Abstract
Retinoic acid (RA) plays important roles in development by modulating gene transcription through nuclear receptor activation. Increasing evidence supports a role for RA and RA receptors (RARs) in synaptic plasticity in the brain. We have recently reported that RA mediates a type of homeostatic synaptic plasticity through activation of dendritic protein synthesis, a process that requires dendritically localized RARα and is independent of transcriptional regulation. The molecular basis of this translational regulation by RA/RARα signaling, however, is unknown. Here we show that RARα is actively exported from the nucleus. Cytoplasmic RARα acts as an RNA-binding protein that associates with a subset of mRNAs, including dendritically localized glutamate receptor 1 (GluR1) mRNA. This binding is mediated by the RARα carboxyl terminal F-domain and specific sequence motifs in the 5′UTR of the GluR1 mRNA. Moreover, RARα association with the GluR1 mRNA directly underlies the translational control of GluR1 by RA: RARα represses GluR1 translation, while RA binding to RARα reduces its association with the GluR1 mRNA and relieves translational repression. Taken together, our results demonstrate a ligand-gated translational regulation mechanism mediated by a non-genomic function of RA/RARα signaling.
Keywords: glutamate receptor 1, nuclear receptor, synaptoneurosome, RNA binding protein, translational regulation
RA is a morphogen that plays key roles in neurogenesis, neuronal development, and cell differentiation (1, 2). However, the molecular components involved in RA synthesis and signaling persist in the adult brain (3). RA is rapidly synthesized in the adult brain (3) and participates in learning-related synaptic plasticity such as long-term potentiation and long-term depression (4, 5). Loss of RA signaling through deprivation of vitamin A (an RA precursor) leads to impaired hippocampal long-term potentiation and long-term depression, both of which can be restored by the administration of vitamin A or RA (6). However, the molecular mechanisms by which RA regulates adult brain function are not understood.
RA signaling is mediated by receptors that are members of the nuclear receptor superfamily of transcriptional regulators. RA receptor α (RARα, NR1B1) regulates the transcription of numerous genes via binding to specific upstream RA-responsive elements (7). RARα is present at high levels in the hippocampus and cortex of adult animals (8). Our recent study revealed an unexpected role in the hippocampus for RA and RARα during homeostatic synaptic plasticity (9). This type of plasticity, manifested as increased synaptic transmission in response to reduced neural activity, requires the translation of new proteins and is characterized by the insertion of newly synthesized homomeric GluR1 receptors (10). RA synthesis is rapidly enhanced upon activity blockade, which in turn potentiates synaptic strength through the activation of glutamate receptor protein translation in neuronal dendrites (9). Surprisingly, this action of RA is mediated by a population of dendritic RARα. Knocking down neuronal RARα expression blocked homeostatic synaptic plasticity induced by either RA or activity blockade. Additionally, the use of an RARα specific agonist recapitulated the effects on both synaptic scaling and local GluR1 synthesis (9). We also observed translation of GluR1 mRNA in dendritic RNA granules containing RARα using immunogold electron microscopy (11). One of the most striking results in these studies was the ability of RA or an RARα agonist to stimulate GluR1 protein synthesis in synaptoneurosomes, a biochemical preparation that lacks somatic components and thus operates in a transcription-independent manner. These results strongly suggest a non-genomic role for RARα and imply that RA/RARα can directly regulate translation. However, the molecular nature of such regulation remains unknown.
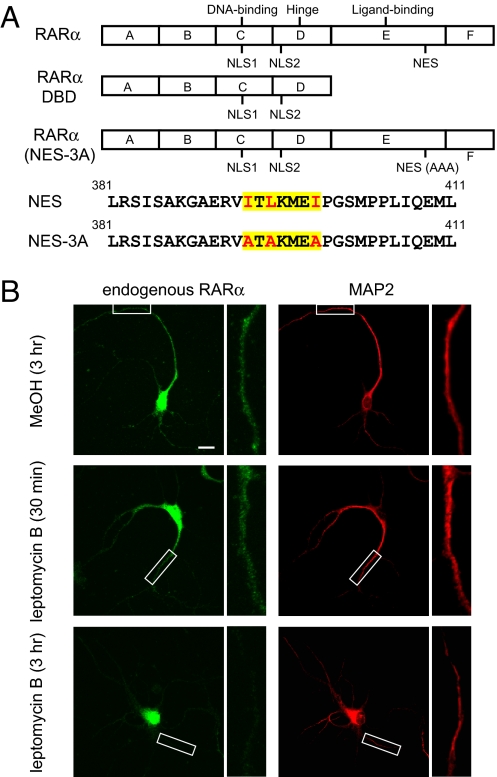
RARα consists of six modular domains (Fig. 1A). The A/B domain, or the N-terminal activation domain, plays a role in transcriptional regulation. The C domain functions as the DNA binding domain and is adjacent to the hinge region (the D domain), which contains nuclear localization signals (NLS). The E domain contains ligand binding and C-terminal activation domains, and the function of the F domain is largely unknown (12). It is thought that RARα resides in the nucleus as a heterodimer with the retinoid X receptor and is associated with corepressors that silence transcription. Upon ligand binding, the RARα ligand binding domain (LBD) undergoes a conformational change, resulting in the release of transcriptional corepressors and the recruitment of co-activators followed by transcriptional activation (13).
Fig. 1.
Dendritic localization of RARα utilizes active export from the nucleus. (A) Schematic of constructs used to examine RARα localization. All constructs encode C-terminal EGFP fusion proteins. (B) Localization of endogenous RARα in hippocampal neurons (14 days in vitro) treated with an active nuclear export inhibitor, LMB. Areas highlighted by white boxes are shown at higher magnification. Green, RARα protein; red, microtubule associated protein 2. (Scale bar, 25 μm.)
Here we investigated the molecular mechanism by which RA/RARα regulates translation of specific mRNAs in neuronal dendrites. We show that RARα is transported by active nuclear export into neuronal dendrites and binds a subset of dendritically localized mRNAs, including the mRNA encoding the glutamate receptor subunit, GluR1. Importantly, this binding is mediated by interactions between specific sequence motifs in the 5′UTRs of target mRNAs and the RARα F-domain. RA activation of RARα reduces its RNA binding affinity, which may underlie the translational de-repression by RA observed both in vivo and in vitro.
Results
RARα Exits the Nucleus via Active Nuclear Export.
We have previously reported that RARα is found in neuronal dendrites and in the nucleus (9). Because many transcription factors contain an NLS, a cytoplasmic delivery mechanism must exist for a transcription factor to be dendritically localized. In the case of CREB, zif 268, and Elk1, this is achieved by mRNA localization to dendrites and local translation (14). The lack of dendritic RARα mRNA by in situ hybridization [supporting information (SI) Fig. S1], however, suggests that a different mechanism transports RARα out of the nucleus.
Nuclear receptors such as nerve growth factor induced clone B (NGFI-B, NR4A1), estrogen receptors, and thyroid hormone receptors contain nuclear export signals (NES), and can be exported from the nucleus in a classical, Crm1-dependent manner (15, 16). To determine whether direct nuclear export participates in RARα localization, we treated cultured rat hippocampal neurons at 14 days in vitro with leptomycin B (LMB), a nuclear export inhibitor (Fig. 1B). Consistent with our previous report (9), endogenous RARα was present in the dendrites of untreated neurons (Fig. 1B). Three hours of LMB led to an 80% decrease in cytoplasmic/dendritic localization of endogenous RARα (n = 9 neurons/group, P < 0.005), indicating that its localization relies on Crm1-mediated export (Fig. 1B). Dendritic localization was also observed when GFP-tagged RARα was expressed for 18 h in cultured hippocampal neurons, and such localization was similarly sensitive to LMB (Fig. S2 A and B). The rapid reduction in cytoplasmic RARα signal following LMB is most likely caused by a shift in the import/export equilibrium towards nuclear import, which is supported by the observation that there is a concomitant ≈60% increase in endogenous nuclear RARα levels after LMB treatment (data not shown). Another possible but perhaps minor factor that may contribute to the reduction in cytoplasmic RARα signal is the relatively short half-life (4 h) of RARα as a result of active turnover by proteasomal degradation (17, 18).
Using NetNES, we identified a putative NES (19) in the ligand-binding domain of RARα (Fig. 1A). To examine the contribution of this putative NES to nuclear export, we tested a truncated RARα GFP construct encoding the DNA binding domain (RARα DBD), which contains both NLS sequences but lacks the putative NES (Fig. 1A). Consistent with NLS function, RARα DBD expression was confined to the nucleus (Fig. S2 A and B). We further examined an NES mutant of RARα, RARα(NES-3A), in which the key residues within the predicted NES motif were replaced with alanines (Fig. 1A). The cytoplasmic/dendritic localization of RARα(NES-3A) was greatly reduced compared to that of the WT RARα, and was comparable to that found when the WT RARα GFP was treated with LMB (Fig. S2 A and B). Taken together, these data suggest that RARα is subject to active nuclear export.
RARα Is Directly Associated with GluR1 mRNA In Vivo.
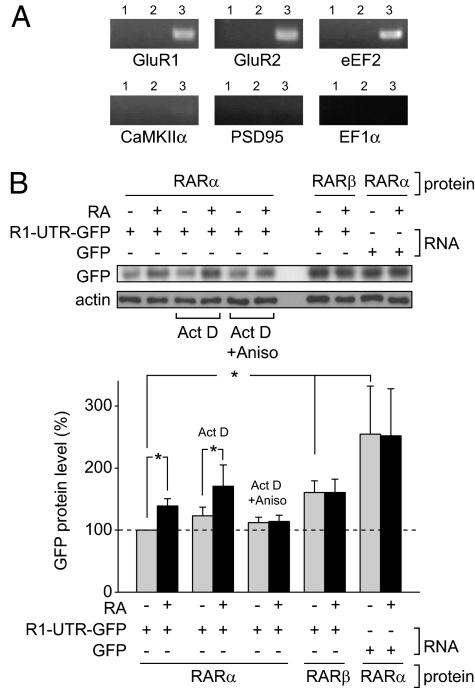
We have previously established that dendritic RA signaling plays an important role in homeostatic synaptic plasticity, which occurs through RARα-mediated translational regulation in dendrites (9, 11). As RNA binding proteins are integral to the intracellular sorting and translational control of mRNAs in dendrites (20), we speculated whether RARα could directly associate with mRNAs. We performed cross-linking followed by immunoprecipitation (CLIP) on 3-week-old rat hippocampal tissue, a method previously used by others to identify interactions between RNA-binding proteins and mRNA (21). CLIP utilizes UV light to cross-link protein-nucleic acid interactions at their contact points, retaining endogenous interactions and allowing for stringent wash conditions during immunoprecipitation. After cross-linking, we immunoprecipitated RARα and identified associated mRNAs by semiquantitative RT-PCR using gene-specific primers spanning two or more exons. We found that RARα protein co-immunoprecipitated with GluR1 mRNA as well as mRNAs encoding GluR2 and eukaryotic elongation factor 2 (eEF2), all of which have been shown to be dendritically localized by in situ hybridization (22, 23) (Fig. 2A and Fig. S3A). This interaction appears to be specific for a subset of mRNAs, as RARα did not co-immunoprecipitate with Ca+2/calmodulin kinase 2, α subunit (CaMKIIα), postsynaptic density protein 95 (PSD95) or eukaryotic elongation factor 1α (EF1α) mRNAs, which are also present in dendrites (23, 24) (Fig. 2A and Fig. S3A). We then performed CLIP using hippocampal synapto-neurosomes as starting material. Similar to our findings with whole hippocampal tissue, we found that RARα associated with GluR1, GluR2, and eEF2 mRNAs, but not those of CaMKIIα, PSD95, and EF1α (Fig. S3B), suggesting that the interaction between RARα and mRNAs is preserved in dendrites.
Fig. 2.
RARα interacts with specific mRNAs in vivo. (A) CLIP in the hippocampus. Triturated rat hippocampal tissue was exposed to UV radiation to cross-link protein-RNA interactions followed by RARα immunoprecipitation and gene-specific RT-PCR. Lane 1, no antibody; lane 2, GFP antibody; lane 3, RARα antibody. (B) Translational regulation of GluR1 5′UTR-GFP-3′UTR reporter by RA in vivo. HEK293 cells were transfected with an untagged RARα or RARβ construct for 24 h, then with a GluR1 5′-GFP-3′ reporter, which consists of the GFP ORF flanked by both 5′ and 3′UTR regions of GluR1 mRNA. Cells were then treated with 1 μM RA for 1 h in the presence or absence of actinomycin D and/or anisomycin. Reporter levels were quantified by immunoblotting (n = 5/group except for RARβ + R1-UTR-GFP ± RA and RARα + GFP ± RA, which were n = 4/group; *, P < 0.05, two-tailed paired t test).
We next examined with CLIP whether another RA receptor family member, RARβ, associated with mRNAs. Although RARα is found in hippocampus, cortex, and striatum, RARβ is specifically expressed in the striatum, hypothalamus, and medulla oblongata, and weakly expressed in the cortex and hippocampus (8). Consistent with results from hippocampal tissue, RARα CLIP with either forebrain or striatal tissue showed association between RARα protein and mRNAs of GluR1, GluR2, and eEF2 (Fig. S4A). By contrast, we observed only minimal association between RARβ and the aforementioned mRNAs in striatum, although RARβ protein is more abundant than RARα (Fig. S3A). Moreover, the RARα and RARβ antibodies were similarly efficient in immunoprecipitation (Fig. S4B).
RARα Regulates the Translation of a Reporter Construct Containing the 5′ and 3′ UTRs of the GluR1 mRNA.
We then sought to determine whether mRNA binding by RARα mediates the RA-dependent translational regulation we observed (9). We transfected HEK293 cells with an untagged RARα or RARβ construct, then, 24 h later, with a reporter encoding GFP flanked by the 5′ and 3′ UTRs of GluR1 (R1-UTR-GFP). GFP reporter expression was significantly lower in cells co-expressing RARα than in cells co-expressing RARβ (Fig. 2B). When the GluR1 5′ and 3′UTRs were not included, RARα-mediated repression was not observed (Fig. 2B), suggesting that the GluR1 UTRs participate in RARα-regulated protein translation. Moreover, addition of 1 μM RA to cells co-transfected with RARα and R1-UTR-GFP increased GFP expression levels by 140%, a regulation not observed with the reporter lacking GluR1-UTRs or in RARβ co-transfected cells (Fig. 2B).
The direct association between RARα and GluR1 mRNA and the lack of an RARβ effect on R1-UTR-GFP expression suggest that RARα regulates GluR1 translation by UTR binding. Indeed, the RA-induced increase in R1-UTR-GFP expression was blocked by the translational inhibitor anisomycin, but not by the transcriptional inhibitor actinomycin D (Fig. 2B). Levels of R1-UTR-GFP transcripts were not altered by RA treatment (Fig. S5), suggesting that RA neither changes RNA stability nor increases GFP protein expression through transcriptional activation. Together, these data indicate that RARα-mediated translational repression of GluR1 is both UTR-dependent and RA-sensitive.
RARα Binds RNA via the F-Domain.
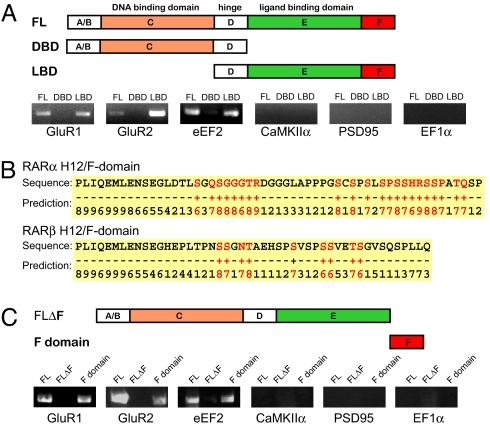
Although RARα binding to DNA has been well characterized, an interaction with RNA has been unclear. Others have shown that DNA binding proteins, such as p53, bicoid, TRA-1, and Xenopus TFIIIA, are able to bind RNA via their DBD, whereas thyroid and estrogen receptors bind RNA via other motifs (25, 26). We sought to identify the RNA interacting domain of RARα with a modified in vitro RNA selection assay in which RARα protein domains were fused to GST and immobilized on glutathione Sepharose beads. A total RNA pool harvested from 3-week-old hippocampal synaptoneurosomes (to enrich for dendritic mRNAs) was used for selection (27). RT-PCR was then used to detect the relative enrichment of selected RNAs, which indicates selective binding to RARα protein domains. Consistent with results from our CLIP experiments, full-length (FL) RARα protein bound GluR1, GluR2, and eEF2 mRNAs, but not CaMKIIα, PSD95, or EF1α mRNAs (Fig. 3A). We tested recombinant proteins containing only the RARα DBD or the RARα ligand-binding domain (RARα LBD), retaining the hinge region in both (Fig. 3A). Specific binding to GluR1, GluR2, and eEF2 mRNA was observed with the RARα LBD, whereas little to no signal was observed for any of the tested mRNAs using the RARα DBD (Fig. 3A), suggesting that either RARα DBD does not bind mRNA or the interaction is not specific.
Fig. 3.
RARα LBD binds RNA through the F-domain. (A) Recombinant full-length and truncated GST fusion RARα proteins used for specific RNA selection from a pool of synaptoneurosome RNA. Lower, RT-PCR of specific RARα-bound mRNAs following selection. (B) RARα (but not RARβ) F-domain contains stretches of RNA binding residues as predicted by BindN. Binding residues (+) are highlighted in red and non-binding residues (−) are black. Numbers indicate prediction confidence, with 0 being the lowest and 9 being the highest. (C) Involvement of F-domain in RNA binding. Upper, recombinant truncated GST-RARα proteins used to identify the F-domain as necessary for mRNA binding. Lower, selection of specific mRNA species from synapto-neurosome total RNA using GST fusion proteins containing full length RARα (FL, lane 1), RARα lacking H12 and F-domain (ΔH12/F, lane 2), and the F-domain alone (H12/F, lane 3).
As the LBD does not have an obvious nucleic acid binding domain, we used two prediction tools, RNABindR and BindN (28, 29), to identify putative RNA binding motifs in the RARα LBD. These tools predicted an RNA-binding region in the C terminus of RARα (i.e., the F-domain) that is immediately downstream of helix 12 (H12; Fig. 3B). To test this prediction, we generated two recombinant proteins, one containing the H12 and F-domain, and the other encoding the entire RARα protein except for the H12 and F-domain (Fig. 3C). When RNA selection was performed with these constructs, binding of GluR1, GluR2, or eEF2 mRNAs was observed with the F domain, but not the protein encoding the entire RARα protein except for the H12 and F-domain (Fig. 3C), indicating that the RARα F-domain is necessary for mRNA binding.
RARα Binds to the 5′UTR of GluR1 mRNA.
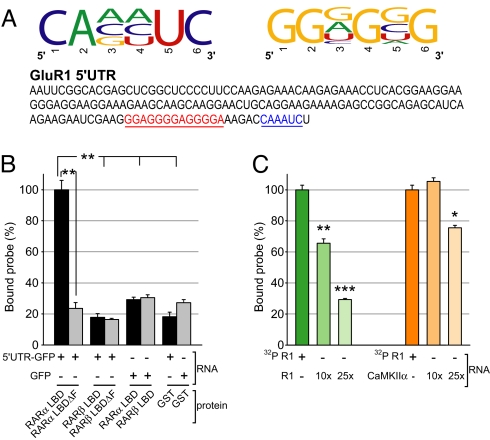
The binding of the RARα LBD to a subset of dendritic mRNAs suggests that RARα binds specific RNA sequences. To identify sequences recognized by RARα, we used an unbiased in vitro selection method called systematic evolution of ligands by exponential enrichment (SELEX). A pool of RNA oligonucleotides containing a stretch of random nucleotides flanked by adapter sequences were synthesized and applied to immobilized recombinant GST-RARα protein. The RARα-bound oligonucleotides were amplified with RT-PCR and transcribed again to generate a new pool of RNAs for subsequent rounds of selection. Five rounds of selection were performed before cloning and sequencing. We analyzed the sequences of positive clones and identified two putative RNA motifs, CAxyUC and GGnGnG (where x represents A, C, or G; y represents A, C, or U; and n represents any nucleotide; Fig. 4A and Fig. S6A). Both motifs were present in the 5′ UTR of GluR1, GluR2, and eEF2 mRNAs, but not in those of PSD-95, CaMKIIα, or EF1α (Fig. S6B). It should be noted that the motifs obtained by SELEX may represent only a subset of possible motifs as a result of inherent biases incurred during oligonucleotide synthesis, PCR, and T7 amplification. Further studies are required to fully understand the properties of the RNAs that are recognized by RARα.
Fig. 4.
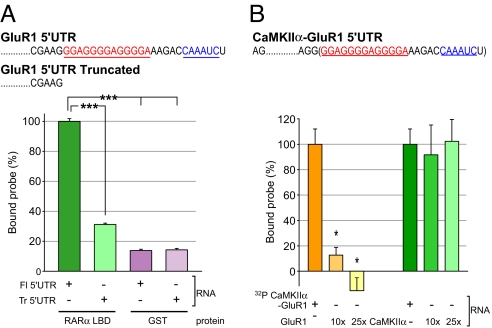
RARα LBD interaction with the GluR1 5′UTR requires the F-domain. (A) Specific consensus RNA motifs identified by SELEX. Both SELEX-selected motifs are present in GluR1 5′UTR. (B) Filter binding assay using RARα or RARβ LBD recombinant protein containing or lacking the F-domain co-incubated with 32P-labeled mRNA encoding GFP or GluR1 5′UTR-GFP as probes (n = 3/group; **, P < 5 × 10−4, single-factor ANOVA). (C) Filter binding competition assay using 32P-labeled GluR1 5′UTR (no GFP sequence) co-incubated with the RARα LBD in the presence of 10- and 25-fold amounts of cold GluR1 or CaMKIIα 5′UTR RNA (n = 3/group; *, P < 0.005; **, P < 5 × 10−4; ***, P < 1 × 10−5, single-factor ANOVA).
We next examined direct binding of RARα LBD to in vitro transcribed RNA using nitrocellulose filter binding assays, a classical test for RNA-protein interaction in vitro (30) We in vitro transcribed radiolabeled (32P-CTP) RNA encoding the GluR1 5′UTR fused to the GFP coding sequence and incubated it with various recombinant GST-fusion proteins. Compared with GST alone, the RARα LBD exhibited significant binding activity to GluR1 5′UTR-GFP RNA, and this binding activity was abolished by F-domain deletion (Fig. 4B). RARβ LBD did not display specific RNA binding activity compared with GST (Fig. 4B). Moreover, a GFP RNA lacking the GluR1 5′UTR did not bind RARα LBD (Fig. 4B), suggesting that RARα binds RNA in a sequence-specific manner. We confirmed this by a competitive binding assay. Addition of non-radiolabeled (i.e., cold) RNA encoding the GluR1 5′UTR significantly reduced the binding of 32P-labeled GluR1 5′UTR in a dose-dependent manner (Fig. 4C). In contrast, addition of cold CaMKIIα 5′UTR RNA did not have any effect at 10-fold and only slightly reduced 32P-labeled GluR1 5′UTR binding at 25-fold excess concentration (Fig. 4C).
We next examined whether the RNA sequence predicted by SELEX conferred binding of the GluR1 5′UTR to the RARα LBD. We generated 32P-CTP RNA probes containing either the entire 5′UTR of GluR1 (Fl 5′UTR) or a truncated GluR1 5′UTR lacking the SELEX-predicted RNA motifs (Tr 5′UTR). Removal of the predicted RNA sequences dramatically reduced affinity for the RARα LBD (Fig. 5A), suggesting that these motifs are required for GluR1 mRNA binding to RARα. We next asked whether the SELEX motifs alone were sufficient for RARα association. We generated a chimeric probe by adding the GluR1 SELEX motif to the 3′ end of the CaMKIIα 5′UTR (CaMKII-GluR1 5′UTR), which does not bind RARα, and subjected it to filter binding (Fig. 5B). This chimeric RNA associated with RARα LBD specifically as addition of cold GluR1 5′UTR at 10- or 25-fold concentrations greatly reduced the binding activity (Fig. 5B). The CaMKIIα 5′UTR did not contribute to the binding activity, as addition of excess amounts of cold CaMKIIα 5′UTR had no effect on the chimera binding activity (Fig. 5B).
Fig. 5.
Specific mRNA sequence motifs in the GluR1 5′UTR convey RARα binding. (A) Filter binding assay using RARα LBD and co-incubating with 32P-labeled full-length GluR1 5′UTR or a truncated (Tr) version lacking both SELEX motifs (n = 3/group; ***, P < 1 × 10−5). (B) Competition filter binding assay using 32P-labeled CaMKIIα probe fused to the GluR1 SELEX motifs. Competition was assayed with excess amounts (10-fold or 25-fold) of cold GluR1 5′UTR or cold CaMKIIα 5′UTR RNA (n = 3/group; *, P < 0.05, single-factor ANOVA).
RARα LBD Represses Translation via the GluR1 5′UTR in an RA-Sensitive Manner In Vitro.
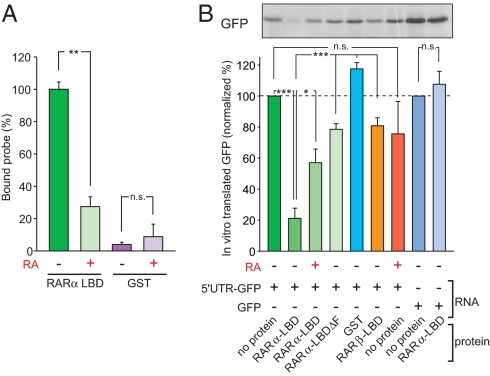
We have established thus far that RARα binds to the 5′UTR of GluR1 mRNA through two RNA motifs that are also present in the 5′UTRs of two other RARα-associated mRNAs. The observed GluR1 translational activation by the addition of RA (9) (Fig. 2B) suggests an RA-sensitive translational repression mechanism, perhaps involving 43S pre-initiation complex scanning. A prediction from this would be that, upon RA binding, a conformational change occurs, leading to dissociation of RARα from the GluR1 5′UTR and permitting translational activation. We tested this hypothesis by first examining the effect of RA on the affinity between the RARα LBD and the GluR1 5′UTR. Indeed, addition of 10 μM RA significantly reduced the association between GluR1 5′UTR and the RARα LBD (Fig. 6A) without affecting RNA stability (Fig. S7A).
Fig. 6.
RARα LBD functions as a retinoic acid-gated translational repressor. (A) Filter binding assay using the RARα LBD and a 32P-labeled GluR1 5′UTR probe in the presence or absence of 10 μM retinoic acid (n = 3/group; **, P < 5 × 10−4). (B) Reconstitution of RARα LBD-mediated translational regulation in an in vitro translation system. RNA encoding GluR1 5′UTR-GFP was added to RARα LBD in the presence or absence of 10 μM RA. Mixture was then added to rabbit reticulocyte lysate and incubated for 90 min at 30 °C in the presence of 35S methionine. Samples were run with SDS/PAGE and analyzed by autoradiography (n = 5/group; *, P < 0.01; ***, P < 1 × 10−6, single-factor ANOVA).
We next asked whether we could reconstitute RA/RARα-mediated translational control by using rabbit reticulocyte lysates in an in vitro translation assay. We preincubated protein, RNA encoding the 5′ UTR-GFP or GFP alone, and/or RA before addition of the reticulocyte lysate. Addition of the RARα LBD to GluR1 5′UTR-GFP RNA repressed baseline translation (as measured by 35S-Met incorporation), whereas addition of the RARβ LBD did not (Fig. 6B). In accordance with the filter binding experiments, the RARα LBDΔF did not repress translation, nor was translation of GFP alone repressed by the addition of the RARα LBD (Fig. 6B and Fig. S7B), presumably because of a lack of protein-RNA interaction. Importantly, however, translation of 5′UTR-GFP incubated with RARα LBD was de-repressed with the addition of 10 μM RA (Fig. 6B). This RA-mediated regulation of translation was not observed when 5′UTR-GFP mRNA was incubated with RARα LBDΔF (Fig. S7B). Moreover, replacing the SELEX-predicted RNA sequence in GluR1 5′UTR with its reverse complementary sequence, as a control, resulted in significantly reduced binding activity with RARα LBD (Fig. S8A) to a level comparable to that of the GFP RNA (Fig. 4B). In addition, translation of the reverse complementary SELEX-GFP RNA sequence was not repressed by RARα LBD when translated in vitro (Fig. S8B). Together, these experiments demonstrate that the three basic components, the RARα LBD, the SELEX-predicted sequence in GluR1 5′UTR, and RA, represent a minimal system of translational regulation.
Discussion
In this article, we provide evidence for a non-genomic role of RARα as an RNA binding protein that directly regulates translation in an RA-gated manner. Specifically, we show that (i) dendritic localization of RARα protein requires active nuclear export; (ii) RARα directly binds specific mRNAs in vivo, including GluR1 mRNA; (iii) RARα binds to RNA via its C-terminal F-domain; (iv) RARα binding to GluR1 mRNA is mediated by consensus sequences in the 5′UTR; and (v) RARα binding to GluR1 mRNA represses translation, and this repression can be relieved by RA.
Our results are partly consistent with another study suggesting that RARα is an RNA-binding protein (31). This study, however, postulates that RARα binds mRNAs in a sequence-independent manner via its DNA-binding domain. It thus remains to be determined whether RARα has two distinct modes of mRNA interaction—the sequence-specific binding described here and the sequence-independent binding described by the other study (31)—or the sequence-independent interaction described in the other study (31) reflects a non-specific nucleic acid-binding reaction.
RARα Binding to the UTRs of Associated mRNAs.
Using SELEX, we have defined two RNA motifs that are preferentially recognized by RARα (Fig. 4A). The fact that both sequence motifs are present in the 5′UTRs of associated mRNAs (GluR1, GluR2, and eEF2) but absent from the 5′UTRs of non-associated mRNAs (PSD95, CaMKIIα, and EF1α) suggests that RARα binds to the 5′UTRs of mRNAs through specific sequence recognition, a notion further supported by the gain-of-binding activity of the CaMKIIα-GluR1 5′UTR chimera (Fig. 5B). A relationship between the 3′UTR and RARα, however, remains to be explored. The 3′UTR is often longer than the 5′UTR and is extensively associated with RNA binding proteins. These interactions regulate mRNA localization as well as translational regulation (22). As a result of the length and complexity of the 3′UTR, the presence of SELEX motifs does not necessarily warrant RARα and 3′UTR binding. However, although RARα alone is sufficient to regulate translation, interactions with other RNA-binding proteins may further tune translation. For example, although GluR2 mRNA binds RARα (Figs. 2 and 3), RA alone is not sufficient to stimulate GluR2 translation in cells (9), suggesting multiple translational control mechanisms (e.g., FMR1 protein-mediated repression [24]). The complexity of translational regulation provided by the differential association of mRNAs with various RNA binding proteins may thus constitute a layer of precise, context-dependent translational control.
RARα F-Domain and RNA Binding.
We show that specific mRNA binding to RARα is mediated by the F-domain, a region of high variability within the RAR family (32). Although unstructured, the RARα F-domain is particularly rich in glycine, proline, and serine residues. The lack of arginine and lysine-rich stretches in the F-domain, however, suggests a non-classical mechanism of RNA binding. Examples of these non-classical glycine- and/or proline-rich RNA binding proteins include cirp (cold inducible RNA binding protein) and mPrrp (mouse proline rich RNA binding protein) (33, 34). We have shown that the F-domain is capable of RNA binding in vitro (Fig. 3C) but it is not clear whether other RARα domains may influence F-domain binding affinity, an issue that requires further examination.
In addition to its ligand-dependent regulation, RARα can undergo many post-translational modifications including phosphorylation, trimethylation, and sumoylation, leading to differential functional outputs. These modifications influence transcriptional control, RARα DNA and/or ligand binding affinity, and possibly protein structure (35). These post-translational modifications are triggered by other regulatory events and may serve as mechanisms for fine-tuning RARα RNA binding and translational control. RARα phosphorylation studies have already identified sites in the LBD and many within the F-domain alone (36). Modification of these residues may thus cause the F-domain to adopt a rigid structure, which could then confer or enhance RNA binding. This also suggests that there may be a distinctly modified form of RARα that can be exported from the nucleus, bind mRNA, and localize to the dendrite. We postulate that cytoplasmic RARα can serve to integrate diverse signaling pathways through post-translational modification, resulting in the regulation of RNA binding and translational control.
A Model for RA-Gated RARα-Mediated Translational Control.
Based on our data, and on the RA-induced conformational change in RARα, we propose a mechanism of translational control whereby un-liganded RARα, of which H12 and the F-domain are extended away from the LBD, binds to the 5′UTR of GluR1 mRNA. This represses translation, possibly by hindering 43S pre-initiation complex scanning in a mechanism similar to ferritin mRNA translational control by iron regulatory protein-1 (37). In this repressed state, the RARα mRNA complex may be mobile, shuttling from the nucleus into the soma (via nuclear export) and dendrites. Upon RA binding, the RARα LBD undergoes a well described conformational change in which H12 (and the F-domain) shifts positions, resulting in mRNA de-repression followed by GluR1 translation (13).
In conclusion, our current findings describe the interactions among RA, RARα, and dendritically localized mRNAs in translation. This expands the scope of the biological function of RARα beyond its role as a regulator of gene transcription. Taken together with the role of RA in homeostatic synaptic plasticity we reported previously (9), RA/RARα signaling in the adult brain may serve to integrate transcriptional and translational events and facilitate cross-talk between cell types (e.g., between neurons and glia), thus making a significant contribution to the regulatory events that occur during normal synaptic function and plasticity.
Materials and Methods
Detailed experimental procedures are described in SI Text. FISH (23) and CLIP (21) studies were performed as described previously. Purified GST fusion proteins and total RNA from synaptoneurosomes were used for in vitro domain-specific RNA selection.
Supplementary Material
Acknowledgments.
We thank Dr. Jennifer Doudna (University of California, Berkeley, CA) for helpful suggestions and discussion. We also thank Jason Aoto for generating the rat GluR1 UTR-GFP construct, Peter Hastie for help with SELEX sequence analysis, Selina Gharasimian for help with GST fusion protein expression, Sandhiya Kalyanasundaram for technical assistance, and the members of the Chen laboratory for discussion and comments on the manuscript. The work was supported by Mabel and Arnold Beckman Foundation, the David and Lucile Packard Foundation, the W. M. Keck Foundation, and the National Institutes of Health (L.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807740105/DCSupplemental.
References
- 1.Germain P, et al. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol Rev. 2006;58:712–725. doi: 10.1124/pr.58.4.4. [DOI] [PubMed] [Google Scholar]
- 2.Jepsen K, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 3.Dev S, Adler AJ, Edwards RB. Adult rabbit brain synthesizes retinoic acid. Brain Res. 1993;632:325–328. doi: 10.1016/0006-8993(93)91170-w. [DOI] [PubMed] [Google Scholar]
- 4.Chiang MY, et al. An essential role for retinoid receptors RARbeta and RXRgamma in long-term potentiation and depression. Neuron. 1998;21:1353–1361. doi: 10.1016/s0896-6273(00)80654-6. [DOI] [PubMed] [Google Scholar]
- 5.Lane MA, Bailey SJ. Role of retinoid signalling in the adult brain. Prog Neurobiol. 2005;75:275–293. doi: 10.1016/j.pneurobio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Misner DL, et al. Vitamin A deprivation results in reversible loss of hippocampal long-term synaptic plasticity. Proc Natl Acad Sci USA. 2001;98:11714–11719. doi: 10.1073/pnas.191369798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umesono K, Giguere V, Glass CK, Rosenfeld MG, Evans RM. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature. 1988;336:262–265. doi: 10.1038/336262a0. [DOI] [PubMed] [Google Scholar]
- 8.Zetterstrom RH, et al. Role of retinoids in the CNS: differential expression of retinoid binding proteins and receptors and evidence for presence of retinoic acid. Eur J Neurosci. 1999;11:407–416. doi: 10.1046/j.1460-9568.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- 9.Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–320. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutton MA, et al. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 11.Maghsoodi B, et al. Retinoic acid regulates RARalpha-mediated control of translation in dendritic RNA granules during homeostatic synaptic plasticity. Proc Natl Acad Sci USA. 2008;105:16015–16020. doi: 10.1073/pnas.0804801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 13.Wurtz JM, et al. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 14.Eberwine J, Job C, Kacharmina JE, Miyashiro K, Therianos S. Transcription factors in dendrites: dendritic imprinting of the cellular nucleus. Results Probl Cell Differ. 2001;34:57–68. doi: 10.1007/978-3-540-40025-7_4. [DOI] [PubMed] [Google Scholar]
- 15.Katagiri Y, et al. Modulation of retinoid signalling through NGF-induced nuclear export of NGFI-B. Nat Cell Biol. 2000;2:435–440. doi: 10.1038/35017072. [DOI] [PubMed] [Google Scholar]
- 16.Maruvada P, Baumann CT, Hager GL, Yen PM. Dynamic shuttling and intranuclear mobility of nuclear hormone receptors. J Biol Chem. 2003;278:12425–14232. doi: 10.1074/jbc.M202752200. [DOI] [PubMed] [Google Scholar]
- 17.Zhu J, et al. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor alpha (RARalpha) and oncogenic RARalpha fusion proteins. Proc Natl Acad Sci USA. 1999;96:14807–14812. doi: 10.1073/pnas.96.26.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boskovic G, Desai D, Niles RM. Regulation of retinoic acid receptor alpha by protein kinase C in B16 mouse melanoma cells. J Biol Chem. 2002;277:26113–26119. doi: 10.1074/jbc.M201185200. [DOI] [PubMed] [Google Scholar]
- 19.la Cour T, et al. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 20.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 21.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 22.Grooms SY, et al. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J Neurosci. 2006;26:8339–8351. doi: 10.1523/JNEUROSCI.0472-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cassiday LA, Maher LJ., 3rd Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu B, Koenig RJ. An RNA-binding domain in the thyroid hormone receptor enhances transcriptional activation. J Biol Chem. 2004;279:33051–33056. doi: 10.1074/jbc.M404930200. [DOI] [PubMed] [Google Scholar]
- 27.Sung YJ, Weiler IJ, Greenough WT, Denman RB. Selectively enriched mRNAs in rat synaptoneurosomes. Brain Res Mol Brain Res. 2004;126:81–87. doi: 10.1016/j.molbrainres.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Terribilini M, et al. RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res. 2007;35:W578–W584. doi: 10.1093/nar/gkm294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyashiro KY, et al. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 31.Chen N, Onisko B, Napoli JL. The nuclear transcription factor RARalpha associates with neuronal RNA granules and suppresses translation. J Biol Chem. 2008;283:20841–20847. doi: 10.1074/jbc.M802314200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farboud B, Privalsky ML. Retinoic acid receptor-alpha is stabilized in a repressive state by its C-terminal, isotype-specific F domain. Mol Endocrinol. 2004;18:2839–2853. doi: 10.1210/me.2004-0236. [DOI] [PubMed] [Google Scholar]
- 33.Hori T, Taguchi Y, Uesugi S, Kurihara Y. The RNA ligands for mouse proline-rich RNA-binding protein (mouse Prrp) contain two consensus sequences in separate loop structure. Nucleic Acids Res. 2005;33:190–200. doi: 10.1093/nar/gki153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiyama H, et al. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137:899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bour G, Lalevee S, Rochette-Egly C. Protein kinases and the proteasome join in the combinatorial control of transcription by nuclear retinoic acid receptors. Trends Cell Biol. 2007;17:302–309. doi: 10.1016/j.tcb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Rochette-Egly C, Adam S, Rossignol M, Egly JM, Chambon P. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 37.Gray NK, Hentze MW. Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and eALAS mRNAs. EMBO J. 1994;13:3882–3891. doi: 10.1002/j.1460-2075.1994.tb06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.