Abstract
Visual system development requires experience-dependent mechanisms that regulate neuronal structure and function, including dendritic arbor growth, synapse formation, and stabilization. Although RNA binding proteins have been shown to affect some forms of synaptic plasticity in adult animals, their role in the development of neuronal structure and functional circuitry is not clear. Using two-photon time-lapse in vivo imaging and electrophysiology combined with morpholino-mediated knockdown and expression of functional deletion mutants, we demonstrate that the mRNA binding protein, cytoplasmic polyadenylation element binding protein1 (CPEB1), affects experience-dependent neuronal development and circuit formation in the visual system of Xenopus laevis. These data indicate that sensory experience controls circuit development by regulating translational activity of mRNAs.
Keywords: circuit integration, experience-dependent plasticity, in vivo imaging, visual system, whole-cell recording
During CNS development, neuronal structure, synaptic connections, and circuit functions change in response to sensory input. Although the regulation of mRNA translation and new protein synthesis have been implicated in some forms of neuronal plasticity in adult animals (1, 2), their role in experience-dependent sensory system plasticity has not been addressed (3). In the brain, the translation of many mRNAs is regulated by their association with ribonucleoprotein (RNP) granules, which are composed of core RNA binding proteins, translational machinery, and mRNAs (4). RNA binding proteins package specific mRNAs within RNP granules, regulate their transport into dendrites, and in principle, could temporally and spatially govern their translation in response to synaptic activity (4). Because of these features, mRNA binding proteins are in a unique position to control developmental events through the coordinated translation of a functionally related cohort of mRNAs (5, 6). Indeed, local protein synthesis affords neurons the ability to stabilize changes in synaptic connections and alter neuronal output (7). Recent studies indicate that cytoplasmic polyadenylation element binding protein 1 (CPEB1) is among a handful of RNA binding proteins that regulate dendritic protein synthesis and synaptic plasticity in vitro (8). Although studies in mutant mice implicate CPEB1 in some forms of synaptic and spine plasticity and in memory extinction (9–11), no studies have examined the potential role of CPEB1 in dendritic arbor development, experience-dependent structural plasticity, or the integration of neurons into a functional circuit in an intact animal.
CPEB1 (Orb in D. melanogaster) is a member of an evolutionarily conserved family of RNA binding proteins that is expressed in the brain (in vertebrates, CPEB1–4). While all members are defined by the presence of RNA recognition motifs in their carboxy-terminals, CPEB1 is the only member that targets mRNAs containing cytoplasmic polyadenylation elements (CPEs) in their 3′ untranslated regions (12). Originally discovered in Xenopus laevis oocytes, CPEB1 binds CPE-containing mRNAs, regulates their microtubule-dependent dendritic trafficking, and represses their translation (13). Unlike vertebrate CPEB3 and 4, ApCPEB in Aplysia (14, 15), or Drosophila Orb2 (16), translational repression by CPEB1 is relieved by phosphorylation by either aurora kinase (17, 18) or calcium/calmodulin-dependent kinase II (CaMKII) (19), following stimulation by progesterone in oocytes or glutamatergic synaptic activity in neurons. Phosphorylation of CPEB1 initiates a biochemical cascade that culminates in polyadenylation of the CPEB1-bound mRNAs and their translational derepression (13).
Distinct requirements for the functional domains of CPEB governing RNA transport and phosphorylation-dependent translational regulation have not been analyzed. Although CPEB mouse knockouts are deficient in some forms of plasticity (9, 10), because both CPEB's translational repression and mRNA transport functions are decreased in the knockout mice, it is unclear whether these distinct CPEB functions contribute specifically to the knockout phenotypes. Expression of a CPEB1 phosphorylation mutant in mouse Purkinje neurons prevented the late protein-synthesis-dependent phase of cerebellar long-term depression and altered dendritic spine morphology (11). This study suggests that CPEB-mediated translational derepression regulates synaptic plasticity, but it remains unclear whether the mRNA transport and translational regulation functions of CPEB play distinct roles in neuronal development and experience-dependent plasticity in vivo.
During Xenopus CNS development, glutamatergic signaling regulates the experience-dependent assembly and refinement of the visual circuitry. Dendritic arbor development of the retinorecipient neurons in the optic tectum is regulated by NMDA receptor and AMPA receptor signaling and affected by the visual experience of the animal (20, 21). The development of the dendritic arbor structure and maturation of tectal cell synaptic physiology progress coordinately and are governed by molecular pathways that include numerous proteins, such as αCaMKII (22) and brain-derived neurotrophic factor (BDNF) (23). Interestingly, the mRNA of both αCaMKII and BDNF are targets of CPEB1 (18, 24, 25), suggesting that CPEB may mediate aspects of activity-dependent visual system development. Indeed, studies in vivo have demonstrated that brief light exposure delivered to dark-reared rats produced CPEB-dependent increases in αCaMKII mRNA polyadenylation and protein synthesis that required NMDA receptor signaling (18, 25). We used a combination of in vivo imaging, electrophysiological measures of synaptic physiology, and evoked visual responses to assess the requirement for CPEB1 in the visual system development and plasticity. Furthermore, by expressing deletion mutants of CPEB1 we determined the relative importance of activity-dependent derepression of protein translation and mRNA trafficking in dendritic arbor development, synaptic physiology, and integration of neurons into functional circuits in vivo.
Results
CPEB is Required for Dendrite Development.
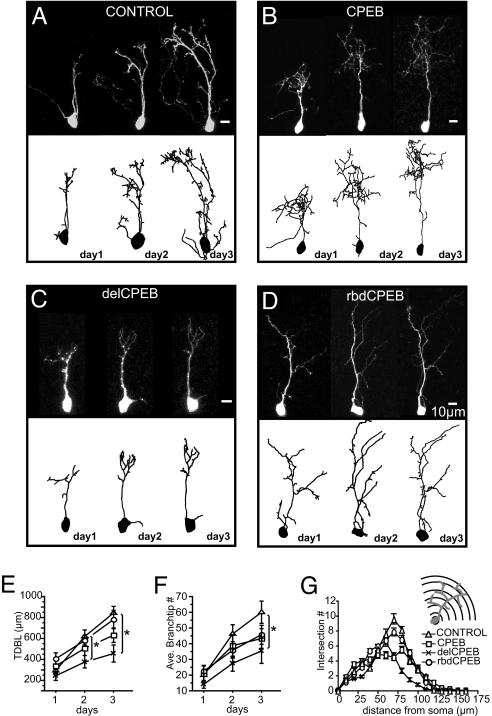
To test whether CPEB1 plays a role in the elaboration of the dendritic arbor, we transfected individual optic tectal neurons with control morpholinos and antisense morpholinos against CPEB1, using electroporation. Time-lapse images of transfected optic tectal neurons in an otherwise normal brain were collected once a day over 3 days, starting 1 day after electroporation [Fig. 1 A–D and supporting information (SI) Fig. S1]. An important feature of this experimental protocol is that neurons were already differentiated at the onset of these experiments; they had a dendritic arbor and synaptic inputs. Consequently, we assessed the effect of CPEB1 on experience-dependent circuit assembly, rather than neuronal differentiation or early stages of neuron development. Three-dimensional (3D) reconstructions show that although the dendritic arbors were matched in size on the first day of analysis, expression of morpholino antisense oligonucleotides directed against CPEB1 significantly decreased arbor development over 3 days and produced cells that were roughly two-thirds the size of the control morpholino group in their total dendritic branch length (TDBL) (see Fig. S1 and Table S1). Compared to the control morpholino cells, which grew 317 ± 79% of the starting size by day 3, the growth rate of the CPEB1 morpholino-expressing neurons was significantly inhibited and their TDBL reached just 156 ± 45% of their initial values (P = 0.05).
Fig. 1.
CPEB is required for dendritic arbor growth in vivo. (A–D) Time-lapse in vivo two-photon images of tectal neurons and their three-dimensional (3D) reconstructions, collected daily over 3 days. (E-G) Quantification of dendritic morphology showing total dendritic branch length (TDBL) (E), branch-tip number (F), and 3D Sholl analysis (G). (E) Compared to control cells, the dendritic growth of delCPEB-expressing neurons, but not CPEB or rbdCPEB-expressing neurons, is significantly impaired by day 2 and 3. (F) By day 3, only delCPEB-expressing neurons have significantly fewer branch tips compared to control neurons. (G) Sholl analysis of 3-day cells illustrating the radial distribution of the branches of the dendritic arbor. Shown are the average number of branch intersections at 10-μm intervals beyond the soma. Control, n = 11; CPEB, n = 12; delCPEB n = 10; rbdCPEB = 12; *, P ≤ 0.05. Values given in Table S2.
Knocking down CPEB1 protein with morpholinos did not allow us to distinguish whether CPEB1's function in microtubule-dependent mRNA transport or in activity-dependent polyadenylation was important for dendritic development. Based on previous studies of CPEB1 protein's functional domains (26–28), we designed experiments to distinguish the role of CPEB1 in the regulation of protein translation independently from its role in mRNA transport. We compared neurons expressing YFP, full-length CPEB1 (CPEB), and two previously characterized deletion mutants (Fig. S2). The first consisted of the CPEB RNA binding domain without the regulatory domains of the protein (rbdCPEB, Δ1–258). This truncated CPEB1 protein interferes with both CPEB-dependent dendritic mRNA transport and protein synthesis (28). The second deletion mutant (delCPEB; Δ124–258) lacks the phosphorylation site targeted by aurora kinase and αCaMKII. Because it is not activated by phosphorylation (26, 27), delCPEB can stably bind and transport mRNA (28) but interferes with the activity-dependent polyadenylation and translation initiation of CPEB mRNA targets. We verified in the retinotectal system that expression of delCPEB interferes with CPEB-dependent protein synthesis in a blind study that compared tissue plasminogen activator (tPA) immunofluorescence in tectal cells expressing delCPEB, CPEB1, or YFP alone with untransfected neighboring neurons (Fig. S3). With these tools, we address the question of whether CPEB1 is required for development of the visual system structure and function in X. laevis tadpoles.
While neurons expressing rbdCPEB, delCPEB, CPEB, and YFP alone were comparable in their TDBL and branch-tip number on the first day of imaging, delCPEB-expressing neurons grew at about half the rate of control cells, increasing only 104 ± 35% of their starting size compared to YFP control cells, which increase their TDBL by 240 ± 40% by day 3 (P = 0.01) (see Fig. 1). By the third day of imaging, delCPEB-expressing neurons had roughly half the TDBL and branch-tip number of control neurons (see Fig. 1 E and F and Table S2). Three-dimensional Sholl analysis (29) revealed that delCPEB-expressing neurons have fewer branches extending beyond 60 μm from the soma (see Fig. 1F). In contrast, while rbdCPEB-expressing neurons did not significantly differ from the control cells in their TDBL or branch numbers over the 3 days (126 ± 39%) (see Fig. 1 E and F and Table S2), their growth rate was significantly slower than controls (P = 0.03). Neurons expressing CPEB were not significantly different from control cells with respect to TDBL, branch-tip number (see Fig. 1 E and F and Table S2) or growth rate (169 ± 54% increase by day 3; P = 0.08). These data indicate that delCPEB, which is incorporated into dendritically targeted RNPs but cannot be phosphorylated and initiate mRNA polyadenylation downstream of extracellular signals, severely impairs dendritic arbor development. By contrast, cells expressing rbdCPEB, which lacks the microtubule-binding domain and therefore is retained in the cell body, can acquire normal dendritic arbor structure, although with slow growth rates.
delCPEB and rbdCPEB Expression Blocks Experience-Dependent Growth.
Optic tectal neurons, like other CNS neurons, show experience-dependent structural plasticity. Exposure to NMDA receptor blockers reduces tectal-cell dendritic-arbor growth rates (30). Enhanced visual stimulation delivered to freely swimming tadpoles for 4 h increases synaptic drive onto optic tectal neurons (31) and significantly increases dendritic-arbor growth rate (21). This activity-dependent dendritic growth operates by an NMDA receptor-dependent mechanism (21). Although NMDA receptor signaling and downstream kinase activity increased CPEB-dependent mRNA polyadenylation and protein synthesis in the rodent visual system (18, 25) and hippocampal neurons (17, 19), CPEB1 has not yet been implicated in experience-dependent structural plasticity.
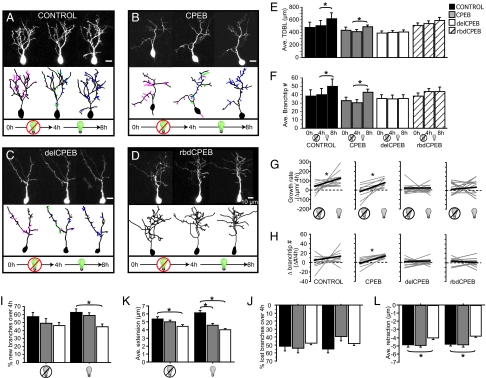
Interfering with CPEB function through the expression of delCPEB or rbdCPEB severely impaired experience-dependent dendritic arbor growth (Fig. 2). Two-photon time-lapse images of neurons in living tadpoles were collected before and after a 4-h period without visual stimulation, and then a final image was collected after the animal was exposed to 4 h of enhanced visual stimulation (21). Images of representative neurons and 3D renderings, color-coded to indicate branch dynamics, are shown in Fig. 2 A to D. The delCPEB- and rbdCPEB-expressing neurons failed to show the experience-dependent increased growth rate seen in control neurons or neurons expressing CPEB (see Fig. 2 E and F; Tables S3 and S4). Nearly all of the control and CPEB-expressing neurons responded to the visual stimulation with increased TDBL (12 out of 12 and 10 out of 10, respectively) and branch-tip number (10 out of 12 and 10 out of 10, respectively). By contrast, only half of the neurons in the delCPEB and rbdCPEB groups increased TDBL (6 out of 14 and 7 out of 11, respectively) or branch tip number (7 out of 14 and 5 out of 11, respectively) (see Fig. 2 G and H) with visual stimulation.
Fig. 2.
CPEB function is required for experience-dependent increased dendritic growth. (A-D) Time-lapse in vivo two photon images collected at 4-h intervals over 8 h, and 3D reconstructions showing terminal branches that are lost (pink), appeared transiently at the fourth hour (green), and are gained (blue) over the 4-h periods without or with visual stimulation (indicated with light-bulb icon) for tectal neurons. (E–H) Quantification of dendritic morphology. During the 4 h of light stimulation, control and CPEB-expressing neurons show a significant increase in TDBL (E) and branch-tip number (F). Individual cell's changes in TDBL (G) and branch-tip number (H) are plotted in gray, averages are plotted in black. Control and CPEB-expressing cells show significant changes in TDBL. Only CPEB-expressing cells show a significant change in branch-tip number. (I–L) Quantification of identified branch dynamics. Proportion of branches newly added (I) and lost (J) during each 4-h. (I) delCPEB-expressing cells add significantly fewer branches during the light-stimulation period compared to control neurons. There were no differences in the proportion of branches lost between groups (J). (K) The average increase in branch length of delCPEB cells is less than that of control cells in both the dark and during the light stimulus. CPEB cells also added significantly less average branch length during the visual stimulation period. (L) The average amount that delCPEB-retracting branches lost was less than control cells in both the dark and during the light stimulation. Control, n = 12; CPEB, n = 10; delCPEB, n = 14; rbdCPEB = 11; *, P < 0.05. Values given in Tables S3–S5.
Because the delCPEB-expressing neurons showed more severe growth defects than rbdCPEB-expressing neurons, we investigated the branching behavior underlying the differences between delCPEB-, CPEB-, and YFP-expressing control neurons. We examined the dynamic growth of these neurons by measuring the same branches at each time point of the experiment (see Fig. 2 I–L and Table S5). This analysis revealed that the decreased activity-dependent dendritic growth seen in delCPEB-expressing cells could be attributed to both a reduction in rates of branch additions and branch-length extension. The delCPEB-expressing neurons added one-third fewer branches during the 4-h visual stimulation period compared to control neurons [see Figs. 2 A, C (blue branches) and I]. There was no significant change in the proportion of lost branches, [see Fig. 2 A, C (pink branches), J]. Branches in the delCPEB-expressing neurons extended and retracted less than control neurons during each 4-h window (see Fig. 2 K and L). Branches in CPEB- and delCPEB-expressing neurons failed to extend as much as control cells during the visual stimulation, but branches in delCPEB-expressing neurons were more severely affected, extending only two-thirds as much as control neurons (see Fig. 2K). These data establish a role for CPEB1 in experience-dependent elaboration of dendritic arbor structure. The analysis also suggests that the mechanism by which CPEB1 affects dendritic arbor elaboration is through an effect on rates of branch additions.
delCPEB Expression Reduces the Strength of Glutamatergic Synapses.
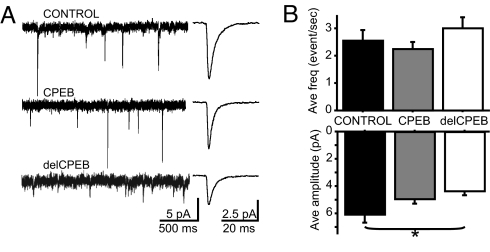
As neurons elaborate a complex dendritic arbor, they also form and strengthen glutamatergic retinotectal synapses by mechanisms that are enhanced by visual stimulation (31) and require NMDA receptor and CaMKII activity (22, 32). Furthermore, AMPA receptor- and NMDA receptor-mediated transmission is required for normal dendritic arbor development (20, 33). Because delCPEB-expressing neurons showed the most severe morphological defects, we tested whether delCPEB expression affects glutamatergic synaptic transmission. We recorded AMPA receptor-mediated miniature excitatory postsynaptic currents (mEPSCs) from control, CPEB-, and delCPEB-expressing neurons, 2 days after electroporation (Fig. 3A). The delCPEB-expressing neurons had smaller amplitude mEPSCs than control neurons (4.4 ± 0.3 pA and 6.0 ± 0.5 pA, P = 0.03) (Fig. 3B), while mEPSCs from CPEB-expressing neurons were not significantly different from control cells (4.9 ± 0.3 pA, P > 0.05) (see Fig. 3B). There were no significant differences in the frequency of mEPSCs recorded from control cells, CPEB- or delCPEB-expressing cells (2.6 ± 0.4, 2.3 ± 0.3, and 3.0 ± 0.4 events per second, respectively) (see Fig. 3B). These data suggest that CPEB activity affects synapse strength but may not alter synapse number.
Fig. 3.
Altering CPEB function reduces amplitude but not frequency of AMPA receptor mEPSCs. (A) Representative traces and examples of average mEPSCs. (B) Quantification of mEPSC amplitude (*, P < Control, n = 32; CPEB, n = 28; delCPEB n = 31.
CPEB Regulates Tectal Cell Performance in the Visual Circuit.
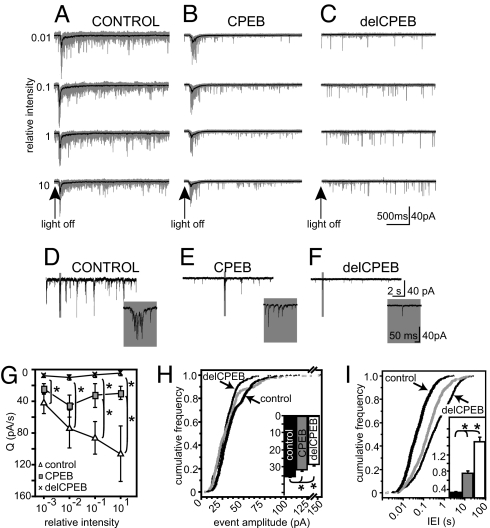
The severe effect of delCPEB expression on dendritic arbor structure, combined with the decrease in mEPSC amplitude, suggests that CPEB1 affects the performance of neurons within brain circuits. To address this, 2 days after electroporation we recorded whole-cell responses from tectal neurons when the intact animals were exposed to full-field visual stimuli of increasing light intensity (Fig. S4). In the optic tectum, neurons show a stronger response to light-off than light-on stimuli (34); therefore, we show data from the light-off responses. These visual responses in control neurons include direct monosynaptic glutamatergic inputs, as well as polysynaptic activity contributed by local tectal inputs (34). To focus our analysis on direct retinal input, we compared the initial 500 ms of the light-evoked compound synaptic currents from neurons expressing CPEB and delCPEB to the responses from control neurons. delCPEB-expressing neurons showed significantly attenuated responses to visual stimuli, as measured as the average charge transfer (Q) (Fig. 4G). By contrast, control neurons displayed increasingly stronger responses as the intensity of the stimulus increased. CPEB-expressing neurons did not show the same magnitude of response as control neurons and were deficient in charge transfer at the two higher stimulus intensities, but responded comparably to control neurons when the stimulus intensities were relatively low (Fig. 4 A–C, G and Tables S6 and S7).
Fig. 4.
Altering CPEB expression interferes with tectal cell performance in the visual circuit. (A–C) Whole-cell recordings of visual responses over the complete 2.5 s of the off responses. Each column shows recordings from the same cell for each 10-dB increase in intensity. Twenty responses are superimposed in gray with the average response in black. (D–F) Spontaneous activity recorded during the dark-adaptation period. Gray areas are expanded in the insets. Bursting activity is only seen in control cells. (G) Quantification of average charge transferred (Q) over 500 ms following light off for each stimulus intensity. Compared to control neurons, delCPEB-expressing cells showed significantly lower Q for each intensity tested. CPEB cells were significantly different at the stronger relative intensities. n = 6–10 cells for each intensity for control, CPEB and delCPEB groups. (H and I) Quantification of spontaneous synaptic activity recorded in the absence of TTX. Distribution and means (inset) for the single event amplitudes (H) and inter-event intervals (IEI) (I) were significantly shifted toward smaller values for both delCPEB and CPEB-expressing neurons compared to control cells. n = 1,190 events and 10 neurons per cell group. *, P < 0.05. Values given in Tables S6 and S7.
As an independent assessment of the integration of tectal neurons into the visual circuit, we recorded their spontaneous activity levels in vivo (Figs. 4 D–F). The average amplitude of the spontaneous EPSCs was significantly smaller in the CPEB- and delCPEB-expressing neurons compared to control neurons, and the distribution of amplitudes was significantly different between the control cells and CPEB- or delCPEB-expressing neurons (Fig. 4H and Table S7). Control neurons also have a significantly greater proportion of responses with short inter-event intervals (IEI) than CPEB and delCPEB-expressing neurons (Fig. 4I). In addition, we recorded bursts (see Materials and Methods) from 8 out of 12 control neurons (see Fig. 4D, inset) and 5 out of 11 CPEB-expressing cells (see Fig. 4E, inset) but failed to detect any bursts in all but 1 out of 13 delCPEB-expressing cells (see Fig. 4F, inset).
Discussion
We used multiple strategies to investigate CPEB1 function in the activity-dependent development of neurons in the optic tectum. Time-lapse in vivo imaging of CPEB morpholino expressing neurons demonstrated that CPEB1 is required for dendritic arbor elaboration, while expression of mutants lacking functional domains of CPEB demonstrated selective roles for translational regulation and mRNA transport in experience-dependent structural plasticity and dendrite development. Whole-cell recordings of visual responses and spontaneous bursting activity in tectal neurons in the intact animal demonstrate that CPEB1 function is required for the integration of neurons into the visual circuit. The results support a model in which synaptic activity leads to phosphorylation of CPEB and translational derepression of target mRNAs (18, 19, 24, 35). The newly translated proteins then act as effectors to control experience-dependent modifications of dendritic arbor structure, synaptic connectivity, and ultimately the function and plasticity of the developing circuit.
Expression of CPEB or the deletion mutants likely sequesters mRNA from endogenous CPEB and the translation machinery. Additionally, because distinct spatial and temporal patterns of input activity may differentially regulate translational derepression by mRNA binding proteins (36), the expression of the CPEB1 constructs may also alter the threshold of input activity necessary for CPEB1-dependent protein synthesis and, therefore, place the transfected neurons at a specific disadvantage with respect to circuit integration compared to neighboring control tectal cells.
It is interesting to note that the deletion mutants of CPEB1 distinguished the requirements for the dendritic mRNA transport and phosphorylation-dependent polyadenylation functions of CPEB in the elaboration of dendritic arbors and in experience-dependent structural plasticity. Because the molecular structure of CPEB has been thoroughly examined (26, 27), further structure/function analysis with previously characterized deletion mutants of CPEB allowed us to identify which CPEB functions are most critical for different aspects of neuronal development and circuit function. Using time-lapse imaging, we found that expressing delCPEB, which interferes with CPEB's activity-dependent mRNA polyadenylation but leaves dendritic mRNA transport intact, impairs dendritic-arbor development and the integration of neurons into the visual circuit. By contrast, neurons expressing rbdCPEB, which have disrupted mRNA transport in addition to altered polyadenylation activity, can acquire normal dendritic arbor structure over 3 days, but fail to show rapid enhanced dendritic arbor growth rates following brief visual stimulation. The results suggest that failure of CPEB1 to transport mRNA into dendrites can be compensated over a timecourse of days, perhaps by transport of CREB mRNA targets by other RNA binding proteins or other means of regulating protein level, but that failure of CPEB1 to respond to synaptic activity is quite devastating for regulation of dendrite development over days, and for short-term structural plasticity in response to sensory input.
About 7% of brain mRNAs are estimated to be targets of CPEB1, although only a relatively small number have been confirmed experimentally (24). Potential mRNA targets include key plasticity genes, such as αCaMKII (18, 25), BDNF (24), tPA (37), engrailed1 (38), Homer (18), and insulin-receptor substrate p53 (11). Proteins from these mRNAs, plus many identified in a recent screen of CPE-containing mRNAs (24), are capable of altering synaptic strength and neuronal structure. Nevertheless, it is unlikely that the effects of CPEB1 manipulations that we document here can be attributed to misregulation of a single CPEB mRNA target. For instance, αCaMKII, BDNF, and Homer proteins have distinct but overlapping functions in the morphological and electrophysiological development and plasticity of optic tectal neurons (22, 23, 32, 39). The role that αCaMKII plays in CNS development and plasticity illustrates how activation of just one of the CPEB1 target mRNAs can have pleiotropic effects that have ramifications beyond its direct mRNA targets. Increasing or decreasing CaMKII activity affects synaptic plasticity and dendritic-arbor development of neurons in the X. laevis optic tectum (32) and mammalian systems, where it is also required for learning and memory (40). CaMKII phosphorylates CPEB1 in response to NMDA receptor activity (19) and its own synthesis increases following plasticity-inducing stimuli (41), NMDA receptor activation, and CPEB1 activation (18). Because αCaMKII both activates CPEB1 and is also a downstream product of CPEB1 activity, αCaMKII may act in a feedback loop where synaptic input that activates αCaMKII can also derepress CPEB-mediated inhibition of αCaMKII synthesis (35). Therefore CPEB1-mediated control of αCaMKII synthesis alone is likely to have both immediate and long-lasting effects on functional and structural plasticity, as well as subsequent regulation of CPEB1, which likely work in concert with the other CPEB1 target mRNAs that are associated with synaptic and morphological plasticity (24).
Specific RNA binding proteins may coordinate the spatial and temporal translation of functionally related subpopulations ofmRNAs (5, 6). Furthermore, different mRNA binding proteins bind different, but potentially overlapping sets of mRNAs, and may independently regulate their translation in response to specific inputs. For example, fragile X mental retardation protein, FMRP, is a dendritically targeted mRNA binding protein, which is activated in response to metabotropic glutamate receptor activity and is thought to regulate forms of long-term depression (42). It is interesting to note that fmr1, the gene encoding FMRP, has a CPE site and colocalizes with CPEB in RNP granules (43, 44), suggesting that synaptic activity that activates CPEB1 may also increase FMRP translation. Combined with evidence for posttranscriptional gene regulation by RNA binding proteins and microRNAs (5, 45), the examples described here show that local regulation of mRNA translation affords considerable spatial and temporal control over synaptic plasticity.
The results presented here demonstrate a requirement for CPEB1-mediated translational regulation for the integration of neurons into a functional circuit in vivo. Not only is CPEB1 function required for neurons to modify their morphological and synaptic development in response to changes in sensory input, but by interfering with CPEB1's ability to interpret activity-dependent signals, tectal neurons do not integrate properly and effectively lose their place in the development of an otherwise unaltered visual circuit.
Materials and Methods
Cell Transfection of Plasmid Constructs and Morpholino Antisense Oligonucleotides.
Stage 46 to 48 albino X. laevis tadpoles were used for all experiments. All protocols were approved by the Cold Spring Harbor Laboratory Institutional Animal Care and Use Committee. For the electrophysiology and morpholino-imaging experiments, cells were transfected by electroporation as described previously (46). For the in vivo imaging experiments, single cells were targeted with electroporation as described previously (47) and in the SI Methods.
The lissamine-tagged morpholino antisense oligonucleotides against CPEB were designed by and purchased from GeneTools (Philomath, OR). The CPEB, delCPEB and rbd-eCFP (referred to as Δ5 and RBD in ref. 28) plasmid constructs were a generous gift of Dr. Joel D. Richter (University of Massachusetts Medical School, Worcester, MA). The original eGFP tags were replaced with eCFP and the genes were transferred to a bidirectional PCS2 plasmid. This plasmid contained two independent CMV promoters, one driving untagged, cytosolic eYFP to visualize the transfected cell (kindly provided by Dr. David Turner, University of Michigan, Ann Arbor, MI).
Two-Photon Imaging and Morphometric Analyses.
Images were collected from anesthetized tadpoles (0.02% MS-222; Sigma) positioned under a glass coverslip in a Sylgard chamber. Animal preparation, laser sources, signal amplification, PMT specifications, and YFP/CFP filter sets have been described previously (48). Using the raw image stacks (1–1.5-μm z interval), manual reconstructions of the dendritic arbor in three dimensions were generated and then analyzed for total dendritic branch length, branch-tip number, and 3D Sholl analysis (29); radius interval 1 μm, averaged over 10-μm bins) using Object-Image macros (written by Dr. E. Ruthazer, McGill University, Quebec).
Electrophysiology.
Whole-cell patch clamp recordings of the AMPA mEPSCs were made from control and transfected cells and analyzed as described (32, 39). Control cells were from unelectroporated animals. For in vivo recordings, anesthetized tadpoles were transferred to the recording chamber where the tectum was exposed by removing the overlying skin and making a dorsal midline cut to uncover the ventricular surface. The tadpole was stabilized with short pieces of tungsten wire (California Fine Wire) and perfused with extracellular saline containing 0.01 mM d-tubocurarine (Sigma) to prevent muscle movement. Cell access and patch quality were monitored throughout the experiment: average input resistance, 1.5 GΩ; average series resistance, 50Ω. Saline compositions are given in SI Methods.
AMPA mEPSC data and spontaneous events were analyzed with template matching (Axograph 4.6 software, or Clampfit 10.0, Molecular Devices). Complex events (bursts, consisting of a minimum duration 50 ms and amplitude of 10pA) were detected using Clampfit 10.0. Light-evoked, -off responses were measured with custom software (Matlab, Mathworks; written by Dr. J. Demas, Cold Spring Harbor Laboratory, NY).
Visual Stimulation.
A Luxeon III LED (530 nm) pigtailed to a 2-mm diameter optic fiber (n.a. = 0.5; Doric Lenses) was used to deliver a full-field stimulus to the eye while the responses from neurons of the contralateral optic tectum were assessed with whole-cell voltage clamp. The LED intensity was controlled with 10-dB neutral density filters that dropped into a housing coupled to the fiber (Oz Optics). The filters and output intensities were measured and verified with an IL 1400 radiometer/photometer, (International Light Technologies) and the radiant flux used in the experiments ranged from 0.06 mW to 64 mW in 10-dB increments (referred to as relative intensities 0.01 to 10 in Fig. 4).
After obtaining whole-cell access and before they were presented with the visual stimulus protocol (described in SI Methods and Fig. S4), the tadpoles were dark-adapted for 6 min, during which spontaneous synaptic activity was recorded. The visual stimulation protocol was repeated up to four times, each repetition with a 10-dB (in some instances, 20 dB) increase in LED intensity.
Statistical Tests.
Mann Whitney U tests were used to make comparisons between groups. Wilcoxon signed rank tests were used for paired data. The data from one cell in the delCPEB group, likely from an untransfected cell, satisfied Grubb's test for outliers (P < 0.01) and was removed from the light-evoked response and spontaneous-response experiments. Kolmogorov-Smirnov tests were used to compare distributions. In the quantification of the spontaneous events (see Fig. 4), 119 events were randomly chosen from each cell. Data presented in bar graphs are mean ± SEM and are presented in Tables S1–S7.
Supplementary Material
Acknowledgments.
The authors thank Kimberly Bronson for expert technical assistance and the past and present members of the Cline Laboratory, and Dr. Robert Darnell for comments on the manuscript. We are grateful to Dr. Jay Demas for developing the Matlab analysis software. Supported by the Fragile X Research Foundation, the Dana Foundation, the Dart Neuroscience LLC, and the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806296105/DCSupplemental.
References
- 1.Kelleher RJ, 3rd, Govindarajan A, Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004;44(1):59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- 4.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 6.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 7.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:1–14. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 8.Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Alarcon JM, et al. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Mem. 2004;11:318–327. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger-Sweeney J, Zearfoss NR, Richter JD. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learn Mem. 2006;13:4–7. doi: 10.1101/lm.73706. [DOI] [PubMed] [Google Scholar]
- 11.McEvoy M, et al. Cytoplasmic polyadenylation element binding protein 1-mediated mRNA translation in Purkinje neurons is required for cerebellar long-term depression and motor coordination. J Neurosci. 2007;27:6400–6411. doi: 10.1523/JNEUROSCI.5211-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Si K, et al. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Schwartz JH. The cytoplasmic polyadenylation element binding protein and polyadenylation of messenger RNA in Aplysia neurons. Brain Res. 2003;959:68–76. doi: 10.1016/s0006-8993(02)03729-0. [DOI] [PubMed] [Google Scholar]
- 16.Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- 17.Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells DG, et al. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkins CM, Nozaki N, Shigeri Y, Soderling TR. Cytoplasmic polyadenylation element binding protein-dependent protein synthesis is regulated by calcium/calmodulin-dependent protein kinase II. J Neurosci. 2004;24:5193–5201. doi: 10.1523/JNEUROSCI.0854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas K, Li J, Cline HT. AMPA receptors regulate experience-dependent dendritic arbor growth in vivo. Proc Natl Acad Sci USA. 2006;103:12127–12131. doi: 10.1073/pnas.0602670103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sin WC, Haas K, Ruthazer ES, Cline HT. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 2002;419:475–480. doi: 10.1038/nature00987. [DOI] [PubMed] [Google Scholar]
- 22.Wu GY, Cline HT. Stabilization of dendritic arbor structure in vivo by CaMKII. Science. 1998;279:222–226. doi: 10.1126/science.279.5348.222. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez AL, et al. BDNF increases synapse density in dendrites of developing tectal neurons in vivo. Development. 2006;133:2477–2486. doi: 10.1242/dev.02409. [DOI] [PubMed] [Google Scholar]
- 24.Du L, Richter JD. Activity-dependent polyadenylation in neurons. Rna. 2005;11:1340–1347. doi: 10.1261/rna.2870505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu L, et al. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 26.Reverte CG, Ahearn MD, Hake LE. CPEB degradation during Xenopus oocyte maturation requires a PEST domain and the 26S proteasome. Dev Biol. 2001;231:447–458. doi: 10.1006/dbio.2001.0153. [DOI] [PubMed] [Google Scholar]
- 27.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 28.Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87(4):387–406. [PMC free article] [PubMed] [Google Scholar]
- 30.Rajan I, Cline HT. Glutamate receptor activity is required for normal development of tectal cell dendrites in vivo. J Neurosci. 1998;18:7836–7846. doi: 10.1523/JNEUROSCI.18-19-07836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aizenman CD, Cline HT. Enhanced visual activity in vivo forms nascent synapses in the developing retinotectal projection. J Neurophysiol. 2007;97:2949–2957. doi: 10.1152/jn.00452.2006. [DOI] [PubMed] [Google Scholar]
- 32.Wu G, Malinow R, Cline HT. Maturation of a central glutamatergic synapse. Science. 1996;274:972–976. doi: 10.1126/science.274.5289.972. [DOI] [PubMed] [Google Scholar]
- 33.Ewald RC, Van Keuren-Jensen KR, Aizenman CD, Cline HT. Roles of NR2A and NR2B in the development of dendritic arbor morphology in vivo. J Neurosci. 2008;28:850–861. doi: 10.1523/JNEUROSCI.5078-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang LI, Tao HW, Poo M. Visual input induces long-term potentiation of developing retinotectal synapses. Nat Neurosci. 2000;3:708–715. doi: 10.1038/76665. [DOI] [PubMed] [Google Scholar]
- 35.Derkach VA, Oh MC, Guire ES, Soderling TR. Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nat Rev Neurosci. 2007;8(2):101–113. doi: 10.1038/nrn2055. [DOI] [PubMed] [Google Scholar]
- 36.Pfeiffer BE, Huber KM. Current advances in local protein synthesis and synaptic plasticity. J Neurosci. 2006;26:7147–7150. doi: 10.1523/JNEUROSCI.1797-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin CY, Kundel M, Wells DG. Rapid, activity-induced increase in tissue plasminogen activator is mediated by metabotropic glutamate receptor-dependent mRNA translation. J Neurosci. 2004;24:9425–9433. doi: 10.1523/JNEUROSCI.2457-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Nardo AA, et al. Dendritic localization and activity-dependent translation of Engrailed1 transcription factor. Mol Cell Neurosci. 2007;35:230–236. doi: 10.1016/j.mcn.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Van Keuren-Jensen K, Cline HT. Visual experience regulates metabotropic glutamate receptor-mediated plasticity of AMPA receptor synaptic transmission by Homer1a induction. J Neurosci. 2006;26:7575–7580. doi: 10.1523/JNEUROSCI.5083-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci. 2002;3:175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- 41.Ouyang Y, Rosenstein A, Kreiman G, Schuman EM, Kennedy MB. Tetanic stimulation leads to increased accumulation of Ca(2+)/calmodulin-dependent protein kinase II via dendritic protein synthesis in hippocampal neurons. J Neurosci. 1999;19:7823–7833. doi: 10.1523/JNEUROSCI.19-18-07823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiler IJ, et al. Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc Natl Acad Sci USA. 2004;101:17504–17509. doi: 10.1073/pnas.0407533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrari F, et al. The fragile X mental retardation protein-RNP granules show an mGluR-dependent localization in the post-synaptic spines. Mol Cell Neurosci. 2007;34:343–354. doi: 10.1016/j.mcn.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Costa A, et al. The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Dev Cell. 2005;8:331–342. doi: 10.1016/j.devcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008;31:20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haas K, Jensen K, Sin WC, Foa L, Cline HT. Targeted electroporation in Xenopus tadpoles in vivo–from single cells to the entire brain. Differentiation. 2002;70:148–154. doi: 10.1046/j.1432-0436.2002.700404.x. [DOI] [PubMed] [Google Scholar]
- 47.Bestman JE, Ewald RC, Chiu SL, Cline HT. In vivo single-cell electroporation for transfer of DNA and macromolecules. Nat Protoc. 2006;1:1267–1272. doi: 10.1038/nprot.2006.186. [DOI] [PubMed] [Google Scholar]
- 48.Ruthazer ES, Li J, Cline HT. Stabilization of axon branch dynamics by synaptic maturation. J Neurosci. 2006;26:3594–3603. doi: 10.1523/JNEUROSCI.0069-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.