Abstract
Here, we define a gene regulatory network for Hoxa2, responsible for temporal and spatial expression in hindbrain development. Hoxa2 plays an important role in regulating the regional identity of rhombomere 2 (r2) and is the only Hox gene expressed in this segment. In this study, we found that a Hoxa2 cis-regulatory module consists of five elements that direct expression in r2 of the developing hindbrain. Surprisingly, the module is imbedded in the second coding exon of Hoxa2 and therefore may be constrained by both protein coding and gene regulatory requirements. This highly conserved enhancer consists of two consensus Sox binding sites and several additional elements that act in concert to direct strong r2 specific expression. Our findings provide important insight into the regulation of segmental identity in the anterior hindbrain. Furthermore, they have broader implications in designing arrays and interpreting data from global analyses of gene regulation because regulatory input from coding regions needs to be considered.
Keywords: cis elements, hindbrain segmentation, Hox genes, regulatory network
Hox genes play an important role in establishing regional identity throughout the anteroposterior (AP) axis of embryo (1–3). The expression of different combinations of Hox proteins (Hox code) is a critical aspect of the regulatory network that provides cells with molecular information about their AP position. The generation of regional diversity in vertebrate hindbrain is achieved through the formation of lineage-restricted segmental compartments called rhombomeres (r) and disruption of the nested domains of Hox expression alters the rhombomere identity and influences other aspects of the segmentation process (4–6). Targeted inactivation of the Hoxa2 gene, which is the only Hox gene expressed in r2, leads to the partial transformation of the segment normally characterized as r2, to an r1 identity (7). This transformation is associated with a changes in the segmental expression patterns and morphological characteristics of the rhombomere (7), demonstrating the key role played by Hoxa2 in control of the r2 gene regulatory network.
Characterizing the gene regulatory networks that control the spatial and temporal expression of Hox genes is essential for understanding the process of hindbrain segmentation and regional identity. Based on experiments in a number of species, there is a considerable body of work on the upstream transcription factors and signaling pathways that regulate segmental expression of Hox genes in r3-r6, and auto- and cross-regulatory interactions between the Hox genes themselves play a key role in control of rhombomere-restricted Hox expression (reviewed in ref. 5, 8–10). However, virtually nothing is known about regulation of Hox expression in r2.
In this study, we used evolutionary comparisons and functional analyses in chicken and mouse embryos to characterize the mechanisms that underlie the regulation of Hoxa2 expression in r2. We identified a Hoxa2 cis-regulatory module capable of directing r2 restricted-expression that is embedded in a coding region of the second exon. This module consists of five elements that act in concert to direct r2 specific expression, and two of these highly conserved elements are binding sites for the Sox family of transcription regulators. These results provide insight into control of r2 identity.
Results
Mapping a Conserved Hoxa2 r2 Enhancer.
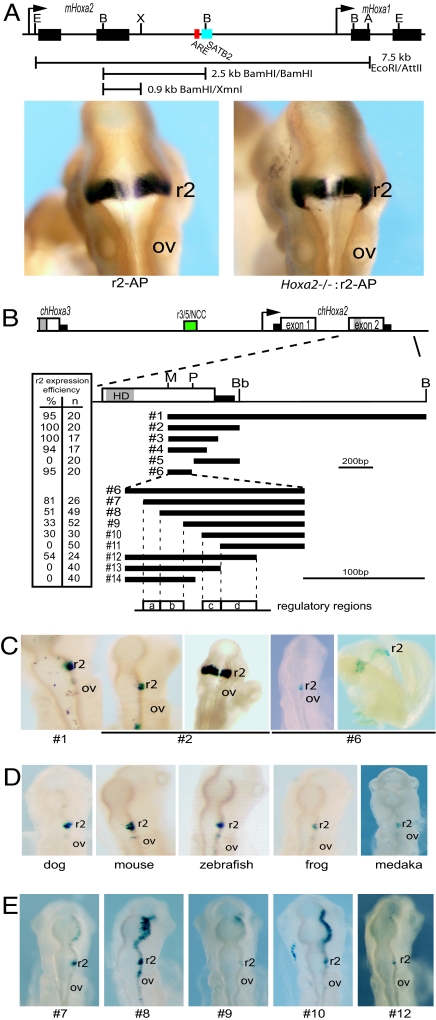
Segmental expression of Hoxa2 in the vertebrate hindbrain involves the combined activities of multiple cis-regulatory modules. It has been shown that the intergenic region between Hoxa2 and Hoxa3 contains conserved enhancers implicated in directing expression in r3 and r5 (r3/5) (11–14) and cranial neural crest cells (ncc) (15). A conserved regulatory module located in the intron of Hoxa2 mediates expression in r4 (16). In mouse, a 7-kb (EcoRI-AatII) fragment, beginning just upstream of the first exon of Hoxa2 to the middle of the first exon of Hoxa1 (17), and a 2.5-kb (BamHI) fragment containing part of the second exon of Hoxa2 and 3′ intergenic region (18) are capable of directing reporter activity in r2 (Fig. 1A). To identify and characterize the r2 control module, we further analyzed regions in and around the 3′ region of the Hoxa2 locus.
Fig. 1.
Identifying the Hoxa2 module directing r2 expression. (A) Genomic map of the mHoxa2 and mHoxa1 genes. The BamHI/XmnI fragment directs r2 expression of an alkaline phosphatase reporter (r2-AP) in a normal (Left) and Hoxa2 knockout (Hoxa2−/−:r2-AP) (Right) mouse embryos (9.5dpc). (B) Map showing the relative location of each fragment used in LacZ constructs. The gray boxes in exon 2 represent the homeodomain (HD). Black rectangles are the 5′ and 3′ untranslated regions. The r2 expression efficiency is calculated from the number of embryos displaying r2 specific expression as a percentage of the total embryos (n) electroporated with the construct. Below the constructs, regions containing regulatory elements (a–d) are defined for reference in subsequent figures. (C and E) Representative transgenic mouse (as labeled) or electroporated chicken embryos (all other embryos) expressing the indicated constructs. (D) Electroporated chicken embryos tested with the Hoxa2 r2 enhancers (equivalent to construct 2) from the indicated species. Restriction enzyme sites in maps: E, EcoRI; B, BamHI; X, XmnI; A, AccI; M, MluI; P, PstI; Bb, BbsI. OV, otic vesicle; r2, rhombomere 2.
Identification of sequence identity between multiple species alignment has been helpful in focusing in on potential regulatory elements (10, 16). Therefore, we cloned, sequenced, and functionally tested the region downstream of chicken Hoxa2 gene. We linked a 1.9-kb MluI-BamHI fragment (Fig. 1B, construct 1), including the second half of Hoxa2 exon 2 and additional 3′ downstream sequence, to a lacZ reporter gene and scored for regulatory activity after electroporation into chicken embryos. This region directs reporter staining specifically in r2 (Fig. 1C). Based on relative position and function, this is the chicken equivalent of the regulatory region contained in the mouse 2.5-kb BamHI fragment (Fig. 1A). In addition to the expected similarity in the coding region, sequence alignments of the mammalian and chicken fragments reveal that the only homologies are located in the 3′ UTR of the gene and sequences surrounding a conserved BamHI site, midway through the Hoxa2-Hoxa1 intergenic region [supporting information (SI) Figs. S1 and S2]. The sequences around the mouse BamHI site contain a putative bipartite Hoxa2-Pbx response element (ARE) (19) and two binding motifs for the nuclear matrix protein, SATB2 (20) (Fig. 1A). Although in chicken we can identify and align a conserved region containing the SATB2 sites, we are unable to identify a Hoxa2-Pbx response element in a similar position (Fig. S2).
In light of this conserved region surrounding the BamHI site and the important role of auto and cross-regulatory mechanisms in control of segmental Hox expression (16, 21–24), we next tested whether these regions are required for activity in r2. The mouse 2.5-kb BamHI fragment was divided into two separate regions by digestion with XmnI, and each region was linked to an alkaline phosphatase (AP) reporter construct to test for regulatory activity in transgenic mice. The 3′ XmnI-BamHI region, containing the Hoxa2 ARE and conserved block, directed no reporter expression (data not shown), whereas the 5′ BamHI-XmnI region (r2-AP) mediated robust reporter staining in r2 (Fig. 1A). We also performed a similar dissection of the chicken MluI-BamHI fragment. The 5′ MluI-BbsI fragment (Fig. 1B, construct 2) directs restricted reporter staining in r2 when tested in both chick and mouse embryos (Fig. 1C, construct 2). No regulatory activity was detected using the 3′ segment of the chicken gene containing the regions up to and including the conserved BamHI site. Therefore, the Hoxa2-Pbx response element (ARE) is not required for r2 activity, consistent with our observation that it is not conserved in the chicken gene (Figs. S1 and S2). To determine whether the activity of the r2 enhancer depends on Hoxa2 mediated through other sites, we crossed the r2-AP transgenic reporter line with mice carrying a null allele of Hoxa2 (25). No changes in reporter staining in r2 were observed in homozygous Hoxa2−/− mutant embryos, compared with control embryos (Fig. 1A). This demonstrates that autoregulatory input from Hoxa2, the only Hox gene expressed in r2, is not required for activity of the r2 enhancer.
The r2 Enhancer Resides in the Second Exon of Hoxa2.
The above results indicate that the cis-regulatory elements directing r2 expression must reside in the second exon or 3′ UTR of Hoxa2. We generated a series of reporter constructs carrying deletion variants of the chicken MluI-BbsI fragment (Fig. 1B, constructs 3–6) to more precisely map the regulatory activity. Surprisingly, analyses of reporter staining with constructs 3–6 demonstrated that the conserved sequence in the 3′ UTR is not required for activity and that the r2 enhancer is contained in an 264-bp MluI-PstI fragment embedded in the coding sequence of the second exon of Hoxa2 (Fig. 1 B and C). Construct 6, containing part of exon 2, efficiently directs r2-specific staining in chicken and mouse embryos (Fig. 1C).
We isolated the equivalent region from other species, including mouse, fish (zebrafish and medaka), dog, and frog (Xenopus tropicalis), and scored for activity in the chicken embryo assay. We find robust reporter staining in r2 when this region of exon 2 is used (Fig. 1D), demonstrating that the r2 module is embedded in the second exon of Hoxa2 and functionally conserved among these different vertebrate species.
Multiple cis-Acting Elements Are Required for Regulating Hoxa2 Expression in r2.
Progressive 5′ and 3′ deletion of the MluI-PstI fragment (construct 6) produced a series of nested fragments (constructs 7–14) that were tested for r2 activity (Fig. 1 B and E). In the 5′ deletion series (constructs 6–10), the efficiency of expression progressively decreased from 95% to 30% (Fig. 1 B and E). Furthermore, the expression patterns became less r2-restricted as other regions of the hindbrain displayed reporter staining. These results suggest that several elements (regions a and b in Fig. 1B) may participate in modulating levels and restricting expression to r2. The region deleted in construct 11 completely abrogates reporter activity, indicating an essential component lies in this sequence (region c in Fig. 1B). The 3′ deletion series (constructs 12–14) revealed that the short region (region d) when deleted in construct 13 also, results in a complete loss of reporter activity (Fig. 1B). These data indicate that the r2 cis-regulatory module combines the activity of two regions in the middle of the fragment (regions c and d) essential for enhancer activity in conjunction with other 5′ elements (regions a and b), which may modulate levels of expression but alone are not sufficient to direct r2 expression.
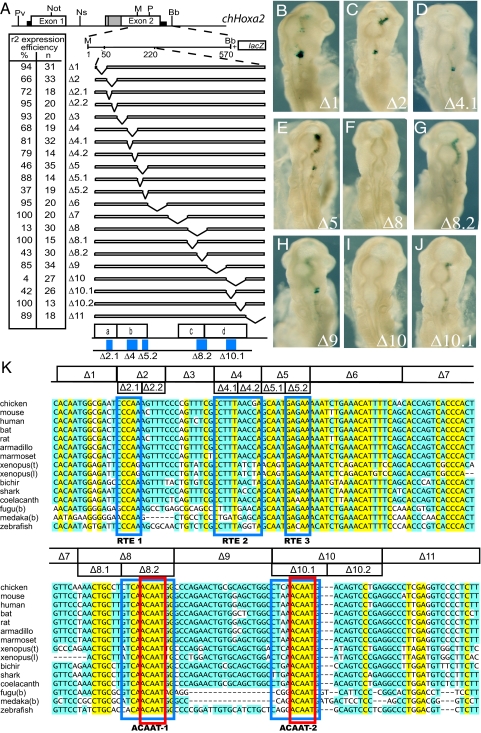
To more precisely map the elements defined by the 5′ and 3′ deletions, we performed 10- to 20-bp internal scanning deletions across regions a–d in the context of the MluI-BbsI fragment (Fig. 2 A and K, constructs Δ1 to Δ11). The deletions in constructs Δ2, Δ4, Δ5, Δ8, and Δ10 lead to a marked reduction in r2 enhancer activity, whereas the other deletions did not lead to any substantial change (Fig. 2 A–J). We performed additional 5- to 10-bp deletions that further refine the five motifs important for enhancer activity (blue boxes at bottom of Fig. 2A).
Fig. 2.
Deletion analysis of the r2 module of Hoxa2. (A) Map of the chHoxa2 locus and the constructs used to further refine the r2 elements. The deletions were performed in construct 2 (see Fig. 1) focusing on the region encompassing the elements (a–d). The refined positions of the r2 enhancer elements (Δ2.1, Δ4, Δ5.2, Δ8.2, and Δ10.1) are shown in blue. The deletion numbers and the efficiency are recorded in the table. Restriction enzyme sites: Pv, PvuI; Not, NotI; Ns, NsiI; M, MluI; P, PstI; Bb, BbsI; B, BamHI. (B–J) Representative electroporated chicken embryos for the indicated constructs. (K) Sequence alignment of the Hoxa2 r2 module. The deletions (Δ1 to Δ11) and the location of the elements (RTE1–3 and ACAAT-1 and -2 motifs, boxed in blue) are positioned on the aligned sequences from various species. The deletions were made in the context of construct 2 (MluI/BbsI). RTE, rhobomere 2 element; ACAAT defines the core bases of the motif (boxed in red).
Interspecies Alignment of the Regions Containing the r2 cis elements.
Having identified specific regions required for Hoxa2 r2 enhancer activity of the chicken gene, we performed a local alignment comparing sequences from various vertebrate species (Fig. 2K). The three rhombomere two elements (RTE1–RTE3) important for modulating the intensity/efficiency of reporter expression in r2 are embedded in the coding sequence of the Hoxa2 gene, so it is not surprising that the alignments are very similar throughout this region. The RTE1 and RTE3 elements show a very high degree of conservation, with consensus core sequences (uppercase letters are conserved in all species) of 5′-cCCAa-3′ and 5′-GAgAA-3′, respectively. The RTE2 motif shows less conservation in frog and fish but in other species has a conserved 5′-CCTTTANC-3′ core.
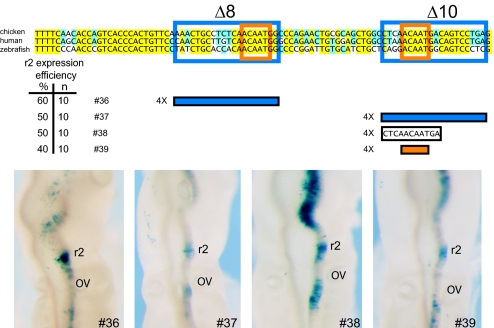
The two elements required for enhancer activity contain very similar, highly conserved 5′-ACAAT-3′ motifs (Fig. 2K, boxed in red), suggesting that multiple binding sites for a similar factor are important for enhancer activity. We used this feature to describe these elements as ACAAT-1 and ACAAT-2. We determined whether the two 5′-ACAAT motifs are also sufficient to direct r2 expression. Multimerized sequences (4 copies) spanning each ACAAT motif were linked to a lacZ reporter and assayed using chick electroporation (Fig. 3). Reporter vectors containing the 20-bp region deleted in Δ8 (construct 36) or in Δ10 (construct 37) show strong expression in r2, although weak reporter staining was also observed in the midbrain and posterior hindbrain (Fig. 3). We tested a shorter sequence (11 bp) encompassing the ACAAT-2 motif (construct 38) and found that it was able to direct lacZ expression in r2 but strong staining was also detected in the midbrain and r4 (Fig. 3). Surprisingly, four copies of the 5′-ACAAT-3′ sequence alone (construct 39), generated a similar patter with staining in r2, r4, and the midbrain (Fig. 3). These results suggest that this motif is sufficient to direct expression in r2 and some adjacent regions but that other factors/sequences may serve to restrict expression specifically to r2.
Fig. 3.
Analysis of regulatory activity of the individual ACAAT motifs. (Upper) A sequence alignment between representative sequences from avian, mammal, and fish. Sequence defined by the deletion experiments (Δ8 and Δ10 in previous figures) is boxed in blue, the location of the core ACAAT motifs is boxed in orange. Four copies of each oligonucleotide containing the ACAAT region from the chicken sequence are fused to the reporter, LacZ. The blue constructs contain four copies of the 20-bp region of defined by the deletion experiments (Δ8.2 and Δ10.1, construct 36 and 37). The black construct contains four copies of a smaller 11-bp region (construct 38). The red construct contains four copies of the 5-bp core ACAAT motif (construct 39). The efficiency of each construct is given. (Lower) Representative chicken embryos, electroporated with the indicated construct stained for reporter activity.
Sox Proteins Bind to the ACAAT Motifs.
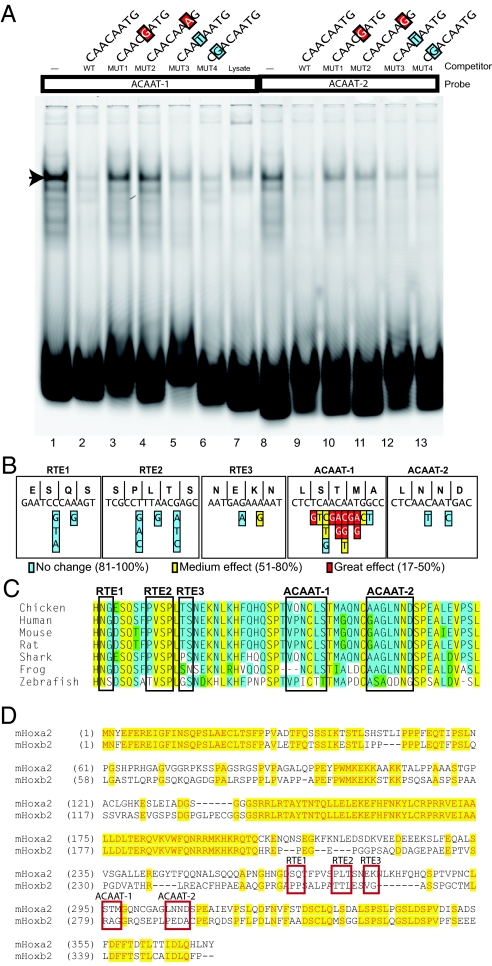
The sequence of the two ACAAT motifs in the r2 enhancer is highly similar to the consensus binding site for several Sox genes (26). Therefore, we performed electrophoretic mobility shifts assays (EMSA), using probes spanning the ACAAT-1 and ACAAT-2 motifs and in vitro translated Sox2 protein to investigate interactions with Sox protein. The EMSA reveal that, in vitro, Sox2 binds to both the ACAAT-1 (lane 1) and the ACAAT-2 (lane 8) probes (Fig. 4A, arrowhead). Furthermore, in competition assays, addition of an unlabeled WT probe significantly decreases Sox2 binding on these sites (Fig. 4A, lanes 2 and 10). In conjunction with functional assays incorporating point mutations in these motifs (see next section and Fig. 4B), we also evaluated the influence specific changes in the ACAAT motifs would have in competing for Sox2 binding to a WT probe. Some alterations strongly reduce or eliminate the ability of these sequences to compete for interactions with Sox2 (Fig. 4A, lanes 3, 4, 10, and 11), whereas others have more mild effects (Fig. 4A, lanes 5, 6, 12, and 13) on binding in agreement with requirements for consensus Sox binding sites from other studies (26, 27).
Fig. 4.
Site-directed mutagenesis of the r2 elements and Sox binding. (A) Double-stranded probes spanning the ACAAT-1 and ACAAT-2 motifs mixed with in vitro translated Sox2 and analyzed with EMSA. The lanes are Cy5-labeled double stranded WT ACAAT-1 (lanes 1–7), WT ACAAT-2 (lanes 8–13) and unlabeled competitor sequences with the mutations indicated, in the context of the appropriate WT ACAAT probe (lanes 2–6 and 9–12). Mutant bases are color coded (blue, no significant change; yellow, 75–80% of all electroporated embryos that showed r2 restricted expression; red, 32–43% of all electroporated embryos that showed r2 restricted expression) to indicate their effect on reporter expression in the electroporation/reporter assays in B. An arrow marks shifted labeled oligomer band. (B) The sequence of the codons and amino acids of the five regulatory elements of the chicken gene. Below are each of the base pair changes introduced in the context of construct 6 (Fig. 1B). The colors illustrate the effect of the individual change as described in A. (C) Sequence alignments of region encoded by the r2 module. No background color, nonsimilar residues; yellow background, amino acids are conserved at a given position; blue background, amino acids are conserved in 50% or more of the species, green background, residues weakly similar to a consensus at given position. (D) A comparison between mHoxa2 and mHoxb2 paralogs to demonstrate that the region of the Hoxa2 protein encoded by the r2 regulatory elements is not conserved. Primers are in Tables S1–S5.
Relationship Between Nucleotide Requirements and Amino Acid Sequence.
A surprising finding of this study is that the elements of the r2 enhancer are embedded in the second exon of Hoxa2. This sequence is highly conserved, as expected for a coding exon. Alteration of this sequence could be constrained by both the requirement to maintain the amino acid sequence of the protein and the requirement to maintain cis-regulatory elements directing r2 expression. The sequence of the five regulatory elements, codons and amino acids encoded by the chicken gene (Fig. 4B) and an interspecies amino acid alignment for the region (Fig. 4C) are shown. To explore the regulatory constraint, we first altered the r2 elements, using synonymous base substitutions that would maintain the encoded amino acids (for example, in the third base wobble position). We subdivided the effect of each nucleotide exchange into three categories: little or no change of r2 activity (blue, 81–100% efficiency); medium effect, in which the number of the tested embryos showing r2 specific reporter staining was reduced to between 51% and 80% (yellow); and great effect, in which <50% (red) of the tested embryos show r2 reporter expression (Fig. 4B). In all cases involving the ACAAT motifs, effects on reporter expression correlated with the ability of these changes to influence binding of Sox2 (Fig. 4A and data not shown).
Site-directed mutagenesis in the ACAAT motifs had the strongest effect, whereas changes in the third base of codons in the RTE motifs had small or negligible effects. The ACAAT-1 motif of chick spans codons for serine (TCA), threonine (ACA), and methionine (ATG) (Fig. 4B). Altering the serine TCA codon to TCG had no effect on the r2 enhancer activity, consistent with sequences of some fishes (fugu, zebrafish, and medaka), which have a G at this position in the ACAAT-2 motif. However, replacement with the serine codons TCC or TCT reduces the ability of the segment to direct r2 expression (Fig. 4B). Replacing the threonine ACA codon with the ACC or ACG alternatives had a profound effect on enhancer activity, whereas the change to ACT had only a medium effect (Fig. 4B).
In the ACAAT-2 motif, the ability to alter nucleotides and maintain the coding information of the sequences is more limited. The first codon, AAC (asparagine), can be changed to AAT, and the second codon, AAT (asparagine), can be changed to AAC without affecting the encoded amino acid. Neither change had any significant effect on the fragments ability to direct r2 expression (Fig. 4B). Several fishes show the consensus sequence 5′-GACAAT-3′ instead of 5′-AACAAT-3′ in the ACAAT-2 motif encoding, an aspartic acid, instead of asparagine. This is consistent with our analysis above (Figs. 1D and 2K), because we found this substitution is tolerated for regulatory activity, whereas a pyrimidine in this position is not.
Next, we made nonsynonymous substitutions in other parts of the ACAAT-1 motif to more extensively test its sequence requirements. There is only one methionine codon so changing the ATG to AAG or AGG resulted in strong effects on enhancer activity, whereas replacing the ATG with ATC has a moderate effect (Fig. 4B). Other substitutions affecting the core 5′-ACAAT-3′ sequence, also decreased its ability to direct efficient r2 expression (Fig. 4B), underscoring the importance of conserving this motif.
Discussion
Hoxa2 is the only Hox gene expressed in r2 and its expression has a crucial function in establishing the identity of this rhombomere in the developing hindbrain (7). In this study, we identify and characterize the regulatory elements that direct r2 expression and demonstrate they are located in a unique position, within the second coding exon of Hoxa2. These findings raise several interesting issues with respect to both the control of hindbrain segmentation and comparative approaches to the analysis of gene expression.
Hoxa2 and Hoxb2 arose by duplication and divergence from a common ancestor; however, Hoxa2 is the only vertebrate Hox gene expressed in r2. Comparison of the predicted proteins of these group 2 members reveals that the RTE and ACAAT regulatory motifs reside on the carboxyl-terminal side of the Hoxa2 homeodomain, in a region that is not conserved with Hoxb2 (Fig. 4D). Hence, this region has diverged at both the nucleotide and amino acid levels in Hoxb2. This domain is not detected in the Drosophila pb gene or in the single amphioxus Hox2 gene. Furthermore, regulatory studies in transgenic mice testing the activity of this and other regions of the AmphiHox cluster revealed no elements with the ability to regulate rhombomere-restricted expression in the hindbrain (28). Together, these observations suggest that the r2 regulatory region embedded in a Hoxa2 coding exon may have arisen early during the evolution of vertebrates in association with either a gain of r2 elements in Hoxa2 or their loss in Hoxb2.
The finding that Sox proteins are implicated in binding to the ACAAT motifs to potentiate r2 activity of the enhancer is interesting in light of the expression patterns of the SoxB class of genes and the patterns of reporter staining we obtained from multimerized ACAAT motifs (Fig. 3). Sox1, Sox2 and Sox3 are expressed in the developing hindbrain and there are generally much lower levels in r3 and r5 with high levels in the midbrain, r2 and r4 (29). These patterns are similar to that observed for reporter staining with the multimerized ACAAT motifs as opposed to the full r2 enhancer (Fig. 2, 3) suggesting that the ACAAT motifs serve as Sox response elements. Our findings underscore the diverse roles this family of proteins has in regulating segmental expression in the vertebrate hindbrain (10, 11). Based on our analyses, it appears that the mechanism for establishing Hoxa2 expression in r2 combines broad activation and specific repression coordinated by multiple elements. In this model, the ACAAT motifs interact with factors (Sox) that potentiate multiple domains of expression in both the midbrain and hindbrain and the RTEs serve to restrict the regulatory potential, specifically to r2.
The presence of cis-transcriptional regulatory elements embedded in a coding exon is a surprising finding and raises some general issues. The overlap between regulatory function and mRNA synthesis may be difficult to conceptualize. However, regulatory modules have been shown to exert their activities on gene expression from a wide variety of positions upstream, downstream, and within a transcriptional unit (introns and UTRs); hence, cis elements in coding regions form a similar paradigm. Cis elements in introns and UTRs may only be constrained based on their regulatory activity whereas those in coding regions may be subject to the two overlapping constraints of regulatory activity and coding potential. In addition to the r2 elements described in this article, other regulatory elements have been found in coding exons. An enhancer has been located in a coding region of the Bcl-2 gene, although its precise sequence characteristics have not been defined (30). Recently, a Hox-Pbx responsive element has been located in the first exon of the Hoxa2 gene (31) and may work with elements located in the intron to regulate expression in r4 (16). Other types of regulatory sequences playing roles in allelic exclusion of odorant receptors (32) or alternative splicing (33) have been identified in exons and would also be subject to multiple sequence constrains.
Regulatory modules contain a number of individual elements that can often vary with respect to both their position and copy number. Phylogenetic studies have provided evidence for large-scale turnover of functional binding sites in Drosophila, presumably due to the gain and loss of small individual elements through random mutation (34). There is no reason why some of these components could not arise or reside in exons and, once there, become fixed, having a dual purpose of regulation and coding information. Most bioinformatic strategies, using evolutionary conservation in the analysis and prediction of cis elements, begin by excluding protein coding regions. This has streamlined experimental analyses needed to empirically define the precise function of conserved elements but ignores input from coding regions. Furthermore, chromatin immune precipitation assays, which map transcription factor binding sites on whole genome tiling arrays, have found many occupied sites, including in exons, which are often ignored and assumed to be background or nonfunctional sites. The emerging evidence that sequences within coding exons can play diverse roles in regulation of gene expression highlights the need to include these regions in regulatory analyses. For example, comparative genomics methods that include a measure of the triplet usage constraints for specific amino acids in protein coding regions was used to identify the potential exonic splicing regulatory sequences (33). A combination of this approach with a screen for known transcription binding sites irrespective of their location could be valuable in future computational analyses.
Materials and Methods
Constructs for in Vivo and Bioinformatic Analyses.
Transgenic mouse (13) and in ovo chicken embryo (35) reporter expression analyses were performed as described. In the in ovo experiments, efficiency is calculated from the number of embryos expressing the pattern of interest, out of the total number of chicken embryos successfully coelectroporated with a pCMV-GFP reporter. The 10.4-kb BamHI-MluI chicken genomic fragment of Hoxa2 was subdivided into a series of smaller regions for functional analysis by restriction enzyme digestion or PCR-directed methodologies, as indicated in Fig. 1 and 2. Fragments from other species were generated by PCR. Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene). Primers are available in Tables S1–S5. Local alignments between the sequences of interest were generated using ClustalW (36) program.
EMSA.
Sox-2 coding sequence was introduced into pDR and the protein was expressed using the TNT system (Promega). The EMSA reaction conditions were described (37). Probes were labeled with Cy-5. Unlabeled competitor was added in 100-fold excess. Probes and competitor sequences are available in the Tables S1–S5.
Supplementary Material
Acknowledgments.
We thank Filippo Rijli (Friedrich Miescher Institute, Basel, Switzerland) for Hoxa2 mutant mice, Akira Murakami (Kyoto University, Kyoto, Japan) for the Sox2 construct, members of the Molecular Biology Facility for sequencing and targeted mutagenesis, Malcolm Cook and our Bioinformatics facility, the Richard Behringer laboratory (University of Texas M. D. Anderson Cancer Center, Houston, TX) for the bat genomic library, and Trey Fondon (University of Texas Southwestern Medical Center, Dallas, TX) for dog DNA. This work was supported by the Stowers Institute for Medical Research. S.T. was supported by Boehringer Ingelheim Funds and this work was done to fulfill, in part, requirements for his Ph.D. thesis research for the Open University, Milton Keynes, U.K.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Gene Networks in Animal Development and Evolution,” held February 15–16, 2008, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS web site at: http://www.nasonline.org/SACKLER_Gene_Networks.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806360105/DCSupplemental.
References
- 1.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 2.Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 3.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 4.Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- 5.Maconochie M, Nonchev S, Morrison A, Krumlauf R. Paralogous Hox genes: Function and regulation. Annu Rev Genet. 1996;30:529–556. doi: 10.1146/annurev.genet.30.1.529. [DOI] [PubMed] [Google Scholar]
- 6.Lumsden A. Segmentation and compartition in the early avian hindbrain. Mech Dev. 2004;121:1081–1088. doi: 10.1016/j.mod.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 7.Gavalas A, Davenne M, Lumsden A, Chambon P, Rijli FM. Role of Hoxa-2 in axon pathfinding and rostral hindbrain patterning. Development. 1997;124:3693–3702. doi: 10.1242/dev.124.19.3693. [DOI] [PubMed] [Google Scholar]
- 8.Trainor PA, Krumlauf R. Patterning the cranial neural crest: Hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci. 2000;1:116–124. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- 9.Nolte C, Krumlauf R. In: HOX Gene Expression. Papageorgiou S, editor. Austin, TX: Landes Bioscience and Springer; 2006. pp. 14–41. [Google Scholar]
- 10.Tümpel S, Maconochie M, Wiedemann LM, Krumlauf R. Conservation and diversity in the cis-regulatory networks that integrate information controlling expression of Hoxa2 in hindbrain and cranial neural crest cells in vertebrates. Dev Biol. 2002;246:45–56. doi: 10.1006/dbio.2002.0665. [DOI] [PubMed] [Google Scholar]
- 11.Nonchev S, et al. Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development. 1996;122:543–554. doi: 10.1242/dev.122.2.543. [DOI] [PubMed] [Google Scholar]
- 12.Nonchev S, et al. The conserved role of Krox-20 in directing Hox gene expression during vertebrate hindbrain segmentation. Proc Natl Acad Sci USA. 1996;93:9339–9345. doi: 10.1073/pnas.93.18.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maconochie MK, Nonchev S, Manzanares M, Marshall H, Krumlauf R. Differences in Krox20-dependent regulation of Hoxa2 and Hoxb2 during hindbrain development. Dev Biol. 2001;233:468–481. doi: 10.1006/dbio.2001.0197. [DOI] [PubMed] [Google Scholar]
- 14.Tümpel S, Cambronero F, Wiedemann LM, Krumlauf R. Evolution of cis elements in the differential expression of two Hoxa2 coparalogous genes in pufferfish (Takifugu rubripes) Proc Natl Acad Sci USA. 2006;103:5419–5424. doi: 10.1073/pnas.0600993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maconochie M, et al. Regulation of Hoxa2 in cranial neural crest cells involves members of the AP-2 family. Development. 1999;126:1483–1494. doi: 10.1242/dev.126.7.1483. [DOI] [PubMed] [Google Scholar]
- 16.Tümpel S, et al. Expression of Hoxa2 in rhombomere 4 is regulated by a conserved cross-regulatory mechanism dependent upon Hoxb1. Dev Biol. 2007;302:646–660. doi: 10.1016/j.ydbio.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 17.Zhang M, et al. Ectopic Hoxa-1 induces rhombomere transformation in mouse hindbrain. Development. 1994;120:2431–2442. doi: 10.1242/dev.120.9.2431. [DOI] [PubMed] [Google Scholar]
- 18.Frasch M, Chen X, Lufkin T. Evolutionary-conserved enhancers direct region-specific expression of the murine. Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development. 1995;121:957–974. doi: 10.1242/dev.121.4.957. [DOI] [PubMed] [Google Scholar]
- 19.Lampe X, Picard JJ, Rezsohazy R. The Hoxa2 enhancer 2 contains a critical Hoxa2 responsive regulatory element. Biochem Biophys Res Commun. 2004;316:898–902. doi: 10.1016/j.bbrc.2004.02.138. [DOI] [PubMed] [Google Scholar]
- 20.Dobreva G, et al. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Manzanares M, et al. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involves auto and cross-regulatory mechanisms. Development. 2001;128:3595–3607. doi: 10.1242/dev.128.18.3595. [DOI] [PubMed] [Google Scholar]
- 22.Gould A, Itasaki N, Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21:39–51. doi: 10.1016/s0896-6273(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 23.Pöpperl H, et al. Segmental expression of Hoxb1 is controlled by a highly conserved autoregulatory loop dependent upon exd/Pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 24.Maconochie MK, et al. Cross-regulation in the mouse HoxB complex: The expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes Dev. 1997;11:1885–1896. doi: 10.1101/gad.11.14.1885. [DOI] [PubMed] [Google Scholar]
- 25.Rijli FM, et al. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell. 1993;75:1333–1349. doi: 10.1016/0092-8674(93)90620-6. [DOI] [PubMed] [Google Scholar]
- 26.Mertin S, McDowall SG, Harley VR. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 1999;27:1359–1364. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanai Y, et al. Identification of two Sox17 messenger RNA isoforms, with and without the high mobility group box region, and their differential expression in mouse spermatogenesis. J Cell Biol. 1996;133:667–681. doi: 10.1083/jcb.133.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manzanares M, et al. Conservation and elaboration of Hox gene regulation during evolution of the vertebrate head. Nature. 2000;408:854–857. doi: 10.1038/35048570. [DOI] [PubMed] [Google Scholar]
- 29.Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86:197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- 30.Lang G, Gombert WM, Gould HJ. A transcriptional regulatory element in the coding sequence of the human Bcl-2 gene. Immunology. 2005;114:25–36. doi: 10.1111/j.1365-2567.2004.02073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lampe X, et al. An ultraconserved Hox-Pbx responsive element resides in the coding sequence of Hoxa2 and is active in rhombomere 4. Nucleic Acids Res. 2008;36:3214–3225. doi: 10.1093/nar/gkn148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131:1009–1017. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goren A, et al. Comparative analysis identifies exonic splicing regulatory sequences—The complex definition of enhancers and silencers. Mol Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Moses AM, et al. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoue T, Krumlauf R. An impulse to the brain: Using in vivo electroporation. Nat Neurosci. 2001;4:6–8. doi: 10.1038/nn1101-1156. [DOI] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami A, Thurlow J, Dickson C. Retinoic acid-regulated expression of fibroblast growth factor 3 requires the interaction between a novel transcription factor and GATA-4. J Biol Chem. 1999;274:17242–17248. doi: 10.1074/jbc.274.24.17242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.