Abstract
From a large-scale screen using splicing microarrays and RT-PCR, we identified 63 alternative splicing (AS) events that are coordinated in 3 distinct temporal patterns during mouse heart development. More than half of these splicing transitions are evolutionarily conserved between mouse and chicken. Computational analysis of the introns flanking these splicing events identified enriched and conserved motifs including binding sites for CUGBP and ETR-3-like factors (CELF), muscleblind-like (MBNL) and Fox proteins. We show that CELF proteins are down-regulated >10-fold during heart development, and MBNL1 protein is concomitantly up-regulated nearly 4-fold. Using transgenic and knockout mice, we show that reproducing the embryonic expression patterns for CUGBP1 and MBNL1 in adult heart induces the embryonic splicing patterns for more than half of the developmentally regulated AS transitions. These findings indicate that CELF and MBNL proteins are determinative for a large subset of splicing transitions that occur during postnatal heart development.
Keywords: CUGBP and ETR-3-like factors, heart development, muscleblind-like, splicing microarray
Coordinated control of alternative splicing (AS) on a genome-wide scale has the potential to drive proteome transitions with wide-ranging and critical biological consequences (1, 2). Disruption of splicing and its regulation, therefore, is implicated in disease causation and susceptibility (3). Splicing is regulated by RNA-binding proteins that bind to cis-regulatory elements near the splice sites. Some of the best-characterized splicing regulators include the serine–arginine (SR)-rich family, hnRNP proteins, and the Nova, PTB, FOX, TIA, CUGBP and ETR-3-like factors (CELF), and muscleblind-like (MBNL) families (4, 5). CELF and MBNL proteins were first characterized as factors involved in the pathogenesis of myotonic dystrophy and were subsequently shown to be direct regulators of AS (6–8). Recent advances in microarray and computational technologies have allowed comprehensive analyses of individual exons on a genome-wide scale, providing the ability to identify commonly regulated splicing events (9–12).
With some exceptions (13, 14), most large-scale analyses of regulated splicing have focused primarily on differences between adult tissues and tissues/cell cultures depleted for a splicing regulator rather than normal physiological transitions within a single tissue (9–11, 15). Developmental transitions provide an excellent opportunity to identify and determine the roles for coordinated splicing regulation associated with normal physiological change. The vertebrate heart is particularly attractive for such analysis because it undergoes extensive remodeling to meet the demands of increased workload in the developing organism (16). In addition, the heart has relatively low cellular complexity and little cell turnover (17) so that developmental splicing transitions reflect changes occurring within individual cells to a greater extent than in many other tissues. The physiological changes that occur before and after birth are particularly important as the fetal heart adapts to birth and converts to adult function. This postnatal remodeling is accomplished through transcriptional and posttranscriptional networks, including AS (16, 18).
In the present study, splicing microarrays and computational screens were used to investigate regulatory networks of AS in vertebrate heart development. A large number of coordinated splicing transitions were identified that undergo dramatic changes during heart development. Strikingly, >60% of the splicing transitions tested were conserved between mouse and chicken, supporting functional relevance to heart development. Computational and expression analyses, coupled with studies using transgenic and knockout mice identified cis elements and associated trans-acting factors that facilitate fetal-to-adult splicing transitions including those regulated by postnatal changes in CELF and MBNL protein expression. Together, these analyses identify and characterize a highly conserved and highly regulated program of AS that supports postnatal growth and maturation of the developing heart.
Results
Global Analysis of AS During Mouse Heart Development.
To identify AS transitions during mouse heart development, we carried out a large-scale screen using alternative splice-event profiling microarrays (described in ref. 15) and literature searches followed by RT-PCR validation. Splicing microarrays were used to detect changes in splicing and mRNA steady-state levels of 10,111 muscle- and heart-enriched genes between embryonic day 17 (E17) and adult mouse heart RNA (supporting information (SI) Fig. S1A). Predicted AS changes were validated by RT-PCR (Fig. S1B). We validated 147 splicing events that showed a > 2-fold change between E17 and adult heart by microarray analysis. We focused on 54 events from microarray analysis and 9 events from the literature validated by RT-PCR as exhibiting ≥20-point change in percent inclusion of the variably spliced region between E14 and adult heart. Among the 63 events collected, 41 (65%) exhibited an increase, and 22 (35%) exhibited a decrease in inclusion of the variable region. Most variable regions (81%) were in-frame (multiples of 3 nt). The breakdown of different splicing modes (e.g., cassette exon, alternative 3′ and 5′ splice site, etc.) is provided in Table S1.
To examine the relationship between transitions in splicing and mRNA levels during heart development, we compared all validated AS events that exhibited either a ≥20-point change in splicing and/or a ≥2.5-fold change in mRNA levels (78 genes total). Linear regression analysis revealed no significant correlation (R2 = 0.19) between the 2 datasets, indicating that different sets of genes are regulated by AS transitions or mRNA levels (Fig. S1C). Comparative gene ontology analysis showed that different but overlapping biological processes are associated with genes that undergo changes in splicing and those that undergo changes in mRNA levels (Fig. S2). Genes that undergo developmental splicing changes were enriched for cell structure and motility functions, but genes exhibiting changes in mRNA levels were also enriched for signal transduction and oxidative (lipid/steroid) metabolism. These results demonstrate that during heart development, distinct subsets of genes are regulated by changes in splicing and by changes in transcript levels. Similar results have been demonstrated in comparisons of splicing and mRNA profiles in adult tissues and during T cell activation (11, 13, 15, 19).
Subsets of Splicing Transitions Are Coordinated at Specific Times During Mouse Heart Development.
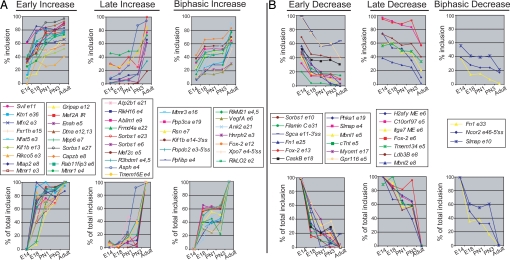
To determine whether splicing transitions are coordinated during mouse heart development, all 63 splicing events were assayed by RT-PCR using E14, E18, postnatal day 1 (PN1), PN3, and adult (3 months) heart RNA. These results were plotted both as percentage inclusion of the variable region(s) (Fig. 1A and B Upper) to show the absolute change and as the percentage of total change (Fig. 1 A and B Lower) to identify the timing of maximal change. This analysis identified groups of splicing transitions that are temporally coregulated. Splicing transitions were grouped as early or late depending on whether the maximal change occurred before or after birth, respectively. For example, 18 AS events exhibited a maximal increase in inclusion before birth (Fig. 1A) whereas 12 exhibited an early decrease (Fig. 1B). Similarly, of 17 postnatal splicing transitions, 10 exhibited increased inclusion, and 7 exhibited decreased inclusion of the variable region. Interestingly, 16 AS events exhibited biphasic transitions in which >20% of the total change took place between E14 and E18, <20% occurred between E18 and PN1, and >20% occurred between PN1 and adult. The complete list of AS events assayed is provided in Table S2.
Fig. 1.
Subsets of AS transitions are coregulated during specific times of mouse heart development. Total RNA was isolated from 6–20 pooled hearts at the indicated time points. RT-PCR analysis was carried out for 63 AS events. Data are expressed as the percentage inclusion (Upper) and as the percentage of total change (Lower) for variable regions that show increased (A) or decreased (B) inclusion during development. Alternative exons are numbered according to Ensembl, and sequences of variable regions are presented in Table S2. In at least 2 independent assays for all E14 and adult samples, the standard deviation was <5 percentage points.
A Large Fraction of Splicing Transitions Are Conserved During Mouse and Chicken Heart Development.
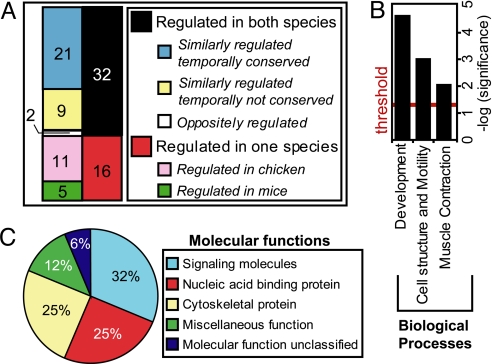
To determine the level of conservation, AS was assayed during chicken heart development by using orthologues of genes that exhibited changes in mouse and splicing events from the literature. A total of 114 AS events were tested, and 51 of them exhibited ≥20-point change between E8 and adult chicken heart (Table S3). Next, we performed a developmental time course of AS in chicken heart using E8, E12, E16, E20, post hatch day 2 (PH2), PH7, and adult hearts (>6 months). Just as most splicing transitions in mouse could be grouped according to maximal change relative to birth, most transitions during chicken heart development could be grouped by maximal change relative to hatching (Fig. S3). Direct comparison of 48 splicing transitions of variable mRNA regions that are conserved between mouse and chicken (≥60% nucleotide identity) revealed that 32 (66.7%) were alternatively spliced and that 30 (62.5%) of these were similarly regulated in that the AS patterns change in the same direction during development (Fig. 2A and Table S4). Furthermore, 21 of the 30 AS transitions switched at comparable times during mouse and chicken heart development (representative examples shown in Fig. S4). Differences include 16 splicing events regulated only in 1 species and 2 transitions regulated in both species but in opposite directions.
Fig. 2.
Most of the splicing transitions regulated during mouse heart development undergo similar transitions during chicken heart development. (A) Conservation of AS in mouse and chicken heart development. (B) Gene ontology of 30 events conserved between chicken and mouse. Enriched biological processes are plotted on a −log (P value) scale with the threshold set to 1.3 [log (0.05)]. (C) Molecular functions assigned to the genes falling in the most significantly enriched process “Development” (P < 0.000005).
Gene ontology analysis of the 30 conserved AS transitions indicated that most genes participate in processes such as development, cell structure/motility, and muscle contraction (Fig. 2B). Genes in the development group were further analyzed for their annotated molecular function, showing that the majority of them encode cytoskeletal, signaling, or nucleic acid-binding proteins (Fig. 2C). Because the majority of the genes in this dataset do not undergo significant changes in mRNA levels or the reading frame, these regulated events represent a conserved qualitative rather than quantitative change in the proteome.
Binding Motifs for Known Splicing Factors Including CELF and MBNL Are Enriched and/or Conserved in the Flanking Introns of Developmentally Regulated Exons.
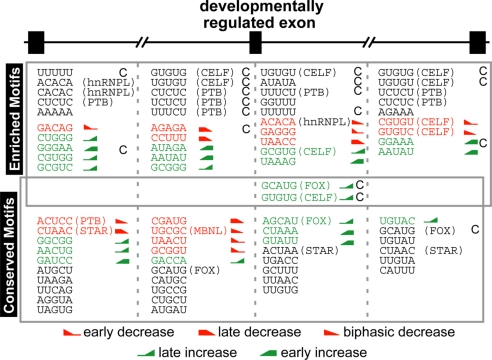
Sequence motif analyses were conducted to identify putative regulatory elements in the flanking intronic regions of developmentally regulated exons in mouse and chicken. The first and last 250 bases of the upstream and downstream introns were used in these analyses with the exclusion of the core splice-site (ss) motifs (8 bases for 5′ss, 30 bases for 3′ss). Enrichment and evolutionary conservation of pentanucleotide motifs were analyzed in the 4 regions comparing to first-order Markov models that account for dinucleotide effects on abundance and conservation (see SI Text). To identify motifs associated with splicing transitions occurring at specific times during heart development, a regression analysis between motif counts and AS changes at each time point was conducted for those pentamers that were significantly conserved or enriched. The most significant pentamers in the above analyses are displayed in Fig. 3 (complete lists of significant motifs are included in Table S5, Table S6, Table S7, Table S8, Table S9, and Table S10).
Fig. 3.
Motifs significantly enriched (Upper) and/or conserved (P < 0.001) among 8 mammalian species (Lower) in the 4 flanking 250-nt intronic regions of developmentally regulated mouse exons. Motifs that are significantly conserved and/or enriched that also exhibit a significant (P ≤ 0.05) association with specific temporal transitions in the regression analysis are indicated in red (decreasing inclusion) or green (increasing inclusion) with icons indicating the transition pattern. Motifs in black are significantly conserved or enriched but not significant the regression analysis. “C” indicates that the motif is also significantly enriched among the developmentally regulated chicken exons. The 5 most significant pentamers are shown (see Table S5, Table S6, Table S7, and Table S8 for the complete lists of motifs). Motifs resembling known binding sites of splicing factors (Table S10) are annotated. Fox (GCAUG) and CELF (GUGUG) motifs were significant in all analyses being enriched in mouse and chicken, conserved in mammals, and associated with a specific temporal pattern.
A number of motifs in each intronic region were identified including some that resemble the binding sites of known splicing factors. Two motifs in particular, GCAUG (Fox) and GUGUG (CELF), located in the immediate downstream intronic region, were significant in all parameters examined: conserved among 8 mammalian species, enriched within this region in both mouse and chicken, and a significant association with specific temporal changes (increased exon inclusion late in development). The regression analysis of additional CELF and Fox motifs enriched or conserved, respectively, in this region also supports a role for these 2 proteins in regulating developmental AS changes, especially in the postnatal stages. Expression of CELF and Fox proteins during postnatal development are consistent with these results (see below). Interestingly, CELF motifs were associated with increased exon inclusion when located in the first 250 bases of the downstream intron and decreased exon inclusion when located in the last 250 bases of the downstream intron. This pattern suggests that, like some other families of factors, the CELF family splicing factors may function to activate or repress splicing in a location-dependent manner (20, 21). The MBNL motif UGCGC was highly conserved in the last 250 bases of the upstream introns and was among the most significant motifs in the regression analysis being associated with decreased exon inclusion compared with both E14 and adult data at all 5 time points. PTB motifs were identified more often in the enrichment analysis (in both mouse and chicken) than in the mammalian conservation analysis, suggesting that PTB sites may evolve more quickly. The regression analysis identified an association between decreased exon inclusion with the ACUCC motif located in the first 250 bases of the upstream intron. Finally, CUAAC and ACUAA were highly conserved in multiple regions among mammals, and CUAAC in the first 250 bases of the upstream intron was associated with decreased exon inclusion. These motifs resemble the motif (A)CUAAC that was found to be associated with skeletal muscle-specific exons (12, 22), and binding motifs for STAR family proteins (23).
A Postnatal Expression of CELF, MBNL, and Fox Proteins Correlates with a Subset of AS Transitions.
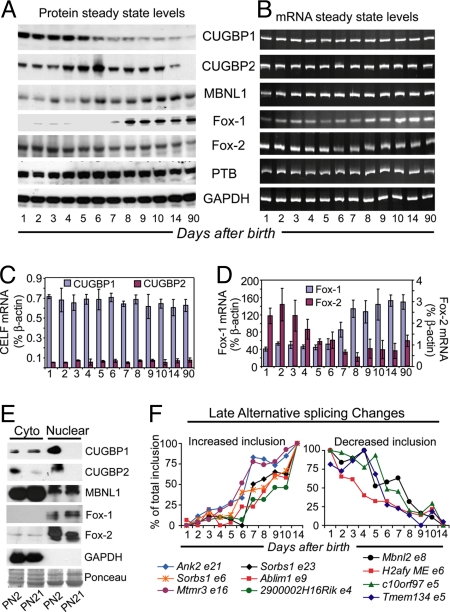
We found a significant enrichment/conservation of binding motifs for the CELF, MBNL, and Fox families flanking the developmentally regulated exons. The two CELF proteins expressed in heart, CUGBP1 and CUGBP2, are expressed at low levels in adult compared with embryonic heart (24). To determine the timing of CUGBP1 and CUGBP2 down-regulation in mouse heart development, we analyzed their protein and mRNA expression during the first 2 weeks after birth. As shown in Fig. 4A, CUGBP1 and CUGBP2 protein levels begin to decrease by PN6 and PN10, respectively. In contrast, MBNL1 protein levels begin to increase at PN5, whereas no consistent change was evident in PTB levels. Strikingly, Fox-1 protein was robustly up-regulated postnatally after PN6, whereas Fox-2 decreased slightly in adult mice when compared with PN1. Western blot analysis of serially diluted PN1 protein samples compared with adult demonstrated ≈10-, 18-, and 3-fold reductions in CUGBP1, CUGBP2, and Fox-2 protein levels, respectively (Fig. S5). On the other hand, 10- and 3.8-fold increases were observed in Fox-1 and MBNL1 proteins, respectively.
Fig. 4.
Postnatal expression of CELF, Fox, and MBNL proteins correlate with AS transitions. (A) Steady-state protein levels. (B) Steady-state mRNA levels by RT-PCR. (C and D) Steady-state mRNA levels determined by real time RT-PCR (TaqMan). (E) Nuclear and cytoplasmic distributions of CUGBP1, CUGBP2, Fox-1, Fox-2, and MBNL1. The distribution of Fox-1 and 2 or GAPDH demonstrate the clean separation of nuclear and cytoplasmic fractions. (F) A tight postnatal time course demonstrates a correlation between the timing of postnatal AS transitions and changes in CELF, Fox, and MBNL expression.
In contrast to the loss of CUGBP1 and CUGBP2 proteins, their mRNA levels remained unchanged during this developmental period (Fig. 4 B and C). These results indicate that steady-state CUGBP1 and CUGBP2 protein levels are regulated posttranscriptionally, either at the level of translation or protein stability (or both). Postnatal expression of Fox-1, Fox-2, and MBNL1 mRNAs paralleled their respective protein expression profiles (Fig. 4 B and D).
MBNL1 has been shown to undergo a cytoplasmic to nuclear transition during the first 3 weeks of postnatal skeletal muscle development (25). To determine whether CUGBP1, CUGBP2, or MBNL1 differed in nuclear-cytoplasmic distribution during postnatal heart development, we prepared nuclear and cytoplasmic fractions from PN2 and PN21 hearts. Loss of CUGBP1 protein at PN21 was seen only in the nuclear fraction, whereas the cytoplasmic levels remained unchanged (Fig. 4E). In contrast, CUGBP2 levels declined sharply in both nuclear and cytoplasmic fractions at PN21. MBNL1 did not show nuclear accumulation as described in skeletal muscle but rather exhibited a small increase in the cytoplasmic fraction at PN21. We conclude that MBNL1 is not primarily controlled by changes in nuclear-cytoplasmic distribution during heart development. In addition, our results suggest selective loss of CUGBP1 from the nucleus during heart development.
CUGBP1 and MBNL1 Regulate Distinct as well as Overlapping Subsets of Postnatal AS Transitions.
Next, we sought to determine whether postnatal splicing transitions coincided with the changes in CELF and MBNL protein expression observed during postnatal development. The timing of 10 postnatal AS transitions selected for detailed analysis (8 late and 2 biphasic) demonstrated a good correlation with changes in CELF and MBNL1 protein expression. For instance, 8 of 10 AS transitions exhibited a maximal change after PN6 (Fig. 4F), the time at which CUGBP1 protein starts to decline and MBNL1 levels begin to rise.
To establish causal relationships between CELF and MBNL protein expression and specific sets of AS transitions, we examined AS changes in adult hearts from mice overexpressing CUGBP1 or deficient in MBNL1 expression. Transgenic mice expressing Flag-tagged human CUGBP1 in a heart-specific and tetracycline-inducible manner were obtained by generating tet-inducible CUGBP1 (TRECUGBP1) lines and crossing these with heart-specific rtTA lines -Myh6-rtTA (see SI Text). TRECUGBP1/Myh6-rtTA bitransgenic animals given doxycycline (designated TgCUGBP1) expressed CUGBP1 protein 5–8 times above the endogenous levels in adult heart (close to levels in PN1 heart) and, importantly, MBNL1 levels were not altered in these heart tissues (Fig. S6A). MBNL1 is required for a subset of postnatal AS changes that occur during skeletal muscle development (25). To investigate the contribution of MBNL1 in regulating AS transitions in the developing heart, we used the previously characterized Mbnl1ΔE3/ΔE3 knockout mice (26). These mice exhibited no change in CUGBP1 protein levels in heart tissue from these mice (Fig. S6B).
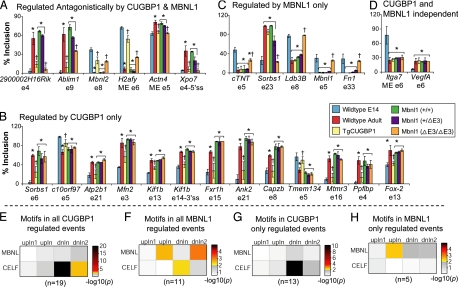
We compared 44 developmentally regulated AS changes in adult TgCUGBP1 mice (+ and − dox), Mbnl1ΔE3/ΔE3 mice and their respective wild-type littermate controls and identified 24 AS events that were altered by either increased expression of CUGBP1 and/or loss of MBNL1 (Fig. 5, all AS events screened are in Table S11). Thirteen AS events exhibited a response only in CUGBP1 overexpressing mice, 5 responded only in Mbnl1ΔE3/ΔE3 mice, and 6 responded in both TgCUGBP1 and Mbnl1ΔE3/ΔE3 mice (Fig. 5 A–C and Fig. S7 A–C). All events reverted to the embryonic/early postnatal splicing pattern strongly suggesting that these events are primarily regulated by elevated CUGBP1 and/or decreased MBNL1 activity in the embryo/early postnatal heart. Interestingly, all splicing events regulated by both proteins exhibited antagonistic responses consistent with previous work (6, 24). Two AS events, Itga7 and VegfA, represent the 20 events that were unaffected by either CUGBP1 overexpression or Mbnl1 depletion (Fig. 5D and Fig. S7D). Further supporting a determinative role for the postnatal expression of CUGBP1 and MBNL1 in regulating postnatal AS transitions, 18 of the 24 exons that responded to either CUGBP1 and/or MBNL1 normally undergo either late or biphasic postnatal transitions (Table S11). In addition, CELF- and MBNL-binding motifs are significantly enriched in the upstream and downstream introns flanking alternative exons regulated by CUGBP1 and MBNL1, respectively (Fig. 5 E–H). These data indicate that over half (24 of 44) of splicing events regulated during heart development are CUGBP1- and/or MBNL1-dependent, with the rest presumably affected by other splicing regulators.
Fig. 5.
A subset of postnatal splicing transitions are regulated by CUGBP1 and/or MBNL1. Forty-four AS events were tested in the hearts of TgCUGBP1 and Mbnl1ΔE3/ΔE3 mice and their respective littermate controls. (A–D) Splicing events were found to be regulated antagonistically by CUGBP1 and MBNL1 (A), regulated by CUGBP1 only (B), regulated by MBNL1 only (C), or CUGBP1 and MBNL1 independent (D). Each bar shows the mean ± SD for the percent inclusion of the specified variable region. Statistical analysis was done by using 1-way ANOVA, followed by Tukey's multiple-range test (P < 0.05). *, significantly different from E14. †, significantly different from wild-type littermates. (E–H) Computational analysis for enrichment of CELF- and MBNL-binding motifs in flanking introns of CUGBP1- and MBNL1-regulated exons. To the right of each plot are heat maps representing the P values for significant enrichment of indicated motifs in positions 12–250 of the upstream intron (upIn1), positions −250 to −31 of the upstream intron (upIn), positions 12 to 250 of the downstream intron (dnIn), and positions −250 to −31 of the downstream intron (dnIn1).
Discussion
Conserved AS Transitions Are Part of the Remodeling Program During Heart Development.
Cascades of transcriptional changes are known to coordinate regulatory networks during heart development (16). Here, we present evidence that ensembles of variably spliced regions are also coordinately regulated during mouse and chicken heart development. Particularly striking is the extent (>60%) to which the regulation of AS is conserved during heart development. This is in contrast to large-scale comparisons of orthologous gene pairs in which <20% of cassette-type events are conserved between human and mouse (27–29). The unusually high level of conservation observed here for both AS and its regulation most likely reflects enrichment of functional splicing events, in contrast to genome wide surveys that are blind to functional consequences. These results illustrate an advantage of performing analysis of splicing events associated with a specific physiological transition. Consistent with studies demonstrating that conserved splicing events tend to maintain reading frame (28, 29), 25 of the 32 (78.1%) splicing events conserved between mouse and chicken maintain the reading frame. Taken together, these results strongly suggest that the majority of these alternatively spliced isoforms have conserved functions. The nonconserved splicing transitions are also of interest as potential contributors to species-specific differences.
Gene Ontology (GO) analysis of conserved splicing transitions revealed particular enrichment for genes that are fundamental to the remodeling of cardiomyocytes during postmitotic growth, such as cytoskeletal rearrangement, nucleic acid binding and signaling. To supplement the GO analysis, we used literature searches to identify functions for 36 of the 44 genes tested in TgCUGBP1 and Mbnl1ΔE3/ΔE3 mice (Fig. S8). Some of the proteins expressed from these genes have diverse cellular functions but many (15 of 36) associate with contractile apparatus either within the Z-disk or via interactions with Z-disk structural or regulatory proteins. In both avian and mammalian hearts, the force generating capacity rises steeply during the first weeks after hatching or birth because of accumulation and structural reorganization of contractile proteins and a decrease in Ca2+ sensitivity (30). The Z-disk forms the structural anchor for the sarcomere as the attachment point of the thin filaments and is crucial for communication of mechanosensory information between cells as the attachment points of costameric filaments (31).
We also found that several proteins (10 of 36) that undergo developmentally regulated splicing transitions have nucleic acid-binding properties. Six of these take part in RNA processing/metabolism, whereas 4 regulate transcription. Consistent with identification of several splicing regulators among the set of developmentally regulated exons, we found examples of autoregulation and cross-regulation of splicing regulators. MBNL1 autoregulates splicing of its own exon 5, which encodes an 18-aa domain located within an ultraconserved segment (32). This exon is included in fetal but not adult hearts of both mouse and chicken. Intriguingly, we find that the analogous exon in Mbnl2 (exon 8) is also within an ultraconserved region, exhibits conserved regulation in mouse and chicken, and is regulated by both CUGBP1 and MBNL1. CUGBP1 also cross-regulates splicing of the splicing regulator Fox-2, which is highly expressed in brain, skeletal muscle, and heart (33). These observations agree with and extend previous findings that auxiliary splicing regulators tend to cross-regulate (10, 25).
Postnatal Switch of CUGBP1 and MBNL1 Protein Expression Controls Fetal-to-Adult Transitions for a Subset of Splicing Events.
CELF- and MBNL-binding sites were found to be enriched/conserved among developmentally regulated AS events in mouse. Only 2 of the 6 CELF genes (CUGBP1 and CUGBP2) are expressed in heart and both proteins are down-regulated during heart development (24, 25). We demonstrated that CUGBP1 and CUGBP2 protein levels drop by PN6 and PN10, respectively, and that regulation of both proteins is posttranscriptional. CUGBP1 protein stability is controlled by phosphorylation through a protein kinase C-dependent pathway, and a change in CUGBP1 phosphorylation during mouse heart development correlates with decreased protein stability in adult tissue (34). Selective loss of nuclear CUGBP1 compared with CUGBP2 in 3-week-old postnatal hearts suggests that CUGBP2 might use a different mechanism of down-regulation. Our results also indicate that MBNL1 expression is up-regulated during postnatal heart development via increased mRNA expression.
Consistent with computational analyses showing enriched CELF- and MBNL-binding sites, we identified 24 developmentally regulated AS events that are sensitive to changes in the steady-state levels of CUGBP1 and/or MBNL1 in mouse heart. Thirteen of these events are CUGBP1-specific, 5 are MBNL1-specific, and 6 are antagonistically regulated by both CUGBP1 and MBNL1. All 24 events revert to embryonic/early postnatal splicing patterns in adult hearts of TgCUGBP1 and/or Mbnl1ΔE3/ΔE3 mice. Eighteen of the 24 regulated exons undergo a postnatal switch, suggesting that they are normally regulated by CUGBP1 and/or MBNL1. Most TgCUGBP1 mice exhibit severe heart failure and arrhythmias, causing high mortality rates. It remains to be determined which, if any, altered splicing event(s) phenocopy the cardiac defects seen in these mice. The cardiac phenotype of MBNL1ΔE3/ΔE3 mice remains to be characterized. Because misregulated expression of both CELF and MBNL is associated with misregulation of alternative splicing in myotonic dystrophy (35), it will be interesting to determine to what extent this postnatal splicing network is disrupted in the disease.
Methods
Details of materials and methods used are in SI Text.
Animals.
Heart tissues were collected from wild-type FvB, Mbnl1ΔE3/ΔE3 (kindly provided by Maurice Swanson, University of Florida College of Medicine, Gainesville, FL), transgenic TRECUGBP mice (design of mice is provided in SI Text) and chickens (White Leghorns) at indicated time points for subsequent analysis.
Splicing Microarrays and RT-PCR Validations.
Microarray studies were performed as described in SI Text. Microarray prediction data were used to design RT-PCR primers around potential splicing regions. Percentage exon inclusion was calculated by using Kodak Gel logic 2200 and Molecular Imaging Software as: [(exon inclusion band/(exon inclusion band + exon exclusion band) × 100].
Computational Motif Analyses.
Four intronic regions (Fig. 4) flanking developmentally regulated exons in mouse and chicken were analyzed for motif conservation, enrichment, and correlation with time-course splicing alteration by using first-order Markov models as described in SI Text.
Western Blot Analysis and Real-Time RT-PCR.
CUGBP1, CUGBP2, MBNL1, PTB, Fox-1, Fox-2, and GAPDH protein expression by Western blot analysis and RNA expression by semiquantitative and real-time RT-PCR for was performed by using standard procedures.
Statistics.
Values are presented as mean ± SD. Statistical significance was determined by using 1-way ANOVA, followed by post hoc Tukey's multiple range test (P < 0.05).
Supplementary Material
Acknowledgments.
We thank C. Armour for sample amplification and microarray hybridization, D. Tran for initial validation, the Human Genome Sequencing Center (Baylor College of Medicine) for sequence confirmation of RT-PCR products, Dr. R. Sorek (Weizmann Institute of Science, Rehovot, Israel) for information on AS in chicken, Dr. D. Black (University of California, Los Angeles) for PTB antibodies, and Dr. M. Swanson for tissues from the MbnlΔE3/ΔE3 line. This project was funded by the National Institutes of Health Grants R01GM076493 and R01HL45565 (to T.A.C.) and HG002439 (to C.B.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809045105/DCSupplemental.
References
- 1.Graveley BR. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 3.Wang GS, Cooper TA. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 4.Gabut M, Chaudhry S, Blencowe BJ. SnapShot: The splicing regulatory machinery. Cell. 2008;133:192–e1. doi: 10.1016/j.cell.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: Towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 6.Ho TH, et al. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 8.Timchenko LT, et al. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blencowe BJ. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fagnani M, et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugnet CW, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ip JY, et al. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castle J, et al. Differential expression of 24,426 human alternative splicing events and predicted cis-regulation in 48 tissues and cell lines. Nat Genet. doi: 10.1038/ng.264. doi: 10.1038/ng64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res. 2002;90:1044–1054. doi: 10.1161/01.res.0000020201.44772.67. [DOI] [PubMed] [Google Scholar]
- 18.Xu X, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation–contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 19.Pan Q, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das D, et al. A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing. Nucleic Acids Res. 2007;35:4845–4857. doi: 10.1093/nar/gkm485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galareau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol. 2005;12:691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- 24.Ladd AN, Stenberg MG, Swanson MS, Cooper TA. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- 25.Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 26.Kanadia RN, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 27.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 28.Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siedner S, et al. Developmental changes in contractility and sarcomeric proteins from the early embryonic to the adult stage in the mouse heart. J Physiol. 2003;548:493–505. doi: 10.1113/jphysiol.2002.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samarel AM. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am J Physiol. 2005;289:H2291–H2301. doi: 10.1152/ajpheart.00749.2005. [DOI] [PubMed] [Google Scholar]
- 32.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 33.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.