Abstract
The initiation of intracellular infection of legume roots by symbiotic rhizobia bacteria and arbuscular mycorrhiza (AM) fungi is preceded by the induction of calcium signatures in and around the nucleus of root epidermal cells. Although a calcium and calmodulin-dependent kinase (CCaMK) is a key mediator of symbiotic root responses, the decoding of the calcium signal and the molecular events downstream are only poorly understood. Here, we characterize Lotus japonicus cyclops mutants on which microbial infection was severely inhibited. In contrast, nodule organogenesis was initiated in response to rhizobia, but arrested prematurely. This arrest was overcome when a deregulated CCaMK mutant version was introduced into cyclops mutants, conferring the development of full-sized, spontaneous nodules. Because cyclops mutants block symbiotic infection but are competent for nodule development, they reveal a bifurcation of signal transduction downstream of CCaMK. We identified CYCLOPS by positional cloning. CYCLOPS carries a functional nuclear localization signal and a predicted coiled-coil domain. We observed colocalization and physical interaction between CCaMK and CYCLOPS in plant and yeast cell nuclei in the absence of symbiotic stimulation. Importantly, CYCLOPS is a phosphorylation substrate of CCaMK in vitro. Cyclops mutants of rice were impaired in AM, and rice CYCLOPS could restore symbiosis in Lotus cyclops mutants, indicating a functional conservation across angiosperms. Our results suggest that CYCLOPS forms an ancient, preassembled signal transduction complex with CCaMK that is specifically required for infection, whereas organogenesis likely requires additional yet-to-be identified CCaMK interactors or substrates.
Keywords: BiFC, map-based cloning, plant-microbe symbiosis, protein phosphorylation, protein–protein interaction
Legume plants can establish endosymbiotic interactions with nitrogen-fixing rhizobia and phosphate-delivering arbuscular mycorrhiza (AM) fungi. Plant root hairs form a tight curl in which rhizobia are entrapped. From this closed infection pocket, the bacteria are guided by plant membrane-delimited infection threads (ITs) into the root nodule, a specialized organ developed by the plant to provide an optimized environment for nitrogen fixation (1). AM fungal hyphae are guided through epidermal and cortical cells toward the inner cortex (2), where arbuscules, highly branched intracellular symbiotic structures, are formed (3). Intracellular infection by rhizobia and AM fungi is preceded by an exchange of specific signaling molecules. Rhizobia produce lipochito-oligosaccharides (Nod factors) that activate host plant responses including root hair deformation, and preinfection thread formation, which are structures that determine the path of IT growth through the root (4), and initiation of cortical cell division (1). One of the earliest plant responses to stimulation by Nod factors is Ca2+-spiking, which consists of perinuclear oscillations of calcium concentration in root cells (5). In the legume Lotus japonicus, a shared genetic program defined by seven “common symbiosis genes” (6), is required for the establishment of both symbioses (7). An LRR-receptor kinase SYMRK (8, 9), the ion channel-like proteins CASTOR and POLLUX (10, 11), and the nucleoporins NUP85 and NUP133 (12, 13) are all required for the generation of Ca2+-spiking, whereas a calcium and calmodulin-dependent protein kinase (CCaMK) is not, suggesting that CCaMK functions downstream of Ca2+-spiking (14–16). CCaMK is composed of a kinase domain, a calmodulin-binding site and 3 EF hand motifs (16). Its catalytic activity is modulated by either free, or calmodulin (CaM)-bound Ca2+ ions (16, 17), suggesting that CCaMK converts the Ca2+ oscillation signal into a protein phosphorylation read-out. Deregulation of CCaMK by either a point mutation in the autophosphorylation site, or the deletion of the C-terminal regulatory domain, results in spontaneous nodule formation in the absence of rhizobia, demonstrating that CCaMK is a central regulator of the nodule organogenesis program (16, 17). CCaMK is also required for root hair curling and IT formation upon rhizobial infection and arbuscule formation during AM (7, 18, 19). However, how CCaMK differentially activates infection- vs. organogenesis-related pathways is still unclear.
CYCLOPS has been positioned downstream of CCaMK, because cyclops mutants exhibit impaired AM and rhizobial infection, but retain Ca2+-spiking and formation of nodule primordia (7, 20–23). In this work, we describe the map-based cloning of CYCLOPS, which we found to encode a nuclear-localized protein with a coiled-coil motif. CYCLOPS is an ortholog of Medicago truncatula IPD3, which was recently identified as Interacting Protein of DMI3 (the M. truncatula CCaMK ortholog) (24). We show that CYCLOPS specifically interacts with kinase-active CCaMK in planta and is phosphorylated by CCaMK in vitro, suggesting that CYCLOPS is a phosphorylation target of CCaMK. Our data position CYCLOPS on an infection-specific branch of the signaling network downstream of CCaMK.
Results
cyclops Mutants Abort Infection by Rhizobia and AM Fungi.
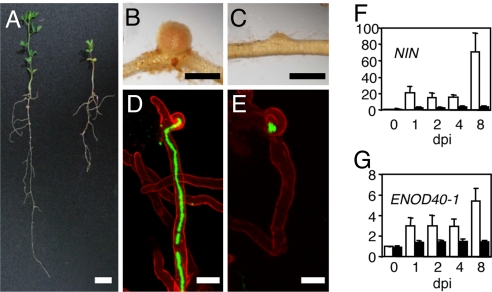
In forward genetic screens, we previously identified an allelic series of mutants impaired in the interaction with both rhizobia and AM fungi [supporting information (SI) Table S1]. Phenotypic analysis revealed that in contrast to WT L. japonicus plants (Fig. 1A), which developed mature nodules upon inoculation with M. loti under nitrogen-limiting conditions (Fig. 1B) on cyclops-3 mutant roots, nodule development was prematurely arrested, and only nodule primordia were observed (Fig. 1 A and C; Table S1). In WT plants, intracellular ITs developed and grew through the root hair toward the nodule primordium (Fig. 1D). On cyclops-3 mutant roots, curled root hair tips were colonized by rhizobia, however no ITs were observed (Fig. 1E). Because of this characteristic mutant phenotype, the corresponding gene was called CYCLOPS. In rare cases ITs were initiated, but elongation was aborted within root hairs (22). On cyclops mutant roots, the passage of AM fungal hyphae through the outer cell layers was characterized by abnormal hyphal swellings forming within epidermal or outer cortical cells, indicating an impairment of the intracellular infection process (Fig. S1 A and B). Where infection events were successful, hyphal growth proceeded toward the inner root cortex, and apoplastic growth along the root axis led to longitudinal spread of the fungal infection. Despite fungal colonization of the root cortex, arbuscules were almost completely absent from cyclops mutants (Fig. S1 A and B, Table S2) (7). These data indicate that CYCLOPS is required for fungal infection of the outer cortical cell layers and for arbuscule development.
Fig. 1.
Phenotypic characteristics of the cyclops-3 mutant. (A) Growth phenotype in nitrogen-limiting conditions of ecotype Gifu WT (Left) and cyclops-3 (Right). (B) WT nodule. (C) cyclops-3 nodule primordium. (D) Infection thread within WT root hair. (E) Aborted infection within curled root hair of cyclops-3. (A–C) Plants were grown for 1 month after inoculation with M. loti MAFF303099. (D and E) Plants were grown for 2 weeks after inoculation with M. loti BN02 expressing GFP. [Scale bars: (A) 1.0 cm; (B and C) 1.0 mm; (D and E) 20 μm.] (F and G) Quantitative RT-PCR analysis of NIN (F) and ENOD40-1(G) expression in noninoculated (0 days) WT (white columns) and cyclops-3 (black columns) roots, and at 1, 2, 4, and 8 dpi with M. loti MAFF303099. Fold increases in expression are shown relative to WT roots at 0 dpi (F and G). Mean values ± SD are shown.
Expression of NIN and ENOD40–1 Is Impaired in cyclops Mutants.
In L. japonicus, transcription of the NIN and ENOD40–1 genes is induced within a few hours during the symbiotic interaction with M. loti (25, 26). The transcript levels of these genes are useful markers for the activity of symbiotic signal transduction processes (26, 27). In the WT, biphasic induction kinetics were observed for both genes upon inoculation with M. loti (Fig. 1 F and G). The first phase occurred at, or before, 1 day after inoculation (dpi), and the second between 4–8 dpi. This two-phase induction most likely reflects epidermal and cortical expression of NIN and ENOD40–1, as reported previously (28). NIN induction was still detectable, but at 8 dpi was 17-fold reduced in cyclops-3 compared to the WT (Fig. 1F). In contrast, slight variations in ENOD40–1 transcript levels in cyclops-3 mutant roots were not statistically significant (Fig. 1G). These data indicate that CYCLOPS is required for full transcriptional activation of NIN and ENOD40–1.
CYCLOPS Is Dispensable for Nodule Organogenesis.
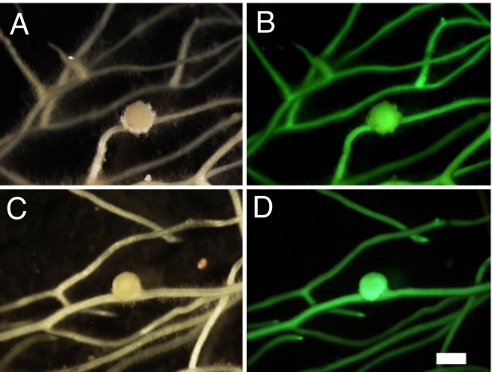
In contrast to previously identified common symbiosis mutants, cyclops mutants retain the ability to initiate cortical cell division. However, it is unclear whether CYCLOPS is necessary for the progression of nodule organogenesis beyond the primordium stage. To answer this question, we took advantage of a derivative of CCaMK conferring spontaneous nodulation. CaMV 35S promoter-driven expression of CCaMKT265D, in which the predicted autophosphorylation site Thr-265 was replaced by Asp, resulted in spontaneous nodule formation in the absence of rhizobia in the WT as well as the cyclops-4 background (Fig. 2 A–D and Table S3). The size of spontaneous nodules formed on cyclops-4 roots did not differ significantly (P < 0.01) from those formed on WT roots (Fig. 2 A and C, Table S3). None of the negative controls including WT and cyclops-4 plants transformed with WT CCaMK developed spontaneous nodules (Table S3). These results indicate that CYCLOPS is dispensable for nodule organogenesis and suggest that the developmental arrest observed in cyclops mutants is an indirect consequence of the aborted bacterial infection.
Fig. 2.
Spontaneous nodule formation on cyclops-4 and WT L. japonicus hairy roots transformed with 35S promoter-driven gain-of-function CCaMKT265D. CCaMKT265D was introduced into cyclops-4 or WT hairy roots. (A–D) Spontaneous nodule development on cyclops-4 (A and B) and WT roots (C and D) induced by CCaMKT265D. Transformed roots were visualized by GFP fluorescence (B and D). Nodules were observed 8 weeks after transformation. (Scale bar, 1 mm.)
Map-Based Cloning of CYCLOPS.
The CYCLOPS gene was isolated from L. japonicus by using a map-based approach (Fig. S2 A–C). Comparison with corresponding cDNA clones originating from nodulated roots (29) revealed that CYCLOPS is composed of 11 exons encoding a protein of 518 aa (Figs. S2D and S3A). Point mutations or single nucleotide deletions result in frame shifts and/or premature stop codons in cyclops-2, -3, -4, and -5 (Table S1). Cyclops-1 carries a transposon insertion in intron 10, resulting in a sequence insertion between exons 10 and 11 of the cDNA (Fig. S2D, Table S1). Hairy root transformation of cyclops-3 with the genomic CYCLOPS sequence, including the native promoter, resulted in the development of WT-like nodules upon inoculation with M. loti (Fig. S4). Similarly, the introduction of CYCLOPS cDNA fused to the native promoter into the same mutant line restored WT-like nodulation, as well as AM formation (Fig. S1, Table S2). The identification of genetic lesions in each of the 5 independent cyclops alleles together with the transgenic complementation of the mutant phenotype is unequivocal evidence for the correct identification of the CYCLOPS gene.
Within the conceptual CYCLOPS gene product, a coiled-coil motif in the C-terminal 67 aa residues and 2 nuclear localization signals (NLSs) were predicted (Figs. S3A and S5). However, no overall similarity to proteins of known function was identified by interrogation of public databases. Genomic DNA gel blot analysis and searches in the complete genome sequence (30) indicated that CYCLOPS is a single copy gene in L. japonicus and that single copy CYCLOPS-related sequences occur in other legume species including Medicago truncatula and Pisum sativum (data not shown). A combination of database searches and PCR amplification of cDNA from the respective species identified putative CYCLOPS orthologs in M. truncatula (IPD3) (24), P. sativum, and Oryza sativa (Fig. S5).
CYCLOPS Expression and Subcellular Localization.
By quantitative RT-PCR, CYCLOPS mRNA was detectable in uninfected roots and increased in abundance between 4–8 dpi with M. loti (Fig. S6A), which is after the initiation of IT development and cortical cell division. CYCLOPS mRNA accumulated in mature nodules, whereas the mRNA level in roots harvested 3 weeks after inoculation, nodules on which were removed, was comparable to uninfected roots (Fig. S6B). In situ hybridization of mature nodules indicated that CYCLOPS mRNA was present in cells of the central tissue (Fig. S6 C and D). CYCLOPS promoter activity during nodule development was monitored in L. japonicus hairy roots transformed with fusions of the CYCLOPS promoter to the beta-glucuronidase (GUS) reporter gene. Upon treatment with M. loti, strong GUS activity was observed in dividing cortical cells during early stages of nodule development (Fig. S6E). In agreement with the in situ hybridization results, strong expression was detected in the central tissue of mature nodules at later stages (Fig. S6F). CYCLOPS mRNA was not detectable in plant shoots, regardless of whether the plants were inoculated with M. loti or not (Fig. S6B).
To determine the subcellular localization of CYCLOPS, we transiently expressed GFP-CYCLOPS fusion constructs driven by the 35S promoter in Nicotiana benthamiana leaf epidermis cells (Fig. S7). GFP-CYCLOPS and a C-terminal truncation lacking the coiled-coil motif (GFP-CYCLOPS 1–449) were exclusively detected in the cell nucleus (Fig. S7 A and B), whereas CYCLOPS deletion mutants lacking either the second or both predicted NLS (GFP-CYCLOPS 1–421 and 1–366) were distributed randomly in the cytosol and nucleus, similar to the pattern observed with GFP alone (Fig. S7 C–E), indicating that at least the NLS proximal to the C terminus is functional. Because CCaMK was also localized in the nucleus (31), we tested for possible colocalization with CYCLOPS in L. japonicus root cells. Fusion constructs of CCaMK-GFP and RFP-CYCLOPS, both under the control of the CaMV 35S promoter, were introduced into WT L. japonicus roots via A. rhizogenes transformation. Transformed hairy roots exhibited GFP and RFP fluorescence in the same nuclei (Fig. S8 A–D). Similar results were obtained with protoplast cells derived from L. japonicus root tissue (data not shown). The observed spatiotemporal expression pattern of CYCLOPS and its nuclear colocalization with CCaMK are consistent with the findings obtained for the M. truncatula CYCLOPS ortholog IPD3 (24).
CYCLOPS Interacts With CCaMK in the Nucleus.
In yeast two-hybrid (Y2H) analysis, full-length CYCLOPS fused to the GAL4 DNA binding domain (BD) showed strong autoactivation in yeast (data not shown). Consequently, a CYCLOPS deletion derivative BD-fusion (residues 1–259) or full-length CYCLOPS fused to the GAL4 activation domain (AD), both lacking autoactivation (data not shown), were used for subsequent experiments. We detected a strong interaction between CYCLOPS and CCaMK (Fig. 3 A and B). A CCaMK point-mutation in the autophosphorylation site (T265I) that causes nodule development in the absence of rhizobia (16) did not disrupt interactions with CYCLOPS (Fig. 3A). However, 2 kinase-defective CCaMK mutants, G30E (as in the ccamk-3 mutant) (16) and a point-mutant of a catalytic lysine residue (K44A) (32), as well as the K44A/T265I double mutant, did not interact with CYCLOPS (Fig. 3A), suggesting that kinase activity of CCaMK is required for interaction with CYCLOPS. The analysis of deletion constructs revealed that the 2 C-terminal EF-hand motifs of CCaMK are not required for a strong interaction with CYCLOPS (Fig. 3A). A CYCLOPS deletion series delimited the region required for CCaMK interaction between CYCLOPS residues 81–366 (Fig. 3B). Using CYCLOPS 1–259 as bait revealed that CYCLOPS forms homodimers, and that the region between residues 81–366 is necessary for self-interaction (Fig. 3B).
Fig. 3.
CYCLOPS–CCaMK interactions and in vitro phosphorylation. (A and B) Detection of protein–protein interactions by Y2H analysis. Representative yeast growth on the selection medium (-leu, -trp, -his, -ade) is shown, and interaction results are indicated on the right. (+, growth; −, no growth; ±, very weak growth; AD, fused to the activation domain of GAL4; BD, fused to the DNA binding domain of GAL4). (A) Schematic illustrations of CCaMK derivatives (gray box, kinase domain; slashed box, calmodulin binding domain; black box, EF-hand; star, amino acid replacement) and their interaction with CYCLOPS. (B) Schematic illustrations of CYCLOPS derivatives (black box, coiled-coil motif; white box, NLS) and interaction of CYCLOPS derivatives with CCaMK and CYCLOPS1−259. (C–F) Detection of protein–protein interactions in N. benthamiana leaves by BiFC. YFP fluorescence of leaves cotransformed with CCaMK-YFPN (N-terminal half of YFP) and YFPC (C-terminal half of YFP)-CYCLOPS (C) or CCaMK K44A-YFPN and YFPC-CYCLOPS (D). Lower Images in (C) and (D) show RFP fluorescence of the corresponding area resulting from cotransformation with a 35S/RFP construct as a positive control for successful transformation. [Scale bars, 100 μm (C and D).] YFP fluorescence of a leaf epidermis cell cotransformed with CCaMK-YFPN and YFPC-CYCLOPS (E) or CYCLOPS-YFPN and YFPC-CYCLOPS (F). Lower Images in (E) and (F) show the cell architecture (differential interference contrast) of the same cell. [Scale bars, 50 μm (E and F).] (G) In vitro phosphorylation of CYCLOPS by CCaMK. (Left Images) Phosphorylation of full-length CYCLOPS and derivatives in the presence of calcium (Ca2+) and calmodulin (CaM). (Right Images) Phosphorylation of full-length CYCLOPS in the presence (+) or absence (−) of Ca2+ and CaM. Arrowheads indicate MBP-CCaMK and asterisks indicate 6×His-CYCLOPS or derivatives. (Upper Images) Autoradiographs of kinase assays (32P) and (Lower Images) Coomassie staining (CBB) of the same gels.
To confirm CYCLOPS protein interactions in planta, we performed bimolecular fluorescence complementation (BiFC) in transiently transformed N. benthamiana epidermis cells (33, 34). Strong fluorescence was observed in the nucleus when CYCLOPS and CCaMK fused to the C- and N-terminal half of YFP, respectively, were coexpressed (Fig. 3 C and E). Consistent with the requirement of kinase-active CCaMK for interaction in Y2H assays (Fig. 3A), no fluorescence was observed in cells cotransformed with constructs encoding CYCLOPS and CCaMK kinase-dead mutants (K44A and G30E) (Fig. 3D and data not shown). Self-interaction of full-length CYCLOPS was also observed in planta (Fig. 3F).
CCaMK Phosphorylates CYCLOPS In Vitro.
Their strong interaction in plant and yeast cells prompted us to analyze whether CCaMK can phosphorylate CYCLOPS in in vitro kinase assays. CYCLOPS, CYCLOPS1−449, and CYCLOPS81−366, which interacted with CCaMK in the Y2H assay, were all phosphorylated by CCaMK (Fig. 3G, Left Image). The N-terminal truncation CYCLOPS255−518, did not interact in the Y2H assay and was not phosphorylated by CCaMK (Fig. 3G, Left Image). The relatively stronger phosphorylation of CYCLOPS81−366 may indicate additional CCaMK phosphorylation sites that are inaccessible in the full-length protein. Ca2+ alone increased autophosphorylation of CCaMK, whereas full-length CYCLOPS phosphorylation was stimulated by Ca2+/CaM (Fig. 3G, Right Image). These results demonstrate that CYCLOPS is a substrate of CCaMK in vitro.
Rice CYCLOPS Is Indispensable for AM and Restores Rhizobial and Fungal Symbiosis in Lotus cyclops-3.
Alignment of the nucleotide and protein sequences of L. japonicus CYCLOPS and rice (Oryza sativa) OsCYCLOPS revealed a 45% overall amino acid sequence identity, and a conserved exon–intron structure (Figs. S3A and S5). To elucidate the function of CYCLOPS in rice, we analyzed the AM phenotype of 4 independent Tos17 lines (35), which each carry an insertion within exon 6 of OsCYCLOPS (Fig. S3A). Upon cocultivation with G. intraradices, arbuscules were not observed in any of the mutant root systems tested, despite the presence of abundant intraradical mycelium (Fig. S3B), indicating that OsCYCLOPS is required for arbuscule and hence, AM development in rice. Hyphal swellings within epidermal and outer cortical cells, resembling aborted infection sites observed in L. japonicus cyclops mutant roots, were also present in rice cyclops mutant roots, but were not indicative of the mutant phenotype in this species as they similarly occurred in rice segregants of the same lines carrying wild-type alleles of OsCYCLOPS (data not shown). By using A. rhizogenes infection, we generated L. japonicus cyclops-3 hairy roots carrying a fusion of the OsCYCLOPS coding sequence to the Lotus CYCLOPS promoter region. In these roots, both AM and nodulation by M. loti were restored (Table S2, Fig. S1), demonstrating that the OsCYCLOPS gene of nonnodulating rice can support not only AM fungal, but also bacterial endosymbiosis.
Discussion
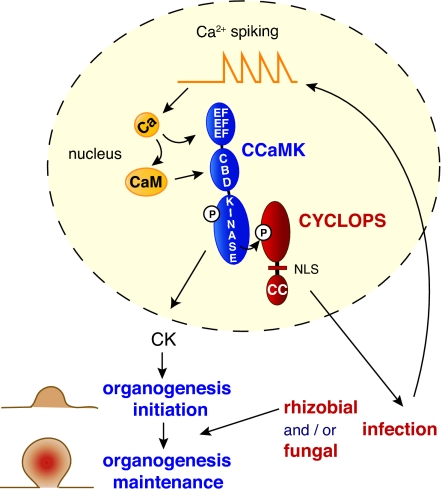
In this study, we identified CYCLOPS by map-based cloning. CYCLOPS encodes a plant-specific protein with a short C-terminal coiled-coil domain and a functional NLS. CCaMK and CYCLOPS are both required for the intracellular accommodation of bacteria and AM fungi. These phenotypes position CCaMK and CYCLOPS on a pathway required for the induction of plant infection structures. In contrast to other common symbiosis genes, both genes act downstream of the generation of Ca2+-spiking (23). CCaMK has previously been localized to the nucleus and the presence of calcium-binding EF-hands and a calmodulin-binding domain made it a prime candidate to be the direct receiver of the calcium signal during spiking (14–16). We found that CYCLOPS and CCaMK colocalize within the nucleus, and we could observe physical interaction between the 2 proteins in yeast, as well as in plant cell nuclei, as was described for the M. truncatula orthologs IPD3 and DMI3 (24). Moreover, CYCLOPS was found to be a substrate for the CCaMK kinase in vitro. It is likely that the molecular mechanism by which CCaMK and CYCLOPS mediate the induction of infection structures involves the activation of symbiosis-related genes downstream (7). In a hypothetical signaling pathway, the perinuclear calcium spiking induced by rhizobial Nod factors or signals from AM fungi (36) modulate CCaMK kinase activity and hence the phosphorylation status of CYCLOPS (Fig. 4). Phosphorylation of CYCLOPS may, in turn, influence the transcription of infection-related plant genes. The identity of signaling components that function directly downstream of CYCLOPS and CYCLOPS-regulated genes remains to be identified.
Fig. 4.
Model for the function of CYCLOPS and CCaMK. Upon Ca2+-spiking generated in the nucleus, CCaMK is activated by binding of calcium (Ca) to the C-terminal EF hands, leading to autophosphorylation (P) and/or binding of calcium-activated calmodulin (CaM) to the CaM binding domain (CBD). Subsequently, signal transduction initiated by CCaMK proceeds via different routes. In the root cortex, CCaMK induces and maintains nodule organogenesis which requires a sustained, elevated concentration of cytokinin (CK). In cells subject to microbial invasion, CCaMK signals via CYCLOPS to mediate infection. Infection thread development and ramification appears to be required for nodule progression, providing a continuous supply of morphogenetic signal, the Nod factor. (NLS, nuclear localization signal; CC, coiled-coil domain.)
Although the physical interaction of CCaMK and CYCLOPS and their requirement for infection are clear cut, there are significant phenotypic differences between corresponding mutants. In ccamk loss-of-function mutants, root-hair curling and cortical cell division were abolished upon symbiotic interaction with M. loti (14–16, 20, 21), whereas both responses were detected in cyclops mutants. Moreover, the introduction of gain-of-function CCaMK into the cyclops mutant background complemented the lack of organogenesis of full-sized (spontaneous) nodules, demonstrating that CYCLOPS is not required for nodule organogenesis. The interaction in Y2H and BiFC occurred in the absence of calcium spiking, indicating that CYCLOPS and CCaMK form a preassembled, stimulus-independent complex. This complex is likely to contain additional signaling components required for nodule initiation. The loss of CYCLOPS may weaken other interactions within the complex. Such reduced stability of the CCaMK complex may explain the reduction of spontaneous nodule numbers in the cyclops mutant background (Table S3).
Our data reveal a bifurcation of signal transduction at or below CCaMK. One CYCLOPS-dependent pathway leads to IT formation and AM infection, whereas a second CYCLOPS-independent pathway mediates nodule organogenesis and root-hair curling (Fig. 4). These 2 pathways may have additional tissue-specific components because a mutant version of CCaMK, snf1, encoding a deregulated kinase, spontaneously induces the expression of NIN in the cortical but not the epidermal cell layer, where nodule organogenesis or infection are respectively initiated (16). The second and CYCLOPS-independent pathway proceeds via hypothetical additional phosphorylation target(s) of CCaMK and likely involves the activation of cytokinin synthesis. Cytokinin is subsequently perceived in a cell nonautonomous fashion by the cytokinin receptor LHK1 (37), an event leading to the induction of cell division in hormonally predetermined cells (38).
If CYCLOPS is not required for nodule organogenesis, why do cyclops mutants not develop mature nodules upon rhizobial infection? Nodule meristem progression mediated by CCaMK may require the continuous and sufficient supply of bacterial morphogenetic signals such as Nod factors being released by rhizobia into the developing ITs that are not provided in cyclops mutants. Thus, the continuation of nodule morphogenesis would rely on continuous IT development that is itself dependent on CYCLOPS. This hypothesis is consistent with the fact that the expression domain of CYCLOPS is congruent with the anticipated developmental region explored by ITs in the developing nodule, as shown in our CYCLOPS promoter-GUS expression analysis (Fig. S6).
The nodule primordia phenotype of cyclops resembles M. truncatula hcl mutants, which induce limited cortical cell divisions in response to rhizobia, and in which spontaneous nodules form on transformation of gain-of-function CCaMK (39). However, in contrast to cyclops mutants, loss-of-function hcl mutants show an aberrant root-hair response toward rhizobia, resulting either in extensive root-hair deformation without curling, or extensive curling and a lack of bacteria within curled root hairs (40). HCL encodes the LysM-type receptor-like kinase LYK3, which is a putative ortholog of the L. japonicus Nod factor receptor NFR1 (27, 41).
Infection and nodule organogenesis are spatially and temporally coordinated, and the CCaMK–CYCLOPS complex may contribute to this coordination. When CCaMK versions in which the C-terminal regulatory domain was deleted were introduced into the M. truncatula dmi3-1 (ccamk) mutant background, spontaneous nodules formed in the absence of rhizobia. Importantly, when these transgenic roots were inoculated with rhizobia, the developing nodules were not infected (17). This specific restoration of the organogenesis, but not the infection program, may be explained by our observation that the site between the CaM-binding domain and the second EF hand is required for the L. japonicus CCaMK–CYCLOPS interaction in yeast (Fig. 3A), a domain that is lacking in the M. truncatula DMI3 1–311 or 1–326 C-terminal deletions.
Interestingly, the allelic series of symbiosis-defective cyclops mutants exclusively comprises frame shifts or premature stop codons, suggesting that they represent loss-of-function alleles (Table S1). No mutants with single amino acid substitutions leading to missense mutations were recovered from forward screens for nodulation defective mutants (42). This is an exceptional bias suggesting that amino acid substitutions are either largely tolerated without significantly impacting CYCLOPS function, or lethal.
Amino acid sequences with high overall similarity to CYCLOPS were only found in legume plants. The C-terminal region of CYCLOPS, comprising the second NLS and the coiled-coil motif, is conserved in a wide range of plant species, including the moss Physcomitrella patens, the primitive angiosperm Amborella trichopoda, and higher plants like Vitis vinifera and Populus trichocarpa. No sequence with significant similarity to CYCLOPS was identified in the genome of the asymbiotic plant Arabidopsis thaliana, in which other common symbiosis genes like SYMRK or CCaMK are also absent (43).
The symbiosis between plants and AM fungi dates back to the earliest land plants (44). Our analysis of rice cyclops mutants has demonstrated the importance of OsCYCLOPS for the establishment of AM. The finding that CYCLOPS is conserved in AM-forming angiosperms is consistent with an ancient and specific role of CYCLOPS in symbiosis. The observed intraradical colonization, but lack of intracellular arbuscules in cyclops mutants, indicates a function predominantly serving the intracellular accommodation of the AM fungi in both L. japonicus and rice. The evolution of plant-derived structures supporting bacterial infection (infection threads) was a critical step during the evolution of root nodule symbiosis (6). In L. japonicus cyclops mutants, arbuscule development and the initiation or elongation of ITs, are aborted (7, 22, this study). This is consistent with the idea that both symbioses share a common genetic program and hence an evolutionary history. Genetic and structural considerations strongly suggest that the IT evolved from the pre-penetration apparatus observed in response to AM fungi (2). The additional and perhaps later invention of nodule organogenesis relies on yet unidentified CCaMK-downstream components that act independently of CYCLOPS.
Materials and Methods
Plant Material, Inoculation, and Growth Conditions.
L. japonicus ecotype Gifu WT and cyclops mutants identified in genetic screens and through TILLING (42) (Table S1) were grown and inoculated with M. loti MAFF303099 or BN02 as described (45).
Expression Analysis.
RNA was quantified by real-time PCR, or detected by in situ hybridization of root sections with CYCLOPS antisense and sense probes as described in (46) and SI Materials and Methods.
Plasmid Construction.
Detailed information is provided in SI Materials and Methods.
Transformation of CYCLOPS and CCaMK Constructs.
T-DNA constructs (described in SI Materials and Methods) were introduced by hairy root transformation as described (47), plants were cocultivated with rhizobia or G. intraradices BEG195, and nodulation or AM formation was scored as described in SI Materials and Methods.
Subcellular Localization, BiFC Analysis, and Microscopy.
T-DNA constructs (described in SI Materials and Methods) were introduced into N. benthamiana leaf cells by A. tumefaciens-mediated transient transformation, and leaf cells were analyzed with an epifluorescence microscope (for detailed description, see SI Materials and Methods). For colocalization analysis, 35S/CCaMK-GFP and 35S/RFP-CYCLOPS T-DNA constructs (described in SI Materials and Methods) were introduced into L. japonicus WT plants by hairy root transformation, and the localization of the corresponding proteins was assessed by confocal laser scanning microscopy.
Yeast Two-Hybrid Analysis.
Y2H analysis was carried out according to standard procedures (Stratagene Product Manual #235702; pBD-GAL4 Cam Phagemid Vector Kit) by using the yeast strain AH109 (Clontech). Synthetic drop-out media (-trp, -leu, -his, 1 mM 3-amino-1,2,4-triazole and -trp, -leu, -his, -ade) were used for selection of protein interactions.
In Vitro Kinase Assays.
Protein purification and kinase assays were done as described (16) with modifications (see SI Materials and Methods).
Genotyping and AM Phenotyping of Oscyclops Lines.
O. sativa subspecies japonica, cv. Nipponbare, was grown as recommended at http://tos.nias.affrc.go.jp/∼miyao/pub/tos17/. The Oscyclops mutant lines ND5032, NC2713, NC2415, and NG0782 were identified by interrogation of a rice-mutant sequence tag library induced by retrotransposon Tos17 (35). For genotyping, primer Tos17 was used with gene-specific primers (Table S4). The presence and location of the insertions were confirmed by PCR and sequence analysis. For AM phenotyping, plants were cocultivated with G. intraradices BEG195 and Allium schoenoprasum nurse plants for 2 or 3 weeks and AM colonization was quantified as described in (48).
Supplementary Material
Acknowledgments.
We thank J. Stougaard (University of Aarhus, Aarhus, Denmark) and K. Szczyglowski (Agriculture and Agri-food Canada, London, Ontario, Canada) for providing cyclops-1, -2, and -3 seeds, respectively. BiFC vectors were kindly provided by T. Lahaye (University of Halle, Halle, Germany). We also thank K.H. Braun, G. Büttner, and G. Strobel for technical assistance. This work was supported by joint research grants of the Japan Science and Technology Agency (to H.I.-A., M.K., M.H., and M.P.). H.I.-A. was supported by the Program of Basic Research Activities for Innovative Biosciences. The Lotus TILLING facility was funded by grants of the Biotechnology and Biological Sciences Research Council. Research at the Sainsbury Laboratory was funded by the Gatsby Charitable Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genomic sequences reported in this paper have been deposited in the DNA Data Bank of Japan [accession nos. LjT10G21 (AP009154), LjT12C04 (AP009155), LjT14N05 (AP009156), LjT12H02 (AP009157), and LjB11B14 (AP009158)] and the GenBank database [accession nos. EF569221–EF569224 (CYCLOPS cDNAs from different species)].
See Commentary on page 20053.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806858105/DCSupplemental.
References
- 1.Oldroyd GE, Downie JA. Calcium, kinases, and nodulation signalling in legumes. Nat Rev Mol Cell Biol. 2004;5:566–576. doi: 10.1038/nrm1424. [DOI] [PubMed] [Google Scholar]
- 2.Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P. Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell. 2008;20:1407–1420. doi: 10.1105/tpc.108.059014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parniske M. Arbuscular Mycorrhiza, the mother of plant root endosymbioses. Nat Rev Microbiol. 2008;10:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 4.van Brussel AAN, et al. Induction of preinfection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science. 1992;257:70–72. doi: 10.1126/science.257.5066.70. [DOI] [PubMed] [Google Scholar]
- 5.Ehrhardt DW, Wais R, Long SR. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- 6.Kistner C, Parniske M. Evolution of signal transduction in intracellular symbiosis. Trends Plants Sci. 2002;7:511–518. doi: 10.1016/s1360-1385(02)02356-7. [DOI] [PubMed] [Google Scholar]
- 7.Kistner C, et al. Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell. 2005;17:2217–2229. doi: 10.1105/tpc.105.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Endre G, et al. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- 9.Stracke S, et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- 10.Ane JM, et al. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science. 2004;303:1364–1367. doi: 10.1126/science.1092986. [DOI] [PubMed] [Google Scholar]
- 11.Imaizumi-Anraku H, et al. Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature. 2005;433:527–531. doi: 10.1038/nature03237. [DOI] [PubMed] [Google Scholar]
- 12.Saito K, et al. NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell. 2007;19:610–624. doi: 10.1105/tpc.106.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanamori N, et al. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA. 2006;103:359–364. doi: 10.1073/pnas.0508883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy J, et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science. 2004;303:1361–1364. doi: 10.1126/science.1093038. [DOI] [PubMed] [Google Scholar]
- 15.Mitra RM, et al. A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc Natl Acad Sci USA. 2004;101:4701–4705. doi: 10.1073/pnas.0400595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tirichine L, et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature. 2006;441:1153–1156. doi: 10.1038/nature04862. [DOI] [PubMed] [Google Scholar]
- 17.Gleason C, et al. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature. 2006;441:1149–1152. doi: 10.1038/nature04812. [DOI] [PubMed] [Google Scholar]
- 18.Catoira R, et al. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell. 2000;12:1647–1666. doi: 10.1105/tpc.12.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulder L. Norwich, England: University of East Anglia; 2003. Genes required for symbioses in Lotus japonicus. PhD thesis. [Google Scholar]
- 20.Schauser L, et al. Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol Gen Genet. 1998;259:414–423. doi: 10.1007/s004380050831. [DOI] [PubMed] [Google Scholar]
- 21.Szczyglowski K, et al. Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol Plant-Microbe Interact. 1998;11:684–697. [Google Scholar]
- 22.Yano K, et al. New nodulation mutants responsible for infection thread development in Lotus japonicus. Mol Plant–Microbe Interact. 2006;19:801–810. doi: 10.1094/MPMI-19-0801. [DOI] [PubMed] [Google Scholar]
- 23.Miwa H, Sun J, Oldroyd GE, Downie JA. Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol Plant–Microbe Interact. 2006;19:914–923. doi: 10.1094/MPMI-19-0914. [DOI] [PubMed] [Google Scholar]
- 24.Messinese E, et al. A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant–Microbe Interact. 2007;20:912–921. doi: 10.1094/MPMI-20-8-0912. [DOI] [PubMed] [Google Scholar]
- 25.Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 26.Takeda N, Okamoto S, Hayashi M, Murooka Y. Expression of LjENOD40 genes in response to symbiotic and nonsymbiotic signals: LjENOD40–1 and LjENOD40–2 are differentially regulated in Lotus japonicus. Plant Cell Physiol. 2005;46:1291–1298. doi: 10.1093/pcp/pci138. [DOI] [PubMed] [Google Scholar]
- 27.Radutoiu S, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 28.Gronlund M, et al. Analysis of promoter activity of the early nodulin Enod40 in Lotus japonicus. Mol Plant–Microbe Interact. 2005;18:414–427. doi: 10.1094/MPMI-18-0414. [DOI] [PubMed] [Google Scholar]
- 29.Asamizu E, Nakamura Y, Sato S, Tabata S. Comparison of the transcript profiles from the root and the nodulating root of the model legume Lotus japonicus by serial analysis of gene expression. Mol Plant–Microbe Interact. 2005;18:487–498. doi: 10.1094/MPMI-18-0487. [DOI] [PubMed] [Google Scholar]
- 30.Sato S, et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smit P, et al. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- 32.Carrera AC, Alexandrov K, Roberts TM. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. Proc Natl Acad Sci USA. 1993;90:442–446. doi: 10.1073/pnas.90.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 34.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 35.Miyao A, et al. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosuta S, et al. Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc Natl Acad Sci USA. 2008;105:9823–9828. doi: 10.1073/pnas.0803499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tirichine L, et al. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315:104–107. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- 38.Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 39.Marsh JF, et al. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007;144:324–335. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Catoira R, et al. The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development. 2001;128:1507–1518. doi: 10.1242/dev.128.9.1507. [DOI] [PubMed] [Google Scholar]
- 41.Smit P, et al. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perry JA, et al. A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 2003;131:866–871. doi: 10.1104/pp.102.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu H, Riely BK, Burns NJ, Ane J-M. Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics. 2006;172:2491–2499. doi: 10.1534/genetics.105.051185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remy W, Taylor TN, Hass H, Kerp H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. 1994;91:11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tansengco ML, Hayashi M, Kawaguchi M, Imaizumi-Anraku H, Murooka Y. crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus. Plant Physiol. 2003;131:1054–1063. doi: 10.1104/pp.102.017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;126:1563–1570. doi: 10.1242/dev.126.8.1563. [DOI] [PubMed] [Google Scholar]
- 47.Diaz CL, Gronlund M, Schlamann H, Spaink HP. In: Lotus japonicus Handbook. Marquez AJ, editor. Dordrecht, The Netherlands: Springer; 2005. pp. 261–277. [Google Scholar]
- 48.McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. A new method which gives an objective measure of colonization of roots by vesicular and arbuscular mycorrhizal fungi. New Phytol. 1990;115:495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.