, and AP-2 expression from the neural plate border, expansion of ZicA expression (caused by the expansion of the neural plate) and loss of Id and n-Myc expression from the posterior neural plate border (MsxA column). Injection of ZicA MO had a similar effect on MsxA, ZicA, and Pax3/7 expression, whereas AP-2 was lost from the neural plate border specifically, and n-Myc and Id were lost or decreased in the entire neural plate border (ZicA column). Phenotypes of AP-2 injected embryos were very similar to those seen in MsxA MO-injected embryos (compare AP-2 column to MsxA column). N-Myc MO resulted in the down-regulation of MsxA and Pax3/7, whereas AP-2 and Id expression appeared unaffected. Id MO caused loss of MsxA, Pax3/7, and AP-2 expression, without affecting ZicA or n-Myc. Black vertical lines indicate the midline. Arrowheads indicate loss of gene expression in the neural plate border. (Magnification: 18×.)
By E4.5, the neural plate border is clearly distinguishable in lamprey embryos, as characterized by increased expression of border and early crest specifier genes (Fig. 2, Control). To analyze the effects of MO knockdowns, spatial information was obtained by in situ hybridization (Fig. 2 and Fig. S1), whereas QPCR analysis was used to quantify the fold change in gene expression (Fig. S2). Because mosaic incorporation of MO can sometimes lead to subtle and difficult to interpret phenotypes, we selected only the embryos that showed strong unilateral incorporation of the MO.
Below, we describe the effects of MO-mediated knockdown of 3 neural border specifiers (MsxA, ZicA, Pax3/7) and 3 early neural crest specifiers (AP-2, n-Myc, Id), on each other in the border territory (Fig. 1; for cumulative results see Table S1).
The specificity of MOs was demonstrated by using a Xenopus laevis oocyte in vitro translation system (Fig. S3). Furthermore, the same set of MOs was used in our previous study, and their specificity was confirmed in a separate set of rescue experiments (7).
MsxA Knockdowns.
MsxA MO-injected embryos exhibited loss or decreased expression of Pax3/7 (66%, n = 15), ZicA (59%, n = 17), and AP-2 (54%, n = 22) at all axial levels, whereas Id and n-Myc expression was lost only in the posterior region on the injected side. As N-myc and Id are expressed in a large ectodermal domain encompassing the future neural plate border at stage E3.5 (Fig. 1 E and F), MsxA is most likely not involved in their regulation at this stage, but could be controlling their expression in the posterior neural plate border at E4. Conversely, the effect of MO-mediated knockdown of MsxA protein on expression of 2 other border specifiers, ZicA and Pax3/7, is different; they are lost at all axial levels, suggesting that MsxA is necessary for their induction in early phases of border specification at E3.5 when MsxA is widely distributed along the anterior/posterior axis, and before its restriction to the posterior border at E4. Interestingly, the effect of MsxA knockdown on AP-2 expression mirrors that on border specifiers, with AP-2 depleted at all axial levels. This finding suggests that MsxA might regulate AP-2 expression in the border as early as E3.5. MsxA MO also affects its own expression (75%, n = 12), suggesting direct or indirect autoregulation (30). These results demonstrate that MsxA plays a critical proximal role in neural crest formation, because it profoundly affects other neural plate border genes and early neural crest specifiers. Intriguingly, its effects on AP-2 (i.e., early up-regulation at the border) differ from those on other neural crest specifiers, suggesting that AP-2 may be controlled by MsxA.
ZicA Knockdowns.
ZicA morphant embryos have an expanded neural plate on the injected side, as visualized by expression of ZicA itself (75%, n = 20). ZicA levels were often visibly reduced on the injected side, although the overall area of expression was larger because of the expanded neural plate. There is loss of Pax3/7 (81%, n = 21), MsxA (85%, n = 33), n-Myc (72%, n = 18), and Id (72%, n = 18) from the neural plate border (Fig. 2). AP-2 transcripts were lost only in the neural plate border, because ZicA is not coexpressed with AP-2 in nonneural ectoderm (72%, n = 25). These results are consistent with our QPCR analysis, because transcript levels of MsxA, AP-2, Id, and n-Myc appear reduced ≈2-fold (Fig. S2). Given that ZicA only appears at E4 and therefore may not participate in the earliest phases of border induction, the effects of its depletion on MsxA and AP-2 in the border territory is likely to occur via a feedback regulatory loop that maintains rather than activates their expression.
Pax3/7 Knockdowns.
Pax3/7 morphant embryos exhibited depletion of all neural plate border and neural crest markers examined. Quantification of the phenotypes by QPCR indicates a 5-fold decrease in MsxA and ZicA levels and 2-fold decrease in expression of all other factors (Fig. S2). The regulation of MsxA and AP-2 by Pax3/7 is likely to occur via a feedback regulatory loop that maintains their expression. In contrast, Pax3/7 may be an activator of n-Myc and Id.
AP-2 Knockdowns.
Our results suggest that AP-2 plays a more upstream role in neural plate border formation than previously thought. It is essential for neural plate border formation at this early stage, where it regulates expression of neural plate border specifiers. Knockdown of AP-2 causes a decrease in MsxA (72%, n = 43) and Pax3/7 (87%, n = 28), whereas there was no change in ZicA expression (Fig. 2). Whereas AP-2 is expressed earlier than and likely controls initial expression of Pax3/7, it is not clear whether AP-2 activates MsxA expression at the border or vice versa, although it is clearly involved in maintenance of its expression at subsequent stages. In AP-2 morphants, MsxA transcripts are lost in the neural plate border, but not in the nonneural ectoderm. In the epidermis, AP-2 directly regulates keratin (31) where it may act downstream of Msx1, which induces keratin while mediating BMP-4 input (32). Conversely, in the neural plate border AP-2 could be positioned upstream of MsxA, because the murine Msx1 promoter contains a consensus AP-2-binding site (30). The effects of AP-2 depletion on n-Myc and Id expression are very similar to the effects of MsxA depletion, with expression lost only in the posterior border between neural and nonneural ectoderm (Fig. 2) in 62% of Id-stained morphants (n = 39) and 78% of n-Myc-stained morphants (n = 38). In contrast, the expression of these genes in the very anterior portion where they are coexpressed with AP-2, but not MsxA, is unaffected. The observed similarity of these phenotypes suggests that AP-2 and MsxA may act in the same pathway to regulate Id and n-Myc expression. This suggests that MsxA (present only in the posterior portion at E4) and AP-2 might activate the transcription of Id and n-Myc in the posterior neural plate border of the embryo at E4.
Id Knockdowns.
MO-mediated Id knockdowns result in the loss of both MsxA and AP-2 expression in the neural plate border and in the ectoderm on the injected side (80%, n = 25 and 75%, n = 16, respectively; Fig. 2). Pax3/7 expression is lost in the neural plate border (79%, n = 14), whereas the expression of n-Myc (100%, n = 20) and ZicA (100%, n = 25) is largely unaffected by Id MO. In ≈20% of cases, loss of ZicA expression is seen on the injected side, which we attribute to nonspecific cell death caused by depletion of Id.
N-Myc Knockdowns.
Loss of n-Myc affects the spatial distribution of ZicA (100%, n = 18), AP-2 (100%, n = 11), and Id (100%, n = 14) transcripts, although we often observed expansion of the neural plate on the injected side in embryos stained for ZicA (Fig. 2). MsxA expression is significantly reduced in both the neural plate border and ectoderm (53%, n = 19), whereas Pax3/7 is reduced but never absent from the neural plate border (53%, n = 31). Quantitative analysis of n-Myc morphants shows that whereas expression levels of n-Myc and Id do not change, there is an increase in ZicA expression (concomitant with neural plate expansion), but also in AP-2 expression, likely corresponding to supplementary transcripts mostly in the nonneural ectoderm (Fig. 2A and Fig. S2).
Proliferation/Cell Death Assays.
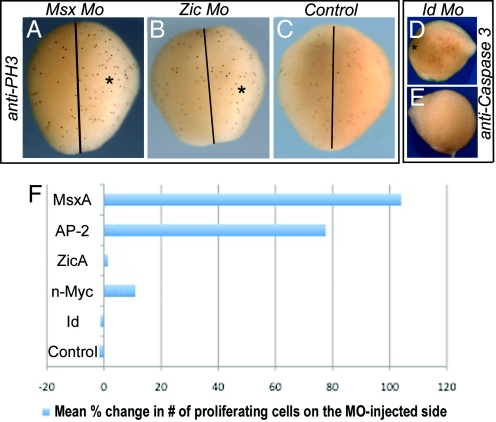
To ascertain whether MO-mediated knockdowns exert their effects via influencing cell cycle progression, we examined expression of the proliferation marker, phosphohistone H3, and the apoptotic marker, cleaved Caspase-3, on the injected versus control, noninjected side (Fig. 3). Interestingly, the results show that knockdown of MsxA and, to a lesser extent, AP-2 protein leads to a prominent increase in cell proliferation within the dorsal field on the MO-injected side versus control side (Fig. 3A). The mean increase in numbers of proliferating cells for MsxA was 104% (range: 38.5% to 243%; n = 8/9 embryos) and 77.6% (range: 37.5% to 113% n = 6/10) for AP-2 (Fig. 3F). N-myc MO-treated embryo exhibited a slight increase in the mean number of proliferating cells on experimental versus control side (11% with range of 6.3% to 14.6% in n = 5/6 embryos), whereas ZicA (mean 1.4%; range: −3.1% to 6.7%; n = 7/8; Fig. 3 B and F) and Id (mean −1.3%; range: −9.1% to 4.5%; n = 7/7; Fig. 3F) had no significant differences from control embryos (mean −1.7%; range: −8.7% to 9.1%; n = 7/7; Fig. 3 C and F).
Fig. 3.
Loss of Msx protein significantly enhances cell proliferation, whereas loss of Id may induce cell death. (A–C) Dorsal view of embryos examined at E4.5 after introduction of indicated MO on the right (*). There is increased proliferation, as assayed by phosphohistone H3 expression (anti-PH3), after Msx MO treatment (A), but not in Zic MO-treated (B) or control embryos (C). Black line indicates the midline. (D and E) Side view of Id MO-treated embryos shows increased cell death on the injected (D) compared with control (E) side, as shown by anti-cleaved Caspase3 expression (anti-Caspase 3). (F) Mean percentage change in number of proliferating cells on the MO-injected versus control side. The MOs used are indicated on the left. (Magnification: A–C, 25×; D and E, 12×.)
WT embryos at the beginning of neurulation show few signs of programmed cell death. Using cleaved Caspase-3 as a marker of cells undergoing terminal apoptosis, we noted a slight increase in the numbers of dying cells in the case of Id MO-treated embryos (n = 5/10; Fig. 3 D and E) and n-Myc treated embryos (n = 3/7).
Taken together, these data indicate that transcription factors present at the neural plate border extensively cross-regulate. As predicted, neural plate border specifiers are positioned upstream. However, AP-2 appears to be aligned with them where it, together with MsxA, appears to play a role activating initial induction. MsxA and AP-2 may function in a common pathway to regulate the later onset of Id and n-Myc in the neural plate border. Based on our cell cycle progression data, an intriguing possibility is that MsxA and AP-2 may play a role in assuring that cells at the neural plate border exit the cell cycle and proceed with expression of the neural crest specifier program. ZicA and Pax3/7 are induced slightly later, at E4, where they may act independently in a separate pathway, involved in maintaining expression of the neural plate border regulatory module through feedback and self-regulatory loops.
AP-2 and ZicA Function Through Independent Pathways.
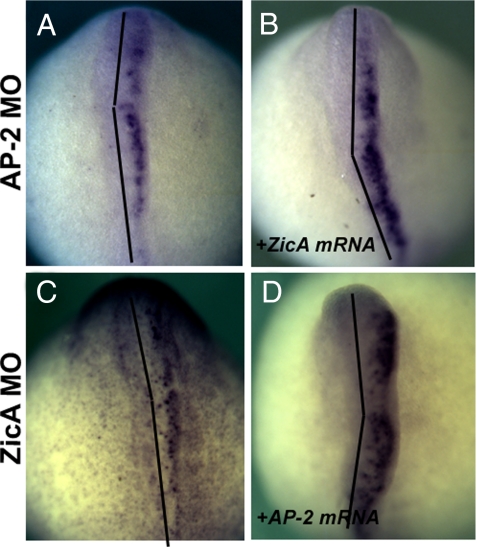
To test possible epistatic relationships within the early neural plate border network, we assessed the ability of ZicA mRNA to rescue the effects of AP-2 MO knockdown and vice versa. Embryos were incubated until E6.5 and the emergence of neural crest was assessed by FoxD-A staining. AP-2 MO completely abolishes the formation of the neural crest (Fig. 4A), in a manner that is not abrogated by coinjection with ZicA mRNA (100%, n = 79) (Fig. 4B). Similarly, in the reciprocal experiment, AP-2 mRNA was not capable of rescuing the effects of Zic Mo (100%, n = 56) (Fig. 4 C and D). In contrast, AP-2 mRNA injection can rescue AP-2 MO-mediated phenotype, as ZicA coexpression rescues the effects of ZicA MO (7). These results indicate that in the process of the initial onset and activation of their expression AP-2 and ZicA act in parallel rather than common pathways.
Fig. 4.
ZicA and AP-2 act in parallel pathways to bring about the formation of the neural plate border. Embryos were injected with AP-2 (A) or ZicA (C) MO or a combination of ZicA MO and AP-2 mRNA (B) or AP-2 MO and ZicA mRNA (D). The effects on neural crest formation were assessed at E5.5 by staining for the neural crest marker FoxD-A. Both AP-2 and ZicA MO abolish the marker expression on the injected side (A and C), whereas coinjection with ZicA mRNA and AP-2 mRNA, respectively, does not rescue this phenotype (B and D). These experiments suggest that these 2 genes act in parallel pathways. Black lines indicate the midline. (Magnification: 40×.)
Functional redundancy and parallel pathways are common features of the few vertebrate gene regulatory networks studied to date and thought to underlie developmental robustness of vertebrate embryos. Because the neural crest is a particularly plastic and highly self-regulating cell population, the complexity of the gene regulatory network controlling its formation may reflect the robustness and regulative ability of this cell population.
Long-Term Effects of AP-2 Knockdown on Formation of Neural Crest Derivatives.
All neural plate border specifiers examined to date are essential for the formation of all neural crest derivatives and some other neural plate border derivatives. To confirm that AP-2 is functioning as a true neural plate border specifier, we examined the long-term effects of its loss on neural crest derivatives at E10–15. Morphant embryos exhibited a significant loss in the neural crest-derived pigment cell population, with a decrease in Neurogenin expression in the cranial ganglia and in SoxE1 expression in the neural crest-derived portion of the branchial cartilage at E11, with many fewer neural crest cells that failed to extend as far distally on the MO-injected side of the embryo (Fig. S4). These observations are consistent with AP-2 acting at the level of a neural plate border specifier. However, we did not detect a significant change in expression of SoxE1 downstream effector, Col2a1, at E14, possibly because of the very subtle penetrance of the cartilage phenotype or to a partial rescue caused by regulative compensation by another factor or crest cells crossing from the contralateral side.
In mouse, loss of AP2 results in a perinatal lethal phenotype characterized by the absence or severe reduction of cranial bones of neural crest origin. In Xenopus, MO-mediated depletion of AP-2 leads to down-regulation of both Sox9 and Slug expression during neural crest induction. Conversely, studies of AP-2 mutants in zebrafish failed to demonstrate an early role for this gene in neural crest specification, as various AP-2 mutants exhibit no change in neural crest specifier expression during early steps of neural crest formation. However, they do show defects in specific subsets of neural crest derivatives (27, 29, 33). The absence of early defects could be caused by compensation by redundant paralogs, endogenously coexpressed with the mutated gene, suggesting all relevant copies of AP-2 gene should be inactivated in zebrafish to replicate the AP-2 mutant phenotype observed in other vertebrates (34).
Conclusion
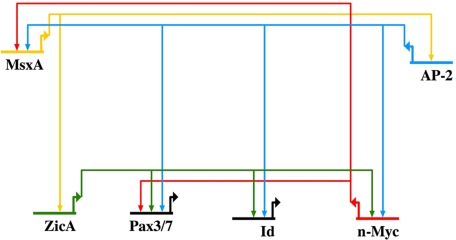
By using the basal vertebrate lamprey as an experimental system, we took advantage of its basal phylogenetic position to examine a NC-GRN that likely bears many characteristics of an ancestral network. Furthermore, lamprey's slow development allows precise temporal resolution at the neural plate border in a manner difficult to accomplish in other model vertebrates. The results reveal some surprising regulatory relationships in the network. Expression analysis together with functional studies suggest that AP-2 and MsxA are upstream of other transcription factors in the NC-GRN (Fig. 5), despite the fact that AP-2 was previously considered to be a neural crest specifier gene. Both have nearly identical spatial distributions and the dynamics of their early expression are similar, whereas Pax3/7 and ZicA are expressed slightly later. Furthermore, inactivation of AP-2 or MsxA causes identical effects on other early regulators. However, n-Myc/Id have different expression dynamics and phenotype. Accordingly, we propose that MsxA and AP-2 are the most proximal regulators in the network and likely act in the same pathway. Ultimate confirmation of direct interactions will require identification and interrogation of neural crest enhancers for AP-2 and MsxA and detailed epistasis experiments.
Fig. 5.
Current status of the NC-GRN in the lamprey.
Materials and Methods
Animal Husbandry/Embryo Culture.
Adult lampreys (Petromyzon marinus) at different stages of sexual maturation were obtained from Hammond Bay Biological Station, Millersburg, MI. Eggs were fertilized, cultured and collected as described (7, 35).
RNA and MO Injections.
FITC-labeled MOs against Pax3/7, ZicA, MsxA, n-Myc, Id, and AP-2 were obtained from Gene Tools and used as described (7). The following amounts of MO were used for injection: 10 ng per cell of Pax3/7 MO, 20 ng/cell of n-Myc, MsxA, and Id MO, and 40 ng/cell of ZicA and AP-2 MO. The injected embryos were allowed to develop for 96 h, selected for unilateral incorporation of MO under the fluorescence microscope, and processed for in situ according to the protocol in ref. 7. For rescue experiments, 1 blastomere of 2-cell lamprey embryos was injected with a mixture of 50–100 pg of lamprey full-length AP-2 or ZicA transcript (7) and 10–20 ng of AP-2 or ZicA Mo.
In Situ Hybridization.
In situ hybridization on lamprey embryos was performed according to the protocol of Sauka-Spengler et al. (7) with several modifications (SI Text).
RNA Extractions and RT-QPCR.
For QPCR assays, both blastomeres of 2-cell lamprey embryo were injected with MO, and embryos were selected for complete MO integration and lysed at 96 hours after fertilization. For timeline of gene expression, WT embryos from the same batch were collected at 6-h intervals from E3 to E4.5. Total RNA was extracted with a RNAqueous kit (Ambion) and DNase-treated with a TURBO DNA-free kit (Ambion), and cDNA was synthesized by using random hexamers and the SuperScript III RT-PCR system (Invitrogen) according to the manufacturer's instructions. Real-time PCR was performed on an ABI7000 QPCR machine using SybrGreen assay. Briefly 25-μL reactions were performed by using the iTaq SYBR green Mix with ROX (BioRad) in the presence of 7–50 pg of cDNA and 450 nM of each primer. The standard curve method was used for quantification, and fold change in expression was determined by dividing the amount of the gene of interest, normalized to RPS9 in MO-injected embryos, by the normalized transcript amount in control embryos (see SI Text).
Cell Proliferation/Cell Death Assays.
Embryos were preselected for unilateral incorporation on either the right or left side, collected at E4.5, fixed in MEMFA (0.1 M MOPS, pH 7.4, 2 mM EGTA, 1 mM MgSO4, 3.7% Formaldehyde), and kept at 4 °C in PBS. Immunocytochemistry with anti-cleaved Caspase 3 (G7481; Promega) and antiphosphohistone H3 (06Ð570; Upstate Biotech) was performed as described (35), except that PBT (PBS, 0.1% Triton X-100, 0.2% BSA) was used in place of PTW (PBS, 0.1% Tween-20).
Supplementary Material
Acknowledgments.
This work was supported by the National Institutes of Health Grant DE017911 (to M.B.F.).
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Gene Networks in Animal Development and Evolution,” held February 15–16, 2008, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS web site at http://www.nasonline.org/SACKLER_Gene_Networks.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806009105/DCSupplemental.
References
- 1.Hall BK, Hörstadius S. The Neural Crest. Oxford: Oxford Univ Press; 1988. [Google Scholar]
- 2.Le Douarin NM, Kalcheim C. The Neural Crest. 2nd Ed. Cambridge, UK: Cambridge Univ Press; 1999. [Google Scholar]
- 3.Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Sato T, Sasai N, Sasai Y. Neural crest determination by coactivation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- 5.Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- 6.Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev Cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Holland LZ, Schubert M, Kozmik Z, Holland ND. AmphiPax3/7, an amphioxus paired box gene: Insights into chordate myogenesis, neurogenesis, and the possible evolutionary precursor of definitive vertebrate neural crest. Evol Dev. 1999;1:153–165. doi: 10.1046/j.1525-142x.1999.99019.x. [DOI] [PubMed] [Google Scholar]
- 9.Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: Toward a comprehensive understanding of gene networks. Development. 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- 10.Langeland JA, Tomsa JM, Jackman WR, Jr, Kimmel CB. An amphioxus snail gene: Expression in paraxial mesoderm and neural plate suggests a conserved role in patterning the chordate embryo. Dev Genes Evol. 1998;208:569–577. doi: 10.1007/s004270050216. [DOI] [PubMed] [Google Scholar]
- 11.Ma L, et al. Expression of an Msx homeobox gene in ascidians: Insights into the archetypal chordate expression pattern. Dev Dyn. 1996;205:308–318. doi: 10.1002/(SICI)1097-0177(199603)205:3<308::AID-AJA10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 12.Meulemans D, Bronner-Fraser M. Amphioxus and lamprey AP-2 genes: Implications for neural crest evolution and migration patterns. Development. 2002;129:4953–4962. doi: 10.1242/dev.129.21.4953. [DOI] [PubMed] [Google Scholar]
- 13.Meulemans D, McCauley D, Bronner-Fraser M. Id expression in amphioxus and lamprey highlights the role of gene cooption during neural crest evolution. Dev Biol. 2003;264:430–442. doi: 10.1016/j.ydbio.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Wada H, Holland PW, Sato S, Yamamoto H, Satoh N. Neural tube is partially dorsalized by overexpression of HrPax-37: The ascidian homologue of Pax-3 and Pax-7. Dev Biol. 1997;187:240–252. doi: 10.1006/dbio.1997.8626. [DOI] [PubMed] [Google Scholar]
- 15.Wada S, Saiga H. HrzicN, a new Zic family gene of ascidians, plays essential roles in the neural tube and notochord development. Development. 2002;129:5597–5608. doi: 10.1242/dev.00156. [DOI] [PubMed] [Google Scholar]
- 16.Yu JK, Meulemans D, McKeown SJ, Bronner-Fraser M. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res. 2008;18:1127–1132. doi: 10.1101/gr.076208.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler AB, Hodos W. Comparative Vertebrate Neuroanatomy: Evolution and Adaptation. New York: Wiley; 1996. [Google Scholar]
- 18.Horigome N, et al. Development of cephalic neural crest cells in embryos of Lampetra japonica, with special reference to the evolution of the jaw. Dev Biol. 1999;207:287–308. doi: 10.1006/dbio.1998.9175. [DOI] [PubMed] [Google Scholar]
- 19.McCauley DW, Bronner-Fraser M. Neural crest contributions to the lamprey head. Development. 2003;130:2317–2327. doi: 10.1242/dev.00451. [DOI] [PubMed] [Google Scholar]
- 20.Ota KG, Kuraku S, Kuratani S. Hagfish embryology with reference to the evolution of the neural crest. Nature. 2007;446:672–675. doi: 10.1038/nature05633. [DOI] [PubMed] [Google Scholar]
- 21.Gess RW, Coates MI, Rubidge BS. A lamprey from the Devonian period of South Africa. Nature. 2006;443:981–984. doi: 10.1038/nature05150. [DOI] [PubMed] [Google Scholar]
- 22.Hong CS, Saint-Jeannet JP. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol Biol Cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrallo-Gimeno A, Holzschuh J, Driever W, Knapik EW. Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development. 2004;131:1463–1477. doi: 10.1242/dev.01033. [DOI] [PubMed] [Google Scholar]
- 24.Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in Xenopus. Dev Cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 25.Kee Y, Bronner-Fraser M. To proliferate or to die: Role of Id3 in cell cycle progression and survival of neural crest progenitors. Genes Dev. 2005;19:744–755. doi: 10.1101/gad.1257405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight RD, Javidan Y, Zhang T, Nelson S, Schilling TF. AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development. 2005;132:3127–3138. doi: 10.1242/dev.01879. [DOI] [PubMed] [Google Scholar]
- 27.Knight RD, et al. Lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755–5768. doi: 10.1242/dev.00575. [DOI] [PubMed] [Google Scholar]
- 28.Light W, Vernon AE, Lasorella A, Iavarone A, LaBonne C. Xenopus Id3 is required downstream of Myc for the formation of multipotent neural crest progenitor cells. Development. 2005;132:1831–1841. doi: 10.1242/dev.01734. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien EK, et al. Transcription factor Ap-2α is necessary for development of embryonic melanophores, autonomic neurons, and pharyngeal skeleton in zebrafish. Dev Biol. 2004;265:246–261. doi: 10.1016/j.ydbio.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 30.Kuzuoka M, Takahashi T, Guron C, Raghow R. Murine homeobox-containing gene, Msx-1: Analysis of genomic organization, promoter structure, and potential autoregulatory cis-acting elements. Genomics. 1994;21:85–91. doi: 10.1006/geno.1994.1228. [DOI] [PubMed] [Google Scholar]
- 31.Luo T, Matsuo-Takasaki M, Thomas ML, Weeks DL, Sargent TD. Transcription factor AP-2 is an essential and direct regulator of epidermal development in Xenopus. Dev Biol. 2002;245:136–144. doi: 10.1006/dbio.2002.0621. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki A, Ueno N, Hemmati-Brivanlou A. Xenopus msx1 mediates epidermal induction and neural inhibition by BMP4. Development. 1977;124:3037–3044. doi: 10.1242/dev.124.16.3037. [DOI] [PubMed] [Google Scholar]
- 33.Luo T, Lee YH, Saint-Jeannet JP, Sargent TD. Induction of neural crest in Xenopus by transcription factor AP2α. Proc Natl Acad Sci USA. 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan YL, et al. A pair of Sox: Distinct and overlapping functions of zebrafish sox9 coorthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- 35.Nikitina N, Bronner-Fraser M, Sauka-Spengler T. Emerging Model Organisms. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.