Abstract
As with other genetically complex common psychiatric and medical conditions, multiple genetic and environmental components contribute to alcohol use disorders (AUDs), which can confound attempts to identify genetic components. Intermediate phenotypes are often more closely correlated with underlying biology and have often proven invaluable in genetic studies. Level of response (LR) to alcohol is an intermediate phenotype for AUDs, and individuals with a low LR are at increased risk. A high rate of concurrent alcohol and nicotine use and dependence suggests that these conditions may share biochemical and genetic mechanisms. Genetic association studies indicate that a genetic locus, which includes the CHRNA5-CHRNA3-CHRNB4 gene cluster, plays a role in nicotine consumption and dependence. Genetic association with alcohol dependence was also recently shown. We show here that two of the markers from the nicotine studies also show an association (multiple testing corrected P < 0.025) with several LR phenotypes in a sample of 367 siblings. Additional markers in the region were analyzed and shown to be located in a 250-kb expanse of high linkage disequilibrium containing three additional genes. These findings indicate that LR intermediate phenotypes have utility in genetic approaches to AUDs and will prove valuable in the identification of other genetic loci conferring susceptibility to AUDs.
Keywords: alcohol use disorders, genetics, quantitative trait locus
Multiple genetic and environmental components contribute to alcohol use disorders (AUDs) (1). As with other genetically complex common psychiatric and medical conditions, it is challenging to match genetic variants with the disease phenotype. Intermediate phenotypes, such as the large numbers of colon polyps seen in the familial polyposis form of colon cancer (2), are often closer to the underlying biology and may provide a better phenotype for genetic analysis (3).
The level of response (LR) to alcohol has been developed as an intermediate phenotype for AUDs (4). Physical responses (e.g., body sway) and subjective feelings (as measured on the Subjective High Assessment Scale [SHAS]) show reproducible individual variation among subjects. Prospective studies have shown that individuals with a low LR are at increased risk for AUDs (5). Alcohol LR is heritable, on a par with alcohol dependence, with genes explaining 40% to 60% of the variance (6–8). Linkage analyses have shown interesting but inconclusive findings (9–11).
Frequent concurrence of alcohol and nicotine use and dependence suggests a shared etiology (12). Twin studies (13–16) conclude there are shared genetic factors that influence alcohol and nicotine consumption and dependence. Functional evidence that nicotinic receptors are involved in alcohol responses is provided by studies showing that mice treated with the smoking cessation drug varenicline, a nicotinic receptor partial agonist, have an attenuated response to alcohol; they drink less and show reduced levels of reinstatement following alcohol abstinence (17). Therefore, genetic loci identified as influencing the consumption and/or dependence of nicotine are potential candidate loci for alcohol dependence.
Association studies have identified several loci associated with nicotine consumption and dependence. A recent case-control study tested 348 candidate genes for association with nicotine dependence (18). The two top loci both subtended nicotinic receptor genes: one on chromosome 8 in and around CHRNB3 and the second on chromosome 15, including the CHRNA5-CHRNA3-CHRNB4 gene cluster. Five genome-wide association studies (19–23) implicated this chromosome 15 cluster as a susceptibility locus for lung cancer and/or nicotine dependence in smokers. The two largest studies, each testing in excess of 10,000 subjects (20, 22), reported that this locus may play a role in quantity smoked and nicotine dependence. The chromosome 15 cluster was also recently tested for genetic association with AUDs using a family-based association test, followed by case-control replication (24); results indicated that the CHRNA5-CHRNA3-CHRNB4 locus is involved in susceptibility to alcohol dependence.
We demonstrate here that two markers in the CHRNA5-CHRNA3-CHNB4 locus showing a strong association with lung cancer/nicotine dependence are also associated with alcohol LR. Because these markers are located in a 250-kb expanse of high linkage disequilibrium (LD), additional markers were analyzed. Although the nicotinic receptors are compelling candidate genes at this locus, the high degree of LD extending across the region does not allow sufficient localization of the susceptibility factor to implicate a single gene.
Results
Nicotine Dependence/Lung Cancer SNPs Are Associated with Alcohol LR Phenotypes.
Two sentinel SNP markers associated with nicotine dependence and lung cancer, RS1051730 and RS8034191, were genotyped with the Illumina HumanCNV370-Duo DNA Analysis BeadChip Illumina Inc. and tested for association with alcohol LR phenotypes in a set of 313 white siblings of the San Diego Sibling Pair cohort. Additional genotyping with TaqMan assays, for verification and subject expansion, resulted in a total of 367 genotyped white siblings, with complete agreement among 294 siblings genotyped by both methods. The number of subjects with both phenotypic and genotypic data ranged from 342 to 365 for the three phenotypic measures.
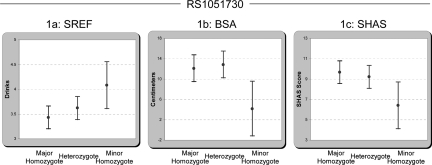
Table 1 shows the results of regression analyses of three LR traits by genotype. BSA (body sway anterior/posterior) and SHAS are alcohol challenge phenotypes measuring physical coordination and feeling of “high,” respectively. The Self-Report of the Effects of Ethanol (SREF) is a questionnaire measuring LR, in which the subjects recall how many drinks it took to reach a given level of intoxication during their first five experiences with alcohol. The SREF incorporates elements of coordination and high. Note that low BSA and SHAS values indicate a low LR, whereas high SREF values also indicate a low LR. Association of the minor allele homozygote of RS1051730 with BSA was highly significant, with a P value of 0.0002, which remains significant after correcting for multiple tests with a q-value of 0.007. Similarly, a P value of 0.0012 (q-value = 0.022) was seen for association of the minor allele homozygote of RS8034191 with BSA. Fig. 1A illustrates the relations at the primary data level between the three genotypes of RS1051730 for BSA. The minor homozygotes demonstrate a lower alcohol LR, with the BSA trait swaying an average of ≈8 cm less at 60 min following the laboratory alcohol challenge.
Table 1.
Results of association tests with alcohol challenge LR phenotypes with the nicotine addiction-associated SNPs RS8034191 and RS1051730
| Marker | Phenotype | n | Genotype | Value | P | q |
|---|---|---|---|---|---|---|
| RS1051730 | BSA | 354 | a/a | −0.52 | 0.0002 | 0.007 |
| RS1051730 | BSA | 354 | g/a | 0.04 | 0.62 | 1.000 |
| RS1051730 | SHAS | 365 | a/a | −0.48 | 0.0074 | 0.092 |
| RS1051730 | SHAS | 365 | g/a | −0.02 | 0.87 | 1.0 |
| RS1051730 | SREF | 353 | a/a | 0.37 | 0.030 | 0.23 |
| RS1051730 | SREF | 353 | g/a | 0.18 | 0.093 | 0.49 |
| RS8034191 | BSA | 353 | c/c | −0.44 | 0.0012 | 0.022 |
| RS8034191 | BSA | 353 | t/c | 0.06 | 0.51 | 1.0 |
| RS8034191 | SHAS | 364 | c/c | −0.45 | 0.010 | 0.097 |
| RS8034191 | SHAS | 364 | t/c | −0.06 | 0.62 | 1.0 |
| RS8034191 | SREF | 352 | t/c | 0.20 | 0.065 | 0.41 |
| RS8034191 | SREF | 352 | c/c | 0.26 | 0.12 | 0.56 |
Genotypes were generated from self-reported white subject DNAs by TaqMan and/or Illumina BeadChip; the phenotypes are described in Methods. Regressions calculate the phenotypic affect and significance of the given genotype as compared with the homozygous major allele for the given marker. The Value column reports the difference in the average phenotypic value for that row's genotype and the major allele homozygote in Z-score units.
Fig. 1.
Average trait values (raw data) and 95% confidence intervals for subjects with all three RS1051730 genotypes. Data for SREF number of drinks (A), body sway measured in centimeters (B), and subjective feeling of high measured by SHAS (C). All traits indicate that the minor homozygotes have a lower response to alcohol. The number of subjects in each genotypic class (major homozygote-heterozygote-minor homozygote) were as follows: SREF (161–156-38), BSA (164–162-38), and SHAS (163–160-38).
Having established the association of the sentinel SNPs with BSA, we asked whether the association extends to two other measures of LR, the SHAS and the SREF (9). As seen in Table 1, both of the sentinel SNPs showed significant nominal P values with these measures, except for RS8034191 with the SREF, which showed a near-significant value of 0.065. All associations were consistent with the minor homozygote being correlated with a lower LR (Fig. 1 A–C).
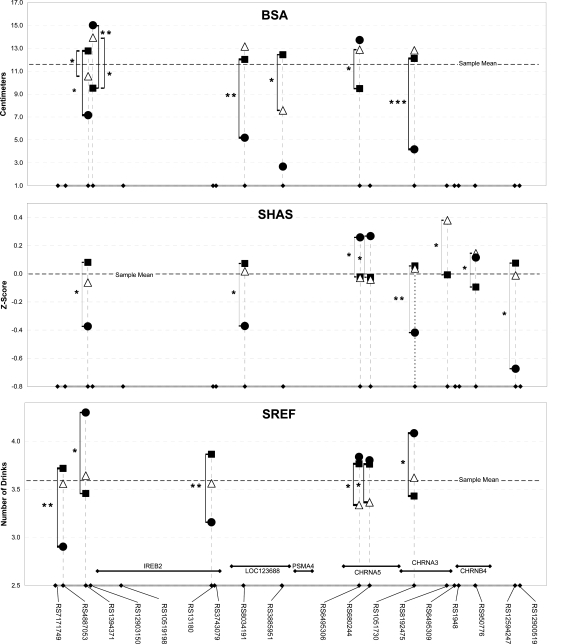
Given the quantitative values for the phenotypic traits shown in Fig. 1, we examined the fit of these data to a dominant genetic model. For BSA, the value of the additive effect for RS1051730 allele a was a −0.26 Z-score (P = 0.0002), and for the dominance deviation, it was a 0.3 Z-score (P = 0.001), indicating that a is recessive to g for BSA. Thus, a/a genotypes had approximately −0.5 SDs less BSA than the a/g and g/g genotypes (see Fig. 2 for genotype mean values with raw data). For the SHAS, the value of the additive effect for allele a was a −0.24 Z-score (P = 0.007), and for the dominance deviation, it was a 0.22 Z-score (P = 0.06), indicating an additive model with a borderline dominance deviation. The a/a genotype was associated with a lower LR, in agreement with that of BSA. For the SREF, the value of the additive effect for allele a was a 0.19 Z-score (P = 0.03), and for the dominance deviation, it was a −0.01 Z-score (P = 0.94), indicating a nominally significant additive genetic model. Although not significant following the correction for multiple testing (q = 0.11), the trend of the test suggests that the a/a genotype had a lower LR, in agreement with the other two phenotypes.
Fig. 2.
Average trait values (raw data) for all genotypes of markers demonstrate significant association with the alcohol LR traits BSA (A), SHAS (B), and SREF (C). Major allele homozygote values are plotted with a square, heterozygotes with a triangle, and minor allele homozygotes with a circle. Genotypes with average trait values that differ significantly from the major homozygote are denoted with a bracket and an increasing number of stars to signify increasing significance (actual P values in Table 2).
Examination of LD in this chromosomal region indicated that RS8034191 and RS1051730 are in strong LD (D' = 0.94, r2 = 0.86). This observation prompted us to search for other markers in the region that might be in LD with RS8034191 and RS1051730. Fig. S1 shows the LD relations among 19 markers in the region. The D' measure is seen to be quite strong across the region. The r2 measures, however, show much more variability among the markers, indicating substantial phase variation between major and minor alleles (i.e., the major allele of one marker system is often coupled with the minor allele of another system). The markers span ≈250 kb of genomic sequence that includes five known genes and one transcript encoding a hypothetical protein (Fig. S1). Genotypes for these markers come from the Illumina HumanCNV370-Duo DNA Analysis BeadChip assay, with markers RS1394371, RS3743079, and RS3885191 confirmed and extended by TaqMan assays. Agreement between the TaqMan and Illumina genotypes was again excellent, with only one discrepancy observed.
The nominally significant associations indicated by the regression analyses of the 19 SNP markers with the three levels of response phenotypes are listed in Table 2. In addition, Table 2 reports the quantitative effect of genotype on phenotype as well as the percentage of the sample's phenotypic variance that is explained by the model. To visualize the quantitative phenotypic effects of the associated genotypes better, they are plotted in Fig. 2 A–C.
Table 2.
Significant associations with alcohol LR traits
| Marker | Phenotype | n | Genotype | Value | P | % phenotypic variance explained | MAF |
|---|---|---|---|---|---|---|---|
| RS1051730 | BSA | 354 | a/a | −0.52 | 0.0002 | 2.9 | 0.33 |
| RS1051730 | SHAS | 365 | a/a | −0.48 | 0.0074 | 1.3 | 0.33 |
| RS1051730 | SREF | 353 | a/a | 0.37 | 0.031 | 0.9 | 0.33 |
| RS12594247 | SHAS | 336 | t/t | −0.80 | 0.010 | 0.9 | 0.16 |
| RS12903150 | BSA | 325 | g/g | 0.46 | 0.0052 | 1.5 | 0.28 |
| RS12903150 | BSA | 325 | a/g | 0.20 | 0.025 | 1.5 | 0.28 |
| RS13180 | SREF | 325 | c/c | −0.41 | 0.0084 | 0.9 | 0.42 |
| RS1394371 | BSA | 354 | c/t | −0.19 | 0.031 | 1.2 | 0.25 |
| RS1394371 | BSA | 354 | t/t | −0.32 | 0.049 | 1.2 | 0.25 |
| RS1394371 | SHAS | 365 | t/t | −0.48 | 0.024 | 1.0 | 0.25 |
| RS1394371 | SREF | 353 | t/t | 0.42 | 0.037 | 0.9 | 0.25 |
| RS3885951 | BSA | 356 | a/g | −0.29 | 0.017 | 1.2 | 0.08 |
| RS4887053 | SREF | 325 | a/a | −0.56 | 0.0052 | 1.0 | 0.26 |
| RS621849 | SHAS | 336 | g/g | 0.32 | 0.037 | 0.8 | 0.40 |
| RS621849 | SREF | 325 | a/g | −0.25 | 0.032 | 0.8 | 0.40 |
| RS6495306 | BSA | 325 | a/g | 0.2 | 0.039 | 0.6 | 0.40 |
| RS6495306 | SHAS | 336 | g/g | 0.31 | 0.047 | 0.7 | 0.40 |
| RS6495306 | SREF | 325 | a/g | −0.27 | 0.020 | 0.9 | 0.40 |
| RS680244 | SHAS | 335 | a/a | 0.32 | 0.036 | 0.8 | 0.40 |
| RS680244 | SREF | 324 | g/a | −0.24 | 0.037 | 0.7 | 0.40 |
| RS8034191 | BSA | 353 | c/c | −0.44 | 0.0012 | 2.4 | 0.33 |
| RS8034191 | SHAS | 364 | c/c | −0.45 | 0.010 | 1.1 | 0.33 |
| RS8192475 | SHAS | 336 | g/a | 0.39 | 0.039 | 0.6 | 0.05 |
| RS950776 | SHAS | 336 | t/c | 0.25 | 0.032 | 0.7 | 0.32 |
All 19 markers in the 250-kb LD region were tested for association by regression; significant associations are reported. Genotypes from self-reported white subject's DNA were obtained by TaqMan and/or Illumina HumanCNV370-Duo DNA Analysis BeadChip; the phenotypes are described in Methods. Regressions calculate the phenotypic effect and significance of the given genotype as compared with the homozygous major allele for the given marker. The Value column reports the difference in the average phenotypic value for that row's genotype and the major allele homozygote in Z-score units. The % phenotypic variance explained reports the percentage of the total phenotypic variance explained by the given genotype in the sample. MAF is the minor allele frequency.
To test which SNPs independently explain phenotypic variation, the joint additive effects of the 19 SNPs were examined in a mixed model framework with a backward elimination procedure. Only RS1051730 remained independently and nominally significant across all phenotypes (BSA P = 0.0003, SHAS P = 0.014, and SREF P = 0.041; data not shown). Furthermore, the proportion of phenotypic variance explained by the markers remains almost constant as markers in addition to RS1051730 are removed or added, although two SNPs, RS12903150 and RS6495306, do seem to explain a small amount of additional variation in BSA (P = 0.005) and SREF (P = 0.04), respectively. This suggests that RS1051730 best tags the underlying genetic trait conferring variation in alcohol LR. Further, because only RS1051730 is consistent across LR phenotypes, greater resolution of the haplotype space is unlikely.
The associated SNPs fall into two groups. The first group shows lower LR associated with minor allele homozygotes and is defined by RS1051730, RS8034191, RS12594247, RS385951, and RS1394371. The best example, RS1051730, demonstrates minor allele homozygote association across all three phenotypes. The other SNPs show the same pattern, but statistical significance does not span all phenotypes. The second group, defined by RS6495306, RS680244, RS8192475, RS950776, RS4887053, RS13180, and RS12903150, consists of SNPs in which the minor allele is associated with an increased LR to alcohol.
Discussion
Association with LR.
Two sentinel SNPs within the chromosome 15q25.1 region, RS1051730 and RS8034191, are significantly associated with a quantitative LR to alcohol. As shown in Table 1, minor allele homozygotes of RS1051730 show a nominal P value of 0.0002 with BSA. The probability that this is attributable to chance, taking into account the multiple phenotypes and genotypes under consideration, was estimated, giving a q-value of 0.007. RS8034191 showed similar results, yielding a nominal P value of 0.0012 and a q-value of 0.022 with BSA. Associations of the two sentinel SNPs with these measures thus leads to the conclusion that the region carries a quantitative trait locus (QTL) affecting body sway in response to alcohol. The nominally significant P values seen with RS1051730 for the SHAS and SREF suggest association with these phenotypes as well (Table 1).
These findings confirm that alcohol LR, an intermediate phenotype associated with alcohol dependence and abuse, is influenced by a genetic component. Together with the previously reported finding of association of genetic variants in this region with alcohol dependence (24), the findings reported here give strong support to the potential utility of alcohol LR as an intermediate phenotype for AUDs.
Both the nominal P values and the phenotypic values of the heterozygotes of the sentinel SNPs, as seen in Table 1 and Fig. 1, suggest a recessive/dominant genetic model rather than an additive genetic model. For SNP RS1051730, regression analysis indicates that the a allele is recessive to the g allele. Similar results were found for the SHAS phenotype, although less support was seen for a recessive/dominant model for the SREF phenotype. These findings may have implications for modeling of the underlying molecular mechanisms linking the causative genetic variant with behavioral phenotypes. Interestingly, the minor allele of RS16969968, a SNP encoding a nonsynonymous amino acid substitution, Asn398Asp, in complete LD with SNP RS1051730, confers nicotine dependence risk recessively (18). Because low LR is an endophenotype for AUDs, these observations are consistent with the same allele(s) conferring increased risk for AUDs and nicotine dependence.
An Extended Region of High LD.
Examination of LD relations in the region revealed a 250-kb 19-SNP block showing high LD values in our sample (Fig. S1). As predicted by the high LD, the majority of these variants show associations with one or more of the LR phenotypes (Table 2). The associated genotypes reported here are estimated to account for a maximum of 2.9% of the total phenotypic variance in the sample; the estimate varies by marker and phenotype. These estimates are for the genotyped markers and may underestimate the quantitative affect of the true QTL if there is imperfect correlation between the marker and the causative variant. Specifically, it appears that virtually all the phenotypic variation explained by the ch15 locus is explained by SNP RS1051730. Based on the strength of association and the percentage of the phenotypic variance explained (Table 2), the marker RS1051730 thus remains, of the markers tested, the best tag for the QTL.
Although the phenotypic subgroups associated with the minor allele homozygotes and heterozygotes of the SNP markers may reflect the presence of more than one QTL in the region, the simplest hypothesis is that there is a single QTL and that the phenotypic subgroups derive from coupling either the major or minor allele of the marker SNP, with the QTL allele conferring low LR.
Multiple Phenotypes Behave in Coordinate Fashion.
The LR phenotypes are generally consistent with one another, suggesting that the same QTL may jointly affect both the coordination and subjective response systems (e.g., the genotype associated with a low LR, as measured by decreased BSA, is also associated with a decrease in the SHAS value [a lower subjective response] and an increase in the number of drinks required to obtain a defined alcohol effect [SREF], which includes both coordination and subjective response elements). Marker RS1051730 provides the clearest example of this pattern. These findings strengthen the hypothesis that the phenotypes making up the observed LR can be coordinate in their response to genetic variants. This is important, because previous linkage studies were ambiguous in this area, often indicating discrepancies among the several phenotypes (9, 11).
Region Carries Multiple Genes.
The 250-kb associated genomic region contains six genes: IREB2, LOC123688, PSMA4, CHRNA5, CHRNA3, and CHRNB4. Because of the high degree of LD among markers observed in our sample, it is not possible to localize the QTL(s) identified by our data to a single gene based solely on genetic considerations.
The three nicotinic acetylcholine receptor genes, however, are the most conspicuous candidates because their known function is the most consistent with nicotine and alcohol dependence phenotypes. The nicotinic acetylcholine receptors are widely distributed throughout the nervous system, controlled by the endogenous ligand ACh and modulated by the exogenous agonist nicotine. In vitro studies indicate that the nicotinic receptors are also modulated by alcohol (25), which, as with nicotine, increases receptor activity. Twin studies have also shown that inherited susceptibilities to nicotine and alcohol dependence are strongly correlated (26).
A frequent amino acid substitution, Asn398Asp, in the CHRNA5 gene has previously been implicated in nicotine dependency (18), although this variant did not show significant association in a subsequent alcohol dependence study (24). The nicotine dependence report, however, did note that there was a high degree of LD between the sentinel marker RS1051730 and the SNP RS16969968, which encodes the Asn398Asp substitution. We therefore examined RS16969968 by DNA sequencing in our sample and found 99% genotype concordance between the two markers (data not shown).
The Asn398Asp substitution is an intriguing candidate for further exploration as a potential causative variant in alcohol LR. Although functional data are not available for the Asn398Asp substitution, a variant in the mouse a4 gene (Ala529Thr) has been shown to alter nicotinic acetylcholine receptor (nAChR) function in mice in response to nicotine exposure (25, 27–29). Both variants are in the second intracellular loop of the protein.
Candidacy arguments for the other three genes are weaker. LOC123688 is a hypothetical protein of no known function, but it may have kinase and transferase activity. IREB2 encodes a posttranscriptional regulator of iron regulatory proteins through binding of a mRNA hairpin structure, the iron-responsive element (30). Iron deficiency during development can have cognitive and behavioral effects (31). PSMA4 encodes a subunit of the proteasome. Genes involved in basic cellular metabolism can have alleles resulting in behavioral phenotypes, such as the self-mutilating behavior caused by hypoxanthine-guanine phosphoribosyltransferase deficiency in Lesch-Nyhan syndrome (32). Determining which of these candidate genes harbors the addiction susceptibility variant will require additional genetic and functional experiments. Functional studies, especially in mouse models, will reveal the properties of the individual genes, the complexes they form, how sequence variation affects function, and, finally, how this variable function affects behavior.
LR to Alcohol as an Intermediate Phenotype for Genetic Studies of AUDs.
The findings reported here, combined with the recently published findings of association of the chromosome 15 locus with alcohol dependence (24), provide further evidence that alcohol LR is an endophenotype of AUDs. Alcohol LR was previously considered to be an AUD endophenotype because it is genetically influenced in animals (33–35) and humans (6–8) and is associated with AUDs in families and the population (5, 36–40). By demonstrating that a single genetic locus is associated with both alcohol dependence and alcohol LR, the importance of alcohol LR as a marker of risk for AUDs is strengthened and indicates that this endophenotype is a suitable target for genetic studies.
LR as an intermediate phenotype has also allowed us to see significant association results with a modest sample size. The 367 white siblings of the cohort characterized for LR phenotypes showed a P value for association of BSA with RS1051730 of 0.0002 and a q-value of 0.007. A recently reported study of alcohol dependence (24) had a P value of 0.016 and an associated q-value of 0.042 for this same SNP from the discovery population of ≈2,000 Collaborative Studies on Genetics of Alcoholism (COGA) family members. Although such P values will fluctuate from sample to sample of the same population, the findings reported here indicate that intermediate phenotypes, such as BSA, can yield associations as strong or stronger than those obtained from much larger sample sizes characterized for AUDs directly.
Methods
Subjects and Testing Protocol.
The San Diego Sibling Pair investigation is described in greater detail elsewhere (9, 11). Under a protocol approved by the Human Subjects Protection Committee of the University of California, San Diego, participants were chosen from among 18- to -29-year-old respondents to a questionnaire mailed to random students at the University of California, San Diego. The initial form, for which they were paid $5, was used to identify pairs of siblings (male and female) within the required age range who had consumed alcohol but did not meet alcohol dependence criteria. To be selected, at least one parent had to have repetitive alcohol-related life problems and meet the criteria for alcohol dependence using the Diagnostic and Statistical Manual of the American Psychiatric Association, Fourth Edition, Text Revision (41). Appropriate subjects were telephoned to review the questionnaire information and invited to a face-to-face interview, where they completed the SemiStructured Assessment for the Genetics of Alcoholism interview (42, 43); participated in an acclamation session regarding the alcohol challenge testing; completed the SREF questionnaire, as described further below; were scheduled for an alcohol challenge protocol; and were asked for 40 ml of whole blood for genetic analyses.
The alcohol challenge began with consumption of a 20% by volume solution of 0.75 ml/kg of ethanol (0.6 g/kg for women, and 0.90 ml/kg for men) within an 8-min period, with doses chosen to produce similar blood alcohol concentrations among individuals. At baseline, 15 min, 30 min, and every half hour after consuming the alcohol, subjects filled out the SHAS, indicating their feelings of intoxication on 13 items, with each rated on a 36-point scale indicating perceived subjective changes from baseline. Using the same time series, body sway was measured using a harness attached to the chest at the level of the axilla from which two perpendicular ropes extended forward and to the left side, passing over pulleys that measured the number of centimeters of movement per minute as gathered through three 1-min evaluations at each time point (44). Postalcohol measures were continued until 210 min after beverage consumption. The key scores used in these analyses include the SHAS score at the time of peak alcohol effect (60 min) and the BSA score at 60 min representing the average of the 3 1-min evaluations.
The SREF score used in this and most evaluations involved the subject's perception of the number of drinks required for up to four different effects obtained during the first five times of drinking. These included the number of standard (10–12 g of ethanol) drinks required to feel any effect, such as slurring of speech, feeling clumsy or unsteady on one's feet, or falling asleep when one did not wish to. Only those events actually experienced early in the drinking career are recorded, with the SREF score generated by adding the number of drinks and dividing the sum by the number of events experienced (45, 46).
For this study, 367 subjects were genotyped and tested for association. The subjects comprise 186 independent families: 38 single-sibling families, 121 two-sibling families, 23 three-sibling families, 3 four-sibling families, and a single six-sibling family. The actual number of subjects per marker-phenotype analysis varied because of missing genotype and phenotype data.
DNA Preparation.
DNA was extracted from blood specimens within 5 days of the draw. DNA was extracted using Gentra Puregene reagents and protocols (Qiagen). Extracted DNA was quantified using the Pico Green method (Molecular Probes/Invitrogen), and all stocks were normalized to a common concentration for genotyping assays.
Genotyping.
Genotyping of self-reported white siblings was carried out using two technologies. All SNPs reported were genotyped on the majority of subjects using the Illumina HumanCNV370-Duo DNA Analysis BeadChip. These genotypes were generated by deCODE Genotyping Service. Six of the SNP markers (RS1051730, RS1394371, RS2036534, RS3743079, RS3885951, and RS8034191) were genotyped again using Applied Biosystems TaqMan assays to confirm genotypes and to add additional subjects. One marker, RS2036534, was removed from analysis because of poor data concordance between the assays.
Analysis.
The pedigree data were examined for misspecifications with Pedigree Relationship Statistical Test (PREST) (47) using a panel of 740 Short Tandem Repeat (STR) marker genotypes. Any suspect pedigrees were removed from the analysis. A few apparent Mendelian errors in the SNP genotypes were detected with MERLIN (48) and set to missing. Phenotypes were corrected for nonnormality using the Box-Cox transformation (49–51) and scaled to mean = 0 and SD = 1. The tests of association were performed in R (51) with the lmekin function of the kinship package (52). This function provides a linear mixed effects model, whereby the genetic relatedness among individuals (based on the kinship coefficient) is incorporated into the covariance structure of the random effects. The fixed effect is used for the tests of association and adjustments for covariates. It included the covariate gender plus the test SNP (a factor of genotypes). Two contrasts were examined each with the Wald test: the major homozygote against the heterozygote and the major homozygote against the minor homozygote. The P values were corrected for multiple testing using the method of Benjamini and Yekutieli (53). The R2 statistic for the mixed models was calculated as RLR2 = 1 − exp(−(log(LM − logL0)), where LM is the maximum log-likelihood of the test model, logL0 is the maximum log-likelihood of the model with the intercept only, and n is the number of individuals (54). RLR2 accounts jointly for the variance attributed to the fixed and random effects. The percentage of phenotypic variance explained by a marker was estimated as the difference in RLR2 between the model with the marker and the model with only the gender term. A backward elimination procedure was used to select models with joint effects in the SNPs with respect to each phenotype. At every step in the selection procedure, SNPs were allowed to leave or rejoin the model on the basis of Akaike's information criterion, AIC = 2logLM + 2k, where LM is the maximum log-likelihood of the current model and k is the number of independently adjusted fixed effects terms (55). LD calculations were performed using the software package Haploview (56). A specific association between a SNP and a phenotype was also tested in the fixed effects component of the mixed model using terms for an additive effect and a dominance deviation. For the additive effect, a SNP genotype (AA, Aa, or aa) was coded as (−1, 0, or 1), and for the dominance deviation, a SNP genotype was coded as (0, 1, or 0), where a is the minor allele.
Acknowledgments.
We thank Andrew Lee, Chris Walker, Raymond Chui, Yabo Su, QianQian Zhu, Larry Kline, Nick Mosekyo, and Tena Sakai for their laboratory and analytical support; David Goldman for his insightful comments; and Claudia Yu for help in preparing the manuscript. This research was supported by funds provided by the State of California for medical research on alcohol and substance abuse through the University of California, San Francisco. This work was also sponsored by the Department of the Army under Award Nos. W81XWH-07–1-0077, W81XWH-08–1-0007, and W81XWH-08–1-0014. The U.S. Army Medical Research Acquisition Activity (820 Chandler Street, Fort Detrick, MD 21702-5014) is the awarding and administering acquisition office. The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810970105/DCSupplemental.
References
- 1.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 2.Gardner EJ. A genetic and clinical study of intestinal polyposis, a predisposing factor for carcinoma of the colon and rectum. Am J Hum Genet. 1951;3:167–176. [PMC free article] [PubMed] [Google Scholar]
- 3.Almasy L. Quantitative risk factors as indices of alcoholism susceptibility. Ann Med. 2003;35:337–343. doi: 10.1080/07853890310004903. [DOI] [PubMed] [Google Scholar]
- 4.Schuckit MA, Smith TL. Assessing the risk for alcoholism among sons of alcoholics. J Stud Alcohol. 1997;58:141–145. doi: 10.15288/jsa.1997.58.141. [DOI] [PubMed] [Google Scholar]
- 5.Schuckit MA, Smith TL. The relationships of a family history of alcohol dependence, a low level of response to alcohol and six domains of life functioning to the development of alcohol use disorders. J Stud Alcohol. 2000;61:827–835. doi: 10.15288/jsa.2000.61.827. [DOI] [PubMed] [Google Scholar]
- 6.Heath AC, Martin NG. Genetic differences in psychomotor performance decrement after alcohol: A multivariate analysis. J Stud Alcohol. 1992;53:262–271. doi: 10.15288/jsa.1992.53.262. [DOI] [PubMed] [Google Scholar]
- 7.Madden PA, Heath AC, Starmer GA, Whitfield JB, Martin NG. Alcohol sensitivity and smoking history in men and women. Alcohol Clin Exp Res. 1995;19:1111–1120. doi: 10.1111/j.1530-0277.1995.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 8.Martin NG, et al. Prodromus to a twin study of sensitivity to intoxication and alcohol metabolism. Aust N Z J Med. 1981;11:140–143. doi: 10.1111/j.1445-5994.1981.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 9.Wilhelmsen KC, et al. The search for genes related to a low-level response to alcohol determined by alcohol challenges. Alcohol Clin Exp Res. 2003;27:1041–1047. doi: 10.1097/01.ALC.0000075551.02714.63. [DOI] [PubMed] [Google Scholar]
- 10.Schuckit MA, et al. A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol Clin Exp Res. 2001;25:323–329. [PubMed] [Google Scholar]
- 11.Schuckit MA, et al. Autosomal linkage analysis for the level of response to alcohol. Alcohol Clin Exp Res. 2005;29:1976–1982. doi: 10.1097/01.alc.0000187598.82921.27. [DOI] [PubMed] [Google Scholar]
- 12.Batel P, Pessione F, Maitre C, Rueff B. Relationship between alcohol and tobacco dependencies among alcoholics who smoke. Addiction. 1995;90:977–980. doi: 10.1046/j.1360-0443.1995.90797711.x. [DOI] [PubMed] [Google Scholar]
- 13.Swan GE, Carmelli D, Cardon LR. The consumption of tobacco, alcohol, and coffee in Caucasian male twins: A multivariate genetic analysis. J Subst Abuse. 1996;8:19–31. doi: 10.1016/s0899-3289(96)90055-3. [DOI] [PubMed] [Google Scholar]
- 14.Swan GE, Carmelli D, Cardon LR. Heavy consumption of cigarettes, alcohol and coffee in male twins. J Stud Alcohol. 1997;58:182–190. doi: 10.15288/jsa.1997.58.182. [DOI] [PubMed] [Google Scholar]
- 15.Hettema JM, Corey LA, Kendler KS. A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population based sample of male and female twins. Drug Alcohol Depend. 1999;57:69–78. doi: 10.1016/s0376-8716(99)00053-8. [DOI] [PubMed] [Google Scholar]
- 16.Hopfer CJ, Stallings MC, Hewitt JK. Common genetic and environmental vulnerability for alcohol and tobacco use in a volunteer sample of older female twins. J Stud Alcohol. 2001;62:717–723. doi: 10.15288/jsa.2001.62.717. [DOI] [PubMed] [Google Scholar]
- 17.Steensland P, Simms JA, Holgate J, Richards JK, Bartlett SE. Varenicline, an alpha4beta2 nicotinic acetylcholine receptor partial agonist, selectively decreases ethanol consumption and seeking. Proc Natl Acad Sci USA. 2007;104:12518–12523. doi: 10.1073/pnas.0705368104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 20.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berrettini W, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bierut LJ, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JC, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.42. Apr 15 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butt CM, et al. A polymorphism in the alpha4 nicotinic receptor gene (Chrna4) modulates enhancement of nicotinic receptor function by ethanol. Alcohol Clin Exp Res. 2003;27:733–742. doi: 10.1097/01.ALC.0000067973.41153.BC. [DOI] [PubMed] [Google Scholar]
- 26.True WR, et al. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56:655–661. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- 27.Butt CM, King NM, Hutton SR, Collins AC, Stitzel JA. Modulation of nicotine but not ethanol preference by the mouse Chrna4 A529T polymorphism. Behav Neurosci. 2005;119:26–37. doi: 10.1037/0735-7044.119.1.26. [DOI] [PubMed] [Google Scholar]
- 28.Stitzel JA, Dobelis P, Jimenez M, Collins AC. Long sleep and short sleep mice differ in nicotine-stimulated 86Rb+ efflux and alpha4 nicotinic receptor subunit cDNA sequence. Pharmacogenetics. 2001;11:331–339. doi: 10.1097/00008571-200106000-00008. [DOI] [PubMed] [Google Scholar]
- 29.Dobelis P, et al. A polymorphism in the mouse neuronal alpha4 nicotinic receptor subunit results in an alteration in receptor function. Mol Pharmacol. 2002;62:334–342. doi: 10.1124/mol.62.2.334. [DOI] [PubMed] [Google Scholar]
- 30.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: Molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 31.Beard J. Iron deficiency alters brain development and functioning. J Nutr. 2003;133(Suppl):1468S–1472S. doi: 10.1093/jn/133.5.1468S. [DOI] [PubMed] [Google Scholar]
- 32.Seegmiller JE, Rosenbloom FM, Kelley WN. Enzyme defect associated with a sex-linked human neurological disorder and excessive purine synthesis. Science. 1967;155:1682–1684. doi: 10.1126/science.155.3770.1682. [DOI] [PubMed] [Google Scholar]
- 33.Baldwin HA, Wall TL, Schuckit MA, Koob GF. Differential effects of ethanol on punished responding in the P and NP rats. Alcohol Clin Exp Res. 1991;15:700–704. doi: 10.1111/j.1530-0277.1991.tb00582.x. [DOI] [PubMed] [Google Scholar]
- 34.Li TK. Pharmacogenetics of responses to alcohol and genes that influence alcohol drinking. J Stud Alcohol. 2000;61:5–12. doi: 10.15288/jsa.2000.61.5. [DOI] [PubMed] [Google Scholar]
- 35.Moore MS, et al. Ethanol intoxication in Drosophila: Genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- 36.Erblich J, Earleywine M. Children of alcoholics exhibit attenuated cognitive impairment during an ethanol challenge. Alcohol Clin Exp Res. 1999;23:476–482. [PubMed] [Google Scholar]
- 37.Pollock VE. Meta-analysis of subjective sensitivity to alcohol in sons of alcoholics. Am J Psychiatry. 1992;149:1534–1538. doi: 10.1176/ajp.149.11.1534. [DOI] [PubMed] [Google Scholar]
- 38.Schuckit MA, Smith TL. An 8-year follow-up of 450 sons of alcoholic and control subjects. Arch Gen Psychiatry. 1996;53:202–210. doi: 10.1001/archpsyc.1996.01830030020005. [DOI] [PubMed] [Google Scholar]
- 39.Schuckit MA, et al. Response to alcohol in daughters of alcoholics: A pilot study and a comparison with sons of alcoholics. Alcohol Alcohol. 2000;35:242–248. doi: 10.1093/alcalc/35.3.242. [DOI] [PubMed] [Google Scholar]
- 40.Schuckit MA, Tsuang JW, Anthenelli RM, Tipp JE, Nurnberger JI., Jr Alcohol challenges in young men from alcoholic pedigrees and control families: A report from the COGA project. J Stud Alcohol. 1996;57:368–377. doi: 10.15288/jsa.1996.57.368. [DOI] [PubMed] [Google Scholar]
- 41.American Psychiatric Association. American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision, (DSM-IV-TR) Arlington, VA: American Psychiatric Press; 2000. [Google Scholar]
- 42.Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA—A comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- 43.Bucholz KK, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: A report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- 44.Schuckit MA, Gold EO. A simultaneous evaluation of multiple markers of ethanol/placebo challenges in sons of alcoholics and controls. Arch Gen Psychiatry. 1988;45:211–216. doi: 10.1001/archpsyc.1988.01800270019002. [DOI] [PubMed] [Google Scholar]
- 45.Schuckit MA, Tipp JE, Smith TL, Wiesbeck GA, Kalmijn J. The relationship between self-rating of the effects of alcohol and alcohol challenge results in ninety-eight young men. J Stud Alcohol. 1997;58:397–404. doi: 10.15288/jsa.1997.58.397. [DOI] [PubMed] [Google Scholar]
- 46.Schuckit MA, et al. The ability of the Self-Rating of the Effects of Alcohol (SRE) scale to predict alcohol-related outcomes five years later. J Stud Alcohol Drugs. 2007;68:371–378. doi: 10.15288/jsad.2007.68.371. [DOI] [PubMed] [Google Scholar]
- 47.Sun L, Wilder K, McPeek MS. Enhanced pedigree error detection. Hum Hered. 2002;54:99–110. doi: 10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- 48.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 49.Box GEP, Cox DR. An analysis of transformations (with discussion) J R Stat Soc B. 1964;26:211–252. [Google Scholar]
- 50.Venables WN, Ripley BD. Modern Applied Statistics with S. New York: Springer; 2002. [Google Scholar]
- 51.Team RDC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 52.Atkinson B, Therneau T. Kinship: Mixed-effects Cox models, sparse matrices, and modeling data from large pedigrees. Rochester, MN: Mayo Foundation for Medical Education and Research; 2008. [Google Scholar]
- 53.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
- 54.Manhattan, KS: Kansas State University; Master's thesis. [Google Scholar]
- 55.Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;AC-19:716–723. [Google Scholar]
- 56.Barrett JC, Fry B, Maller J, Daly M J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]