Abstract
DEAD-box proteins, the largest helicase family, catalyze ATP-dependent remodeling of RNA–protein complexes and the unwinding of RNA duplexes. Because DEAD-box proteins hydrolyze ATP in an RNA-dependent fashion, the energy provided by ATP hydrolysis is commonly assumed to drive the energetically unfavorable duplex unwinding. Here, we show efficient unwinding of stable duplexes by several DEAD-box proteins in the presence of the nonhydrolyzable ATP analog ADP-beryllium fluoride. Another ATP analog, ADP-aluminum fluoride, does not promote unwinding. The findings show that the energy from ATP hydrolysis is dispensable for strand separation. ATP binding, however, appears necessary. ATP hydrolysis is found to be required for fast enzyme release from the RNA and multiple substrate turnovers and thus for enzyme recycling.
Keywords: helicase, RNA, Ded1, Mss116, eIF4A
Virtually all aspects of RNA metabolism require DEAD-box proteins, a large family of RNA-dependent ATPases that are involved in the localized, ATP-dependent manipulation of RNA and RNA protein complexes (1–3). DEAD-box proteins are highly conserved from bacteria to human and share at least 8 characteristic sequence motifs, one of which reads “D-E-A-D” in single amino acid letter code (2). DEAD-box proteins, the largest family of the “helicase” superfamily 2, are structurally similar to DNA helicases, enzymes that unwind DNA helices in an ATP-driven fashion (2, 4, 5). Seemingly analogously, DEAD-box proteins separate RNA duplexes in an ATP-dependent manner, but in contrast to DNA helicases, duplex unwinding is not based on translocation of the enzymes on one of the nucleic acid strands (2). Instead, DEAD-box proteins unwind by directly loading onto helical regions and then locally prying the strands apart (6–8). This unwinding mode enables DEAD-box proteins to disrupt duplexes without directionality and explains why efficient strand separation by these enzymes is restricted to RNA duplexes with <2 helical turns (6, 8).
How ATP is used for the unwinding process remains a central, unresolved mechanistic question. It is commonly inferred that DEAD-box proteins use the energy from ATP hydrolysis to drive the thermodynamically unfavorable strand separation, because the enzymes hydrolyze ATP in an RNA-dependent fashion (2), and because unwinding of stable duplexes has been seen only with ATP and hydrolyzable analogs, but not with nonhydrolyzable ATP analogs (1, 2, 9). However, only very few nonhydrolyzable analogs have actually been tested in unwinding reactions, and it has therefore remained possible that previously untested analogs promote strand separation without ATP hydrolysis.
It is well established that the ATP hydrolysis cycle involves a series of distinct steps, which include, at the most basic level, ATP binding, hydrolysis, and subsequent release of the hydrolysis products (10). For several ATPases, steps other than the actual hydrolysis have functional relevance (11, 12). For DEAD-box proteins, ADP is known to reduce their affinity for RNA, compared with ATP (13), but it has not been experimentally studied which steps in the ATPase cycle steps are linked to strand separation. Notwithstanding, speculations about a possible role for ATP binding in the strand separation process have been sparked by a recent structure of Vasa, bound to ssRNA and the nonhydrolyzable ATP analog adenylyl imidodiphosphonate (ADPNP) (14). However, ADPNP does not promote unwinding for any DEAD-box protein tested to date, and the functional interpretation of the structural data are thus complicated. Nevertheless, delineating how DEAD-box proteins use ATP for duplex unwinding and other functions is essential for understanding mechanism and regulation of these ubiquitous enzymes.
To approach this problem, we probed whether DEAD-box proteins indeed require the energy from ATP hydrolysis to unwind stable duplexes. We examined whether DEAD-box proteins can unwind duplexes with the nonhydrolyzable ATP analogs ADP-beryllium fluoride (ADP-BeFx) and ADP-aluminum fluoride (ADP-AlF4). These ATP analogs are extensively used in mechanistic and structural studies of other ATPases. ADP-BeFx generally mimics the ATP prehydrolysis state in these proteins (15–17), whereas ADP-AlF4 resembles the transition or posthydrolysis state (15, 17). Structures of DEAD-box proteins with ADP-BeFx are not currently available, but in the DEAD-box protein eIF4A-III, ADP-AlF4 also resembles the transition state (18).
Here, we show that DEAD-box proteins readily unwind RNA duplexes with ADP-BeFx, but not with ADP-AlF4 or ADPNP. The data indicate that the energy from ATP hydrolysis is dispensable for duplex unwinding, whereas ATP binding appears necessary. We further show a requirement for ATP hydrolysis for substrate turnover and fast enzyme release from the RNA, indicating that DEAD-box proteins use ATP hydrolysis for enzyme recycling. The results of this study, in conjunction with previous data, suggest a basic mechanism linking ATP utilization by DEAD-box proteins to duplex unwinding.
Results
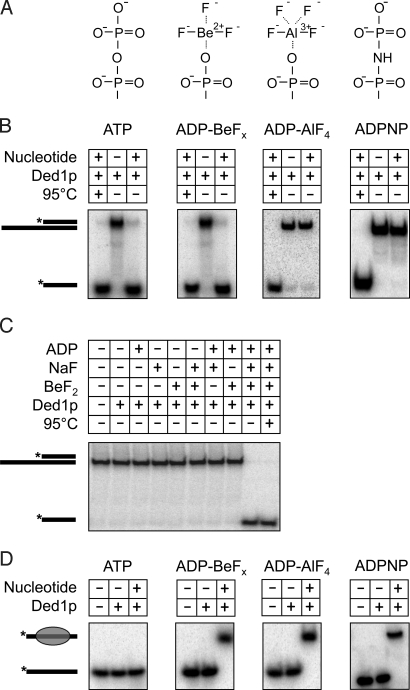
To examine previously untested nonhydrolyzable ATP analogs for their capacity to promote duplex unwinding by DEAD-box proteins, we investigated whether ADP-BeFx and ADP-AlF4 (Fig. 1A) promoted duplex unwinding by the DEAD-box protein Ded1p from Saccharomyces cerevisiae (13, 19). Unwinding reactions under presteady-state conditions, i.e., with an excess of enzyme over substrate, were first performed with ATP, using a substrate comprising a 13-bp duplex with a 3′ 25-nt unpaired region (Fig. 1). As expected, strand separation was seen with, but not without ATP (Fig. 1B Left). Strikingly, clear strand separation was also seen with ADP-BeFx (Fig. 1B). ADP-AlF4 did not promote strand separation (Fig. 1B). As expected, no unwinding was observed with ADPNP (Fig. 1B). Unwinding reactions with all ATP analogs were performed in the presence of hexokinase to eliminate spurious ATP contamination (ref. 20 and Fig. S1). No significant strand separation was measured with ADP alone (Fig. 1C) or with ADP plus inorganic phosphate (data not shown). Unwinding with ADP-BeFx depended on the entire ADP-BeFx compound and not only on parts of the noncovalent composite (Fig. 1C and Fig. S2).
Fig. 1.
ADP-BeFx promotes duplex unwinding by Ded1p. (A) Chemical structures of the terminal phosphate groups of ATP and the analogs used. (B) Unwinding reactions of 0.1 nM substrate (13 bp with 3′ 25-nt unpaired RNA) with 1.1 μM Ded1p in the absence or presence of 0.5 mM ATP or analog (0.5 mM ADP for ADP-BeFx and ADP-AlF4) as indicated. Reactions proceeded for 60 min. Diagrams indicate the RNA species; asterisks mark the radiolabel. (C) Unwinding reactions (60 min) with individual components of the noncovalent ADP-BeFx. Components were present at identical concentrations in the experiments. (D) Binding of Ded1p to a 25-nt ssRNA (sequence of the unpaired RNA region of the substrate in Fig. 1A) under conditions identical to those in the unwinding reactions above. Components were incubated for 60 min. Before application to PAGE, excess ssRNA (1 μM final, 73-nt scavenger RNA; see Methods) was added and incubated for 1 min to bind free and remove loosely bound Ded1p from the substrate RNA. Diagrams indicate bound and free RNA.
We next examined whether the strand separation with ADP-BeFx resulted from a mere ability of the compound to promote stable binding of Ded1p to RNA. Ded1p formed a stable complex with RNA in the presence of ADP-BeFx, but stable complexes were also formed with ADP-AlF4 and ADPPNP (Fig. 1D). Dissociation rate constants for Ded1p-RNA complexes with either ADP-BeFx, ADP-AlF4, or ADPPNP all were below kdiss < 10−3 min−1, compared with a dissociation rate constant of kdiss > 10 min−1 for Ded1p alone (data not shown). Thus, all 3 ATP analogs induced exceptionally tight binding of Ded1p to the RNA, indicating that duplex unwinding with ADP-BeFx was not simply caused by formation of stable Ded1p–RNA complexes.
Collectively, the data above clearly established that at least one nonhydrolyzable ATP analog promoted duplex unwinding by Ded1p. The energy from ATP hydrolysis is thus not essential for strand separation. Yet, ATP or ADP-BeFx are required, suggesting that ATP binding is obligatory for the reaction with the duplex substrates used here.
It was next important to probe whether ADP-BeFx promoted duplex unwinding by the same means as ATP and not in a completely different manner. To that end, we tested whether unwinding with ADP-BeFx displayed characteristics similar to the reaction with ATP. First, we examined unwinding with ADP-BeFx for a blunt-end duplex, which is unwound with ATP with decreased efficiency, compared with substrates with single-stranded regions (ref. 6 and Fig. 2A Left). Similarly, ADP-BeFx promoted unwinding of the blunt-end duplex with decreased efficiency, compared with the unwinding of tailed substrates (Fig. 2A Right). No significant separation of a DNA duplex was detected with ADP-BeFx (data not shown), again paralleling the characteristics seen with ATP (6).
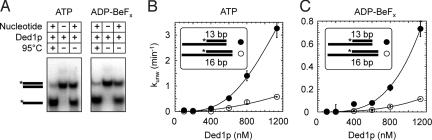
Fig. 2.
Duplex unwinding with ADP-BeFx and ATP display similar characteristics. (A) Unwinding reactions (60 min) of a 13-bp blunt-end substrate (0.1 nM) with 1.1 μM Ded1p in the absence or presence of 0.5 mM ATP (Left) or ADP-BeFx (Right). (B and C) Dependence of unwinding rate constants on the Ded1p concentration with 0.5 mM ATP (B) and 0.5 mM ADP-BeFx (C) for substrates with 13-bp (●) and 16-bp (○) and identical 25-nt unpaired RNA regions at the 3′ end. Asterisks indicate the radiolabels. Unwinding rate constants were determined as described (21). Error bars indicate the SD of 2 or more independent measurements; curves signify a trend through the data points.
To more quantitatively compare unwinding reactions with ADP-BeFx to those with ATP, we measured unwinding rate constants as function of the Ded1p concentration for 2 substrates containing a 25-nt single-stranded overhang and duplexes with 13 and 16 bp, respectively (Fig. 2 B and C). With ATP, unwinding rate constants were lower for the longer duplex (Fig. 2B), consistent with previous data (21). With ADP-BeFx, both duplexes were unwound as well, indicating that strand separation with this ATP analog was not limited to short helices (Fig. 2C). Unwinding rate constants with ADP-BeFx were lower for the longer duplex by a factor comparable with that seen with ATP, further highlighting the similarity of the unwinding reactions with ATP and ADP-BeFx. Notwithstanding, at comparable nucleotide concentrations, the unwinding rate constants with ADP-BeFx were lower than with ATP by a factor of ≈4.5 at the highest experimentally accessible Ded1p concentrations (Fig. 2 B and C). Further investigation of the unwinding kinetics under single cycle conditions suggested that the difference between the rate constants with ADP-BeFx and ATP was at least in part because the unwinding–competent complex of Ded1p, RNA, and nucleotide formed slower with ADP-BeFx than with ATP (Fig. S3). This slower complex formation with ADP-BeFx is most likely caused in large part by competition from free ADP in solution, which is a necessary component of the noncovalent ADP-BeFx. In addition, Ded1p might not bind ADP-BeFx as fast as ATP. Considering these possibilities, the difference between the rate constants measured for ATP and the noncovalent ADP-BeFx is remarkably small. Despite the difference in unwinding rate constants, the data clearly show that duplex unwinding with ADP- BeFx is highly similar to unwinding with ATP in major reaction characteristics.
Next, we tested whether other DEAD-box proteins also catalyzed strand separation with ADP-BeFx. We measured unwinding with ADP-BeFx for 2 different DEAD-box proteins, Mss116p (22, 23) (S. cerevisiae), and mammalian eIF4A (24). Notable strand separation was observed with these enzymes, too (Fig. 3). The lower unwinding efficiency by eIF4A with ADP-BeFx, compared with reactions with ATP, most likely reflects the low affinity of the enzyme for RNA (24), and corresponding stronger inhibitory effects of free ADP. As seen with Ded1p, neither ADP-AlF4 nor ADPNP facilitated comparable strand separation for Mss116p and eIF4A (data not shown). The non-DEAD-box RNA helicase NPH-II (Vaccinia virus), which unwinds duplexes by translocation along the nucleic acid (25, 26), could not separate the duplex with ADP-BeFx (Fig. 3 Right). This finding suggests that DEAD-box proteins and non-DEAD-box RNA helicases use ATP for unwinding reactions in different ways.
Fig. 3.
ADP-BeFx promotes duplex unwinding by different DEAD-box proteins, but not by a non-DEAD-box RNA helicase. Unwinding reactions of 0.1 nM substrate with 13-bp and 25-nt unpaired RNA at the 3′ end with 1 μM Mss116p (19 °C), 2 μM eIF4A (37 °C), and 100 nM NPH-II (19 °C) at 0.5 mM ATP and 0.5 mM ADP-BeFx as indicated. Protein was present in all reactions. Reactions proceeded for 60 min. Diagrams indicate the RNA species; the asterisks mark the radiolabel.
Although the above data established that ATP hydrolysis was not required for duplex unwinding by DEAD-box proteins, we wanted to better understand the role of the ATP hydrolysis step for these enzymes. We had observed that ADP-BeFx induced tight binding of Ded1p to RNA, whereas ATP did not (Fig. 1D). It was thus reasonable to assume that ATP hydrolysis affected the release of the enzyme from the RNA. Therefore, Ded1p should remain tightly bound to the RNA upon strand separation in the presence of ADP-BeFx, but not in the presence of ATP. To test this prediction, we directly measured tight binding of Ded1p to RNA during unwinding reactions, using a substrate comprising a DNA loading and an RNA top strand (Fig. 4A). This substrate was chosen because it was readily unwound with both ATP (6, 8) and ADP-BeFx, and because Ded1p forms tight complexes only with RNA, but not with DNA (6). Therefore, tight binding of Ded1p would be restricted to the RNA strand.
Fig. 4.
ATP hydrolysis is necessary for fast enzyme release from RNA. (A) Simultaneous monitoring of duplex unwinding and Ded1p–RNA complex. Diagrams indicate the substrate and Ded1p. The DNA strand is gray, RNA is black, and Ded1p is depicted as semitransparent circle. The asterisk indicates the radiolabel on the RNA strand. DNA–RNA substrate (0.1 nM final) with 16-bp and 25 nt unpaired RNA at the 3′ end was incubated with 1.13 μM Ded1p for 10 min. The unwinding reaction was then started by addition of ATP or ADP-BeFx (0.5 mM final) and allowed to proceed for 60 min. Subsequently, the reaction was divided in 2 parts. One part was terminated with SDS (see Methods) to remove the protein from the RNA; the other part was terminated with excess ssRNA (1 μM final, 73-nt scavenger RNA; see Methods) and EDTA. Both samples were applied to 2 different nondenaturing PAGE to visualize the unwound duplex (SDS) and the RNA protein complex (EMSA). (B) Nondenaturing PAGE to simultaneously monitor Ded1p–RNA complex (EMSA) and strand separation (SDS) during and unwinding reaction. Diagrams indicate the substrate species as described in A.
To simultaneously follow strand separation and Ded1p-RNA complexes, we performed unwinding reactions with the DNA–RNA substrate but terminated the reaction by 2 different means (Fig. 4A). Half of the reaction was terminated by the addition of SDS, which removes the protein from the RNA, to allow monitoring of strand separation. The other half of the reaction was terminated without removing the protein from the RNA, through addition of scavenger RNA and EDTA, to visualize Ded1p–RNA complexes. Both terminated reactions were applied to 2 different nondenaturing PAGEs (Fig. 4A). Stable binding of Ded1p to the RNA strand was evident with ADP-BeFx, but not with ATP (Fig. 4B Upper). As expected, the substrate was unwound with both ATP and ADP-BeFx (Fig. 4B Lower). Monitoring unwinding and formation of the stable Ded1p–RNA complex with ADP-BeFx over time revealed that the stable Ded1p–RNA formed at a faster rate than the strand separation, indicating that tight binding of Ded1p to the RNA strand preceded the actual strand separation (Fig. S4). Taken together, these results strongly suggest that hydrolysis of ATP is necessary for efficient release of Ded1p from the RNA.
To confirm and extend these observations, we measured unwinding reactions with excess RNA substrate over Ded1p. Under these conditions, repeated release of unwound substrate is critical for efficient unwinding. Turnover of substrate excess by Ded1p was observed with ATP, but not with ADP-BeFx, which only promoted unwinding of a small fraction of the substrate (Fig. 5). Virtually identical observations were made for Mss116p (Fig. S5). These results indicate that ATP hydrolysis is needed to accomplish substrate turnover. Thus, efficient enzyme recycling requires ATP hydrolysis to promote the dissociation of DEAD-box proteins from the RNA.
Fig. 5.
Turnover of excess substrate requires ATP hydrolysis. Representative reactions for unwinding of excess RNA substrate (1 μM, 13 bp with 25-nt single-stranded region at 3′; Fig. 1) by Ded1p (100 nM) with 0.5 mM ATP (Upper) and 0.5 mM ADP-BeFx (Lower). Diagrams indicate the substrate species. The rate of substrate unwinding with ATP corresponds to vobs = 0.125 μM·min−1. After 20 min 2.5 μM substrate is unwound. With ADP-BeFx, ≈0.05 μM substrate is unwound after 20 min.
Discussion
In this study, we have shown that DEAD-box proteins readily unwind stable duplexes with the nonhydrolyzable ATP analog ADP-BeFx and thus without the need for ATP hydrolysis. However, ATP hydrolysis is necessary for the fast release of DEAD-box proteins from the RNA and for multiple substrate turnovers. Our results indicate that ATP hydrolysis is needed for DEAD-box protein recycling and that DEAD-box proteins do not require energy from ATP hydrolysis for strand separation.
If the energy from the ATP hydrolysis is dispensable for strand separation, what drives the reaction? ATP binding must clearly occur, simply because either ATP or ADP-BeFx have to be present to promote unwinding. But although the energy from ATP hydrolysis is not essential, it is formally possible that unwinding requires the ATP hydrolysis cycle to progress to transition or posthydrolysis states or even beyond. However, several lines of evidence render this possibility unlikely. First, no unwinding is seen with ADP-AlF4, which resembles the ATP posthydrolysis or transition state in the DEAD-box protein eIF4A-III and in other ATPases (17, 18). ATP posthydrolysis or transition states are therefore unlikely to trigger strand separation. Second, it is unlikely that the dissociation of inorganic phosphate causes the strand separation, because unwinding rate constants with ADP-BeFx (e.g., kunw = 3 min−1; Fig. 2C) are orders of magnitude greater than dissociation rate constants for any of the ADP-BeFx components from the enzyme RNA complex (kdiss < 10−3 min−1; Table S1). Finally, no unwinding is seen with ADP or inorganic phosphate alone.
Collectively, these observations are consistent with a scenario where ATP binding elicits strand separation. Accordingly, ADP-BeFx resembles the ATP prehydrolysis state in P-loop ATPases (17), and, because of the conservation of the ATPase active site in P-loop proteins, ADP-BeFx most likely resembles the prehydrolysis state in DEAD-box proteins as well. However, the absence of an ADP-BeFx-bound DEAD-box protein structure currently precludes a definitive statement to this effect. Further support for the notion that ATP binding elicits strand separation is provided by quantitative comparisons of kinetic parameters for ATPase and unwinding reactions (27). These data show for several DEAD-box proteins that strand separation requires only the presence, but not the hydrolysis of ATP (ref. 27 and unpublished results).
With several lines of evidence suggesting that ATP hydrolysis is not required for strand separation, it is curious that no unwinding is seen with ADPNP, which is generally considered a prehydrolysis state analog. However, ADPNP is also known to not always faithfully mimic ATP in other ATPases (28). The same may be true for DEAD-box proteins. The oxygen–nitrogen substitution in ADPNP may induce an inhibited ATP prehydrolysis conformation, an ATP transition state, or a posthydrolysis state.
Irrespective of the state induced by ADPNP, the insight that ATP hydrolysis facilitates enzyme dissociation, but not strand separation, and previous results regarding DEAD-box protein loading and unwinding kinetics (2) now suggest a model for a mechanism by which DEAD-box proteins couple ATP binding and hydrolysis to duplex unwinding (Fig. 6). As shown previously, DEAD-box proteins are loaded directly on the duplex region, aided by single-stranded or structured nucleic acid regions (6, 7). Multiple protomers are known to participate (8), also in the presence of ADP-BeFx (Fig. 2C), but the exact mechanism of the loading process has not yet been elucidated. However, it is clear that enzyme loading to the duplex either requires or is accompanied by ATP binding. In the ATP-bound state, the DEAD-box protein locally opens the duplex strands. This local strand opening may occur either by capturing transiently frayed base pairs, actively inducing helix opening, or a combination of both scenarios.
Fig. 6.
The basic mechanism by which DEAD-box proteins couple ATP binding and hydrolysis to duplex unwinding. RNA strands are depicted as lines, and Ded1p is semitransparent circles. ATP and ADP are small squares. The single-stranded substrate region is gray, and the dotted line emphasizes that this region does not need to be physically connected to the RNA duplex to aid enzyme loading (6). The asterisk on the partially unwound RNA species without protein indicates the transient existence of this species. Unwinding reactions described here are started with the enzyme loading step. More than 2 Ded1p protomers may participate in the unwinding reaction (8). The mode of association of the 2 protomers to the RNA substrate is speculative.
It is unclear whether the local strand opening is accompanied or caused by a bending of the RNA seen in the structures of the DEAD-box proteins eIF4A-III and Vasa bound to single-stranded RNA and ADPNP (14, 29, 30). Based on the latter structure, it had been hypothesized that the conformation of the ssRNA, which is incompatible with a perfect helix, could represent a partially opened duplex (14). However, as shown here and by a host of other studies, ADPNP does not support duplex unwinding by DEAD-box proteins (6, 8, 31, 32), indicating that the RNA–Vasa–ADPNP complex is not competent for strand separation. Further structural work is necessary to elucidate which conformational changes in the RNA accompany duplex unwinding by DEAD-box proteins. Our study may be useful for guiding the choice of ATP analogs for structural examination of the respective unwinding steps.
Although the helix opening does not require ATP hydrolysis, it is nevertheless possible that ATP hydrolysis occurs after the helix has been opened by ATP binding, but before the strands have separated. Although ATP hydrolysis promotes enzyme dissociation, strand separation may take place before the enzyme actually dissociates, and in these cases, unwinding can occur upon ATP hydrolysis. ATP hydrolysis may even disrupt additional base pairs and thus accelerate duplex unwinding, compared with the reaction driven by ATP binding. This scenario may contribute in part to the higher unwinding rate constants with ATP, compared with those with ADP-BeFx (Fig. 2).
The local helix opening without or with ATP hydrolysis reduces the number of base pairs in the duplex, and the remaining base pairs dissociate without further action from the enzyme (Fig. 6). Unwinding rate constants therefore decrease with duplex length and stability, because more or more stable base pairs dissociate slower (2, 8, 24, 32, 33). However, not every ATP-driven local helix opening necessarily leads to complete strand separation. If ATP hydrolysis occurs before the strands are completely separated, notwithstanding the scenario discussed above, the enzyme has a propensity to dissociate from the duplex. If the enzyme dissociates before complete helix separation, the strands quickly reanneal (27, 34). Such “nonproductive” ATP hydrolysis events are likely to be more prevalent for longer and more stable duplexes (27, 34).
The actual ATP hydrolysis presumably is induced by the emergence of ssRNA in the locally unwound RNA duplex. ssRNA is more flexible than duplex RNA, and a certain flexibility of the RNA backbone may be needed by the enzyme to attain the conformation necessary to cleave the ATP γ-phosphate while bound to RNA. It may thus be instructive to view ATP hydrolysis as a consequence of helix opening. Although our results clearly show that ATP hydrolysis promotes enzyme dissociation, it is not yet clear which molecular event triggers the substrate release. Recent experimental evidence suggests that phosphate release might play a role in substrate release by the DEAD-box protein DbpA (10). This notion is consistent with our data and with previous findings showing a strongly decreased RNA affinity in the presence of ADP (35–37).
The reaction scheme in Fig. 6 reconciles the available functional data for duplex unwinding by DEAD-box proteins. It is not immediately clear whether the ATP-driven displacement of proteins from RNA by Ded1p (38) also follows this mechanism. The tight binding of Ded1p to RNA with ADPNP does not promote protein displacement (38), but further studies are necessary to determine whether the dislodging of proteins occurs with ADP-BeFx or requires actual ATP hydrolysis. Although it is interesting to speculate that different activities of DEAD-box proteins might use the steps of the ATP hydrolysis cycle in a distinct fashion, our results clearly demonstrate that DEAD-box proteins use ATP for duplex unwinding in a manner that fundamentally differs from canonical helicases. Canonical helicases couple ATP binding and hydrolysis to translocation along the nucleic acid (39, 40). DEAD-box proteins appear to use ATP primarily to modulate their affinity for RNA during the unwinding reaction. The main energy for duplex unwinding is derived from the association of the ATP-bound DEAD-box protein to the RNA, not from ATP hydrolysis.
Therefore, it seems appropriate to view DEAD-box proteins not as poorly processive helicases, but as ATP-dependent RNA binding proteins. In this sense, DEAD-box proteins bear a striking functional resemblance to G proteins (3, 21, 31). Both DEAD-box- and G proteins tightly bind their substrates with NTP, but only weakly with NDP (33, 35, 41). NTP hydrolysis promotes substrate release from both enzyme groups (10, 41). Our results now show that DEAD-box proteins even perform duplex unwinding, i.e., “work” on RNA, based on ATP-driven affinity changes, and without the requirement for energy provided by the hydrolysis reaction.
Methods
Materials.
Proteins used in this study (Ded1p, Mss116p, eIF4A, and NPH-II) were expressed and purified as described (22, 24, 38). RNA oligonucleotides were purchased from Dharmacon, and radiolabeled duplex substrates were prepared as described (26). Substrate sequences were as follows: 13-bp duplex with 25-nt 3′ overhang (underlined regions correspond to the duplex region), 5′-AGCACCGUAAAGA-3′ (radiolabeled strand) and 3′-(A4C)4AAAAUUCGUGGCAUUUCU-5′; 13-bp blunt-end duplex, 5′-AGCACCGUAAAGA-3′ (radiolabeled strand) and 3′-UCGUGGCAUUUCU-5′; 16-bp duplex with 25-nt 3′ overhang, 5′-AGCACCGUAAAGACGC-3′ (radiolabeled strand) and 3′-(A4C)4AAAAUUCGUGGCAUUUCUGCG-5′. The 73-nt ssRNA used as scavenger was 5′-CCGUACAGGCUCUGGGUACAAUGCUUGUUUUUUUUCUGUCUGGGACGUA CUGCAUCAAUGACAUCAGCAUCAA-3′. ADP-BeFx and ADP-AlF4 were a mixture of ADP with 5-fold molar excess of the corresponding metal fluoride and 25-fold molar excess of NaF.
Unwinding Reactions.
Unwinding reactions with enzyme excess.
All reactions with ATP analogs were conducted in the presence of hexokinase (5 units/mL; Roche) to remove traces of ATP from ADP preparations (20, 22). Reaction mixtures (30 μL) contained 40 mM Tris·HCl (pH 8.0), 50 mM NaCl, 0.5 mM MgCl2, 2 mM DTT, 1 unit/μL−1 RNasin, 0.01% (vol/vol) Nonidet P-40, 10.7 mM (NH4)2SO4 (to account for the ammonium sulfate introduced with the hexokinase storage buffer in the reactions with the ATP analogs), 1 mM d-glucose (substrate for the hexokinase), 0.1 nM radiolabeled RNA substrate, and hexokinase (5 units/mL; Roche). This amount of hexokinase was sufficient to inhibit reactions to virtual completion even with 0.5 mM ATP (Fig. S1a). Radiolabeled substrate was present in the reaction at 0.1 nM. Unwinding rate constants for Ded1p under these reaction conditions were slower by a factor of 3.2 than reactions without (NH4)2SO4 and d-glucose under otherwise identical conditions (data not shown). Unwinding reactions were performed with enzyme excess to ensure presteady-state conditions. The comparably low concentrations of ATP analogs in the reactions (0.5 mM) required high enzyme concentrations, because the functional affinity of Ded1p decreases with decreasing ATP concentrations (21).
Before the reactions, RNA in reaction buffer was incubated with the respective enzyme at the concentrations indicated for 5 min at the reaction temperatures. Reactions were performed at 19 °C for Ded1p, Mss116p, and NPH-II, and at 37 °C for eIF4A. Unwinding reactions were initiated by adding an equimolar mixture of 0.5 mM ATP or analog and MgCl2. In the reactions with ADP-BeFx and ADP-AlF4, ADP was present at 0.5 mM. Aliquots were removed from the reaction at the time points indicated, and the reaction in those aliquots was quenched with 5 vol of stop buffer [50 mM EDTA, 1% SDS, 0.01% bromophenol blue, 0.01% xylene cyanol, 10% (vol/vol) glycerol]. Aliquots were then applied to 15% nondenaturing polyacrylamide gel, and duplex and single-stranded RNA were separated at room temperature at 10 V·cm−1. Gels were dried and the radiolabeled RNAs were visualized and quantified with a PhosphorImager and ImageQuant 5.2 (Molecular Dynamics). Unwinding rate constants were determined as described (21). Unwinding with ADP-BeFx was also observed when the reaction was monitored by fluorescence changes of dye-labeled RNA substrates (data not shown).
Unwinding reactions with substrate excess.
Reaction mixtures were identical to those described above. The RNA substrate comprised the 25-nt unpaired region 3′ to a 13-bp duplex (sequence identical to the substrates used above). In the reaction, the RNA substrate was at 1 μM (including 0.5 nM radiolabeled RNA). In addition, the reactions contained 5 μM ssRNA identical to the labeled top strand, to ensure an appreciable reaction amplitude. Reactions were conducted with 100 nM enzyme at 25 °C and analyzed on nondenaturing PAGE as described above. Because of the high concentration of RNA, separated duplexes reformed immediately (21), which resulted in an essentially unchanged duplex concentration. The given rate constant was calculated by multiplying the observed rate constant with the substrate concentration (34).
RNA Protein Binding Reactions.
Reaction mixtures (10 μL) contained the same components as the corresponding unwinding reactions. Radiolabeled RNA (0.1 nM final concentration) was incubated with 800 nM Ded1p and, where indicated, with 0.5 mM ATP or analog (plus 0.5 mM MgCl2) for 60 min in a temperature-controlled aluminum block at 19 °C. Reactions with ATP analogs were conducted in the presence of hexokinase. After the incubation time, 1 μM (final concentration) of the 73-nt ssRNA and glycerol (25% vol/vol, final concentration) were added to each sample and incubated for 1 min. The ssRNA sequestered excess and nonstably bound Ded1p, thus leaving only Ded1–RNA complexes that did not dissociate within 1 min (6). Those Ded1p–RNA complexes are termed stable. Samples were then applied to 7% nondenaturing PAGE gels, and Ded1p–RNA complexes, duplex, and ssRNA were separated at 4 °C. Gels were dried, and the radiolabeled RNAs were visualized and quantified with a PhosphorImager and ImageQuant 5.2 (Molecular Dynamics).
Supplementary Material
Acknowledgments.
We thank Margaret Fairman (Case Western Reserve University) for supplying purified NPH-II; Mark Del Campo and Alan Lambowitz (University of Texas, Austin, TX) for Mss116p; William Merrick (Case Western Reserve University) for eIF4A; and Timothy Nilsen, William Merrick, Jeff Coller, Rick Russell, and Gregers Andersen for comments on the manuscript. This work was supported by National Institutes of Health Grant GM067700 (to E.J.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811115106/DCSupplemental.
References
- 1.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky E, Fairman M. RNA helicases: One fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 4.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 6.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 7.Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci USA. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Peck ML, Herschlag D. Adenosine 5′-O-(3-thio)triphosphate (ATPγS) is a substrate for the nucleotide hydrolysis and RNA unwinding activities of eukaryotic translation initiation factor eIF4A. RNA. 2003;9:1180–1187. doi: 10.1261/rna.2103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henn A, Cao W, Hackney DD, De La Cruz EM. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J Mol Biol. 2008;377:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 12.Vale RD, Milligan RA. The way things move: Looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 13.Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 14.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 15.Fisher AJ, et al. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP·BeFx and MgADP·AlF4−. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 16.Kagawa R, Montgomery M, Graig K, Leslie AG, Walker JE. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004;23:2734–2744. doi: 10.1038/sj.emboj.7600293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B, et al. ATP ground- and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, sigma54. Structure (London) 2007;15:429–440. doi: 10.1016/j.str.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen KH, et al. Mechanism of ATP turnover inhibition in the EJC. RNA. 2008 doi: 10.1261/rna.1283109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linder P. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol Cell. 2003;95:157–167. doi: 10.1016/s0248-4900(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 20.Yang Q, Fairman ME, Jankowsky E. DEAD-box-protein-assisted RNA structure conversion toward and against thermodynamic equilibrium values. J Mol Biol. 2007;368:1087–1100. doi: 10.1016/j.jmb.2007.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 22.Halls C, et al. Involvement of DEAD-box proteins in group I and group II intron splicing: Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang HR, et al. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers GW, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 25.Shuman S. Vaccinia virus RNA helicase: An essential enzyme related to the DE-H family of RNA-dependent NTPases. Proc Natl Acad Sci USA. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, et al. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci USA. 2008 doi: 10.1073/PNAS.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fak JJ, et al. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry. 2004;43:7307–7327. doi: 10.1021/bi0357208. [DOI] [PubMed] [Google Scholar]
- 29.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Andersen CB, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 31.Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 32.Del Campo M, et al. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linder P. Dead-box proteins: A family affair–active and passive players in RNP remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 35.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 36.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry. 1998;37:2194–2206. doi: 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- 37.Cordin O, Tanner NK, Doere M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fairman M, et al. Protein displacement by DExH/D RNA helicases without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 39.Mackintosh SG, Raney KD. DNA unwinding and protein displacement by superfamily 1 and superfamily 2 helicases. Nucleic Acids Res. 2006;34:4154–4159. doi: 10.1093/nar/gkl501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.