Abstract
Motivation
Properly annotated sequence data for flaviviruses, which cause diseases, such as tick-borne encephalitis (TBE), dengue fever (DF), West Nile (WN) and yellow fever (YF), can aid in the design of antiviral drugs and vaccines to prevent their spread. Flavitrack was designed to help identify conserved sequence motifs, interpret mutational and structural data and track evolution of phenotypic properties.
Summary
Flavitrack contains over 590 complete flavivirus genome/protein sequences and information on known mutations and literature references. Each sequence has been manually annotated according to its date and place of isolation, phenotype and lethality. Internal tools are provided to rapidly determine relationships between viruses in Flavitrack and sequences provided by the user.
Availability
Contact
chschein@utmb.edu
Supplementary information
1 INTRODUCTION
Flaviviruses (FV), (+)-strand RNA viruses in the same genus as hepaci- and pestiviruses, are responsible for many emerging human encephalitic and hemorrhagic diseases (Barrett and Higgs, 2007). The ∼10.5 kb genome encodes a single polyprotein that is cleaved into 10 viral proteins (Fig. 1). A large amount of sequence and structural data is now available for FV such as WN and dengue (DV), which have spread worldwide due to increasing intercontinental travel, relaxation in vector control and lack of effective antiviral drugs (Kuno and Chang, 2005; Mackenzie et al., 2004). Although a successful vaccine against YF has existed for many years, vaccine design for other FV is more complicated (Adams and Boots, 2006; Seligman and Gould, 2004; Thomas et al., 2006). For example, a primary infection with one strain of DV may predispose an individual to Dengue Hemorrhagic Fever, a more severe disease, if infected subsequently with a different DV strain. It is thus critically important to distinguish the common features of these viruses, as well as differences that may be associated with lethality. Flavitrack was designed to ease the identification of conserved functional areas, using methodology previously developed in this group (Negi et al., 2006; Schein et al., 2005a), and to group viruses according to their phenotypic characteristics. The database contains all publicly available full-genome flavivirus sequences and provides access to sequence analysis tools. Flavitrack will eventually also contain structures or 3D models for all flavivirus proteins, allowing combined sequence/structure analysis to characterize common B- and T-cell epitopes, account for the functional effects of mutations, and determine highly conserved areas.
Fig. 1.
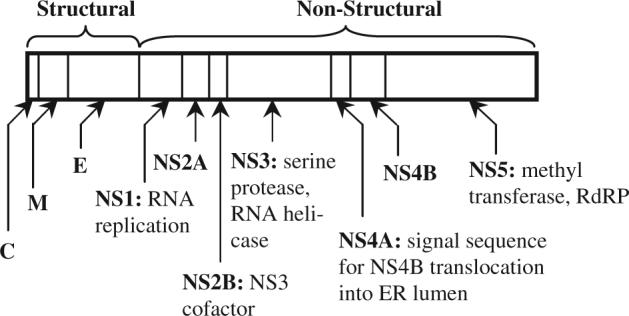
Polyprotein common to FV with preferred abbreviations for the structural [capsid (C), membrane (M) and envelope (E)] and non-structural (NS) proteins, whose function is noted where known.
2 FEATURES
The major features of Flavitrack are sequence retrieval, BLAST comparison of a given RNA or protein sequence to those in the database, and sequence alignment of user selected sequences. Precalculated sequence alignments, integrated with the Jalview (Clamp et al., 2004) multiple alignment editor, are provided for all polyproteins in Flavitrack (currently 544 sequences), an overview subset of 49 distinct FV, and one that combines all the FV, pestivirus and hepacivirus polyproteins in the NCBI (923 sequences). Principal components analysis (PCA) of aligned sequences (using Jalview) allows rapid derivation of phylogenetic viral groupings, most obviously according to vector (Fig. 2). To aid data analysis of these large alignments, each sequence has been manually assigned a unique identifier, a ‘license plate’, which concisely highlights important information about the virus. Strain names such as ‘New Guinea C’ are not transparent and NCBI numbers do not reveal even the viral strain. To allow maximum flexibility, license plates are a concatenation of eight identifying characteristics: virus abbreviation (2−3 letters) according to the Center for Disease Control (CDC), phenotype (encephalitic/hemorrhagic/vaccine), year and country (ISO code) of isolation, vector (mosquito, tick or none), lethality, host type (human, bird, rodent, etc.), age (adult or juvenile) and gender. For example, the license plate for the Genbank gi:28453847 sequence is TBEe85RUtFhM which clearly marks it as a tick-borne encephalitis strain isolated in 1985 in Russia that resulted in a fatal encephalitis of a human male.
Fig. 2.
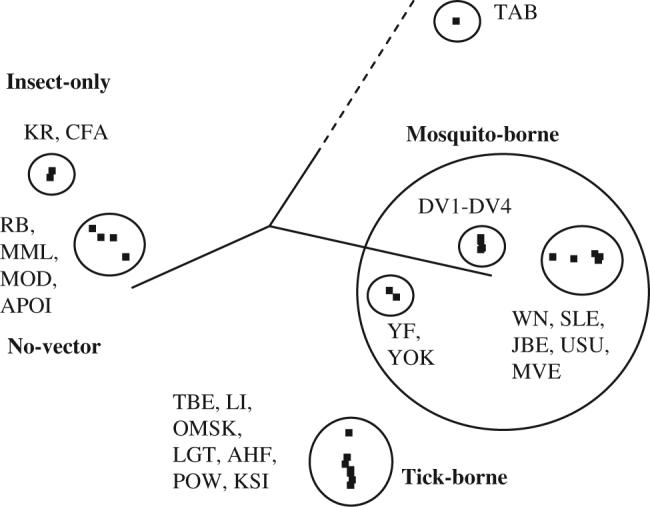
Principal components analysis (PCA) of major flavivirus reference sequences, with varying identity to DV1 (Supplementary material, Table S1), using the sum of pairwise BLOSUM scores for eigenvector decomposition, separates them according to vector.
A list of mutants and variant sequences, tabulated according to derived strains, location of mutations, corresponding altered phenotypes, and the references for each mutation has been included in Flavitrack. Flavitrack also provides access to our in-house program, PCPMer (http://born.utmb.edu/BinZhou/ PCPMer; Schein et al., 2005b), which can be used to automatically visualize areas that are highly conserved on structures of FV proteins. PCPMer results, coupled with the mutation data, aid in identifying structural motifs associated with viral function or lethality. This enables a user to correlate the genotype with viral characteristics.
Searches can be done for individual proteins in the annotated sequences or by any of the terms described above for license plates. Terms can also be combined to identify, e.g. all WN sequences from the United States since 1999.
Flavitrack is currently updated every 3 months by uploading the latest Genbank sequences for FV deposited since the last update and the corresponding manually assigned license plates. The mutations list is also updated after periodic literature searches.
3 DESIGN AND IMPLEMENTATION
The major challenge was to cross-reference the sequences of the FV with consistent annotation in such a way that protein sequence motifs corresponding to phenotype could be obtained automatically. The sequences were downloaded from Genbank and processed before inclusion. A MySQL/PHP design with embedded Bioperl functions was used to construct the database. The design extends the BioSQL model, which is a generic relational model for storing annotated biological sequences. Additional tables have been included for the license plate and mutation information.
The non-uniform terminology for virus features in Genbank annotations required very comprehensive identifier definitions in the scripts for extracting data from the NCBI headers. For example, there are about 30 different ways the capsid protein is identified, ranging from ‘C’ (our preferred notation, Fig. 1) to ‘nucleocapsid’ to ‘NC’. To automatically populate the database with accurate information, we first identified object classes that included all synonyms for all virus proteins and wrote comprehensive regular expressions to parse the files. The files were then hand edited, during the assignment of the license plates, to check for other novel synonyms. This detailed ontology or controlled vocabulary for virus features can be used directly with the underlying BioSQL model's generic ontology tables.
4 CONCLUSION
We have archived the sequences of flaviviruses in a relational database designed to aid in identifying surface-exposed clusters of conserved amino acids and correlating these with data on mutational data for altered phenotypes, vector type and disease characteristics. We have used Flavitrack to generate multiple sequence alignments of the polyproteins, which indicate a phylogeny similar to that previously determined based on individual FV proteins (Kuno et al., 1998). The integrated Jalview tools also enable more rigorous viral groupings, e.g. a principle component analysis (PCA) based on pairwise BLOSUM scores for reference flavivirus polyproteins separates them according to vector (Fig. 2). Analysis of even larger groups of sequences will be aided by the manual editing of the sequences and their license plates, to enable extraction of features related to disease severity. The complete sequence alignment of all FV polyproteins identifies a set of conserved regions (PCPMer motifs) that can be used to determine their relationship to more distantly related (+)-strand viruses, such as the pesti- and hepaciviruses, and even the alphaviruses, which cause similar diseases but have a different genome organization (Strauss and Strauss, 1994). With continued refinement, Flavitrack will be a useful tool in vaccine design and identifying conserved areas that could be targeted by inhibitors of Flaviviruses.
Supplementary Material
ACKNOWLEDGEMENTS
We thank summer interns Viet Tran (mutation list and object identifiers) and Michael Zhang (license plate assignment), and Alan D.T. Barrett and Werner Braun at the UTMB for advice. Support: NIH grant AI064913 (to Dr Werner Braun and C.H.S.).
Footnotes
Conflict of Interest: none declared.
REFERENCES
- Adams B, Boots M. Modelling the relationship between antibody-dependent enhancement and immunological distance with application to dengue. J. Theor. Biol. 2006;242:337–346. doi: 10.1016/j.jtbi.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Barrett ADT, Higgs S. Yellow fever: a disease that has yet to be conquered. Annu. Rev. Entomol. 2007;52:209–229. doi: 10.1146/annurev.ento.52.110405.091454. [DOI] [PubMed] [Google Scholar]
- Clamp M, et al. The Jalview Java alignment editor. Bioinformatics. 2004;20:426. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Kuno G, Chang GJ. Biological transmission of arboviruses: reexamination of and new insights into components, mechanisms, and unique traits as well as their evolutionary trends. Clin. Microbiol. Rev. 2005;18:608–637. doi: 10.1128/CMR.18.4.608-637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G, et al. Phylogeny of the genus Flavivirus. J. Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS, et al. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile, and dengue viruses. Nat. Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Negi SS, et al. Determining functionally important amino acid residues of the E1 protein of Venezuelan equine encephalitis virus. J. Mol. Model. 2006;12:921–929. doi: 10.1007/s00894-006-0101-7. [DOI] [PubMed] [Google Scholar]
- Schein CH, et al. Stereophysicochemical variability plots highlight conserved antigenic areas in Flaviviruses. Virol. J. 2005a;2:40. doi: 10.1186/1743-422X-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein CH, et al. Molego-based definition of the architecture and specificity of metal-binding sites. Proteins. 2005b;58:200–210. doi: 10.1002/prot.20253. [DOI] [PubMed] [Google Scholar]
- Seligman SJ, Gould EA. Live flavivirus vaccines: reasons for caution. Lancet. 2004;363:2073–2075. doi: 10.1016/S0140-6736(04)16459-3. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, et al. Antibody-dependent enhancement and vaccine development. Expert Rev. Vaccines. 2006;5:409–412. doi: 10.1586/14760584.5.4.409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


