SUMMARY
Evidence for active DNA demethylation in vertebrates is accumulating, but the mechanisms and enzymes remain unclear. Using zebrafish embryos we provide evidence for 5-methylcytosine (5-meC) removal in vivo via the coupling of a 5-meC deaminase (AID, which converts 5-meC to thymine) and a G:T mismatch-specific thymine glycosylase (Mbd4). The injection of methylated DNA into embryos induced a potent DNA demethylation activity, which was attenuated by depletion of AID or the non enzymatic factor Gadd45. Remarkably, overexpression of the deaminase/glycosylase pair AID/Mbd4 in vivo caused demethylation of the bulk genome and injected methylated DNA fragments, likely involving a G:T intermediate. Furthermore, AID or Mbd4 knockdown caused the remethylation of a set of common genes. Finally, Gadd45 promoted demethylation and enhanced functional interactions between deaminase/glycosylase pairs. Our results provide evidence for a coupled mechanism of 5-meC demethylation, whereby AID deaminates 5-meC, followed by thymine base excision by Mbd4, promoted by Gadd45.
INTRODUCTION
DNA methylation is associated with gene silencing, and plays important roles in mammalian development and genomic imprinting (Reik, 2007). Misregulation of DNA methylation also contributes to cancer development by causing genomic instability and inappropriate silencing of tumor suppressor genes (Esteller, 2008). Although the DNA methyltransferase (DNMT) enzymes that generate 5-methylcytosine (5-meC) in vertebrates are well studied (Goll and Bestor, 2005), evidence for a vertebrate enzyme exhibiting reproducible DNA demethylation either in vitro or in vivo is lacking. However, various studies have provided evidence for a replication-independent (active) mode of DNA demethylation. For example, in activated T cells a promoter-enhancer element of interleukin-2 gene undergoes demethylation within 20 min of stimulation (Bruniquel and Schwartz, 2003). Also, targets of the estrogen receptor (ERα) show cyclical DNA demethylation uncoupled to replication (Metivier et al., 2008). During mouse embryonic development, the paternal genome undergoes demethylation in a replication-independent manner (Mayer et al., 2000; Oswald et al., 2000; Reik, 2007). Furthermore, in zebrafish embryos, the demethylation of injected methylated DNA can occur in a replication-independent manner (Collas, 1998), and quantification of methylation levels suggests active demethylation at fertilization (Mhanni and McGowan, 2004). Notably, pioneering work in plants provides strong evidence for a set of glycosylase/lyase enzymes (Demeter, ROS, DML2, DML3) in the removal of the 5-meC in various biological contexts (Gehring et al., 2006; Gong et al., 2002; Morales-Ruiz et al., 2006; Penterman et al., 2007). However, this mechanism is only one of many proposed but unproven mechanisms for DNA demethylation in vertebrates, which include: (1) direct removal of the methyl group, regenerating cytosine, (2) direct removal of the base (via glycosylase/lyase base excision activity, as in plants), followed by repair/replacement with cytosine, (3) conversion of the base to thymine (via deamination), followed by removal and subsequent repair, and (4) excision of one or more nucleotides surrounding 5-meC, followed by repair.
In regard to the first mechanism, vertebrates contain orthologs of the bacterial AlkB demethylases (dioxygenases) which operate through direct methyl removal in prokaryotes (Morgan et al., 2005; Sedgwick, 2004). However, none of the vertebrate orthologs have yet displayed direct 5-meC demethylation activity. Although the jumanji family of histone demethylases (HDMs) share homology to bacterial AlkB (Ozer and Bruick, 2007) none have been reported to direct DNA demethylation. Earlier work suggested that methyl-binding domain protein 2b (Mbd2b) could directly remove the methyl group, leaving cytosine and methanol as products (Bhattacharya et al., 1999). However, as Mbd2b lacks an enzymatic domain, this activity remains controversial. The second mechanism (observed in plants) is also plausible, as vertebrates contain two demonstrated glycosylases: thymine DNA glycosylase (TDG), and methyl-binding domain protein 4 (Mbd4). However, both TDG and Mbd4 have only weak 5-meC base excision activity, relative to their activity on thymine (Cortazar et al., 2007; Zhu et al., 2000). For the third mechanism, certain enzymes in the cytidine deaminase family (Activation Induced deaminase (AID) and Apolipo-protein B RNA-editing catalytic component-1 (Apobec-1)) have been shown to deaminate 5-meC in single-stranded DNA (generating thymine and yielding a G:T mismatch), establishing their candidacy in vivo (Morgan et al., 2004). Furthermore, AID and Apobec-1 are coexpressed with pluripotency genes in oocytes, embryonic germ cells and embryonic stem cells (Morgan et al., 2004). Finally, an intriguing recent study provided evidence that the DNA methyltransferase DNMT3B may function either as a DNMT, or alternatively as a 5-meC deaminase in conditions of low cofactor (S-adenosylmethionine, see Discussion) (Metivier et al., 2008).
Elegant work by many groups has shown that enzymes of the AID and Apobec family are structurally related zinc-dependent cytidine deaminases that can operate on both RNA or DNA, with some members showing moderate deamination of 5-meC (Conticello et al., 2007; Morgan et al., 2004). Hereafter, we will refer to their zebrafish orthologs collectively as AID/Apobec proteins. However, the use of 5-meC deamination as the first of two steps for 5-meC demethylation has been reasonably questioned, due to the creation of a mutagenic G:T intermediate. However, the physical coupling of a 5-meC deaminase with a thymine glycosylase could, in principle, rapidly remove the mutagenic thymine, enabling cytosine replacement by base excision repair (BER). Two potential glycosylase candidates are TDG and Mbd4, discussed above. Notably, MBD4 contains a methylated-DNA binding domain as well as a glycosylase domain that prefers to remove the thymine from a G:T mismatch (Hendrich et al., 1999). Although mice lacking Mbd4 are viable, they show a higher frequency of mutations at CpG sites (Millar et al., 2002; Wong et al., 2002).
Recently, a role for the non-enzymatic factor Gadd45α in promoting demethylation was reported (Barreto et al., 2007). Gadd45α knockdown lead to hypermethylation of bulk genome in human cells and its overexpression clearly reduced the DNA methylation status of the bulk genome and Oct-4 promoter (Barreto et al., 2007). However, a recent study questioned the sufficiency of Gadd45 overexpression to elicit DNA demethylation in human cells (Jin et al., 2008). Here we provide data that may help reconcile these observations, both supporting a role for Gadd45 while revealing Gadd45 as only one factor in a system of proteins involved in the demethylation process. Currently, the enzymes that may cooperate with Gadd45 to perform demethylation have remained unknown, though a link to DNA repair has been suggested (Barreto et al., 2007), and here candidate cooperating enzymes are evaluated. Taken together, we provide evidence for a coupled mechanism for 5-meC demethylation; deamination by AID/Apobec followed by thymine base excision by MBD4, can occur in zebrafish embryos, and is promoted by Gadd45.
RESULTS
A DNA Demethylation/Remethylation Activity in Zebrafish Embryos
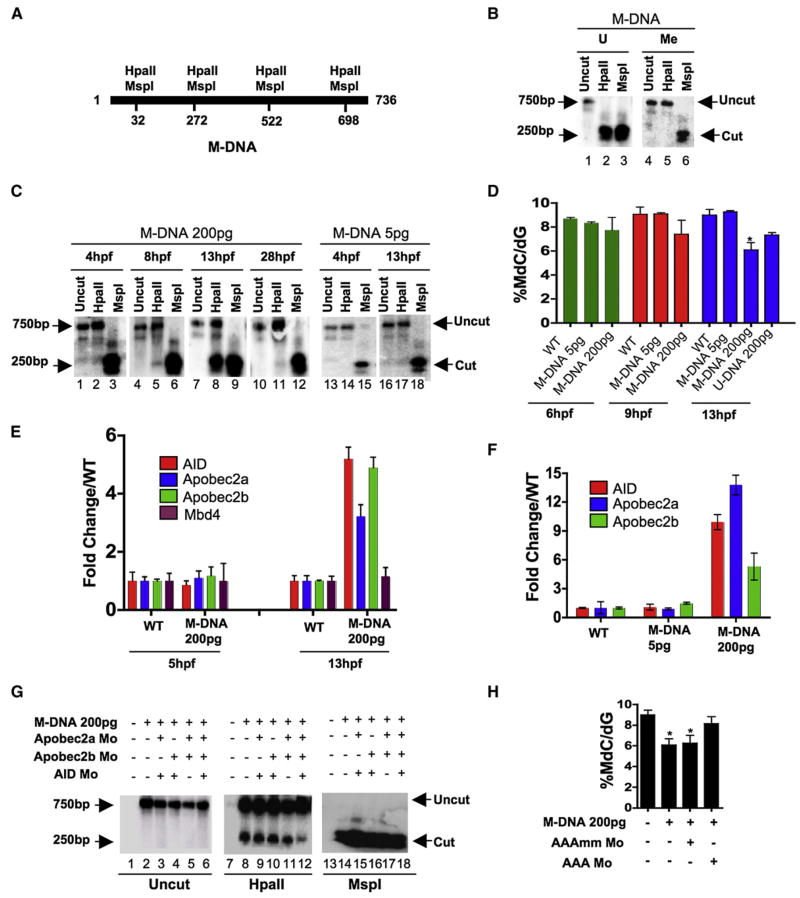
Previous work in zebrafish embryos revealed the in vivo demethylation of an in vitro-methylated DNA fragment (or plasmid) occurring during a particular window of embryo development (Collas, 1998), suggesting the presence of regulated DNA demethylation activity. In that study, and in our initial assay, the steady-state methylation status of a methylated DNA fragment (M-DNA, 736 bp, injected at the single-cell stage) was assessed by susceptibility to the restriction enzyme HpaII (which is methylation-inhibited). Four HpaII/MspI sites (CCGG) are present (Figure 1A), with HpaII or MspI digestion of the unmethylated (U) 736 bp DNA fragment generating five smaller fragments: two comigrating fragments of 250 bp and 240 bp, one of 176 bp, and two that are too small for detection (32 and 38 bp). MspI digestion (which is methylation insensitive) generates this spectrum from either unmethylated or fully methylated M-DNA (Figures 1A and 1B). Full methylation of the DNA fragment by HpaII methylase (which methylates the internal cytosine of an HpaII/MspI restriction enzyme site, CmeCGG) and the digestion behavior of the substrate was verified in vitro (Figure 1B). Following M-DNA injection into single-cell fertilized embryos, M-DNA was reisolated from the embryo at different developmental time points, treated with HpaII or MspI, and cleavage assessed via Southern analysis (note: the cleaved products (176–250 bp) transfer much more efficiently to the membrane that does the intact 736 bp M-DNA substrate, providing greater signal). We find that M-DNA remained largely methylated at ~4 hr postfertilization (hpf), was slightly demethylated at ~8 hpf (75% epiboly, Figure 1C, lane 5), became clearly demethylated at 13 hpf (early somite stage, Figure 1C, lane 8) and then became largely remethylated by 28 hpf (prim stage, Figure 1C, lane 11). This temporal pattern of demethylation/remethylation was also observed with an injected methylated closed circular plasmid (Figure S1 available with this article online). In addition, we noticed that a threshold level (~50–100 pg) of M-DNA was required to elicit demethylation (Figure 1C and data not shown). Unexpectedly, M-DNA injection (or a methylated plasmid, > 100 pg) caused the demethylation of 20%–40% of the bulk genome at 13 hpf, as determined by mass spectrometric analysis of 5-meC content (Figure 1D) and HpaII sensitivity of the bulk genome (Figure S1). This genome-wide demethylation could also be elicited by the injection of the unmethylated 736 bp DNA fragment (U-DNA, Figure 1D), though not to the same extent as M-DNA. However, bisulphite sequencing revealed the methylation of > 50% of the CpGs on this fragment by 6 hpf (data not shown), in keeping with previous observations that injected DNA acquires methylation in early zebrafish embryos (Collas, 1998). Notably, remethylation of the DNA fragment and the bulk genome subsequently occurred by 28 hpf (Figures 1C and S1), showing that DNA methylation systems remain functional. Taken together, genome-wide demethylation can be induced in zebrafish embryos by the injection of methylated DNA in a time and concentration dependent manner.
Figure 1. Involvement of Cytidine Deaminases in DNA Demethylation in Zebrafish.
(A) Schematic of the M-DNA (Methylated DNA) fragment injected into fertilized embryos at the single-cell stage. The four HpaII/MspI sites are indicated. HpaII-resistant (methylated CmeCGG) and HpaII-cleaved species (unmethylated) run at ~750 bp and ~250 bp, respectively (B).
(B) M-DNA methylation status was assessed by HpaII susceptibility, with cutting observed on unmethylated (U) (lane 2) but not methylated (Me) DNA (lane 2). MspI-digested (lanes 3 and 6) and uncut DNA (lanes 1 and 4) served as controls. Fragments were detected by Southern blot analysis probed with full-length M-DNA probe.
(C) M-DNA methylation status during development. Total DNA was isolated at the time points shown from embryos injected at the single-cell stage with M-DNA (5 pg or 200 pg), and treated as in (B). M-DNA induced demethylation peaks at ~13 hpf (Compare lanes 2, 5, 8, and 11).
(D) LC-MS quantitation of 5-meC content of the bulk genome (normalized to total deoxyguanosine content) in genomic DNA isolated from embryos (hpf indicated) injected with methylated M-DNA or unmethylated U-DNA at the single-cell stage.
(E and F) qRT-PCR determinations from embryos injected with M-DNA (E), and at different fragment concentrations (F).
(G and H) Methylation status of M-DNA assessed by HpaII digestion and Southern blotting (G), or LC-MS quantitation of total 5-MeC (H) in total genomic DNA isolated from embryos at 13 hpf, injected at the single-cell stage with M-DNA (200 pg) and morpholinos as indicated. Lanes 1, 7, and 13 correspond to wild-type sample.
AAAmm refers to a set of three control morpholinos against AID (4 pg), Apobec2a (4 pg), and Apobec2b (2 pg) (AAA), which each contain five mismatched (mm) bases (of 25 total to prevent binding) relative to the efficacious morpholino (same amount as controls). For HpaII/MspI susceptibility, one representative of at least three biological repeats is shown. LC-MS measurements; two biological replicates. Asterisks (*) depict statistical significance (p < 0.05). Error bars: +/− one standard deviation.
Upregulation of AID/Apobec2a/b Proteins Coincident with Demethylation
To help focus our studies on particular enzyme candidates, we tested whether candidate enzymes were transcriptionally upregulated during or just prior to the peak of M-DNA demethylation (though a lack of upregulation does not exclude their involvement). First, we tested for upregulation of the three annotated members of the AID/Apobec deaminase family (AID, Apobec2a, and Apobec2b), all six annotated members of the AlkB dioxygenase family, four of the six Gadd45-family proteins, and the sole zebrafish homolog of human Mbd4, zMbd4. We note that zebrafish lack Apobec1/3 families as they are restricted to placental mammals. M-DNA injection upregulated the three AID/Apobec members (at 13 hpf), but not AlkB-related factors or zMbd4 (Figure 1E and data not shown), in a time and concentration-dependent manner (Figure 1F). Finally, RT-PCR analysis revealed exceptionally high levels of AID and zMbd4, and very low levels of zTDG, in the single-cell-stage embryo (30 min postfertization, Figure S2), where bulk DNA demethylation has most clearly been demonstrated in other studies (Mhanni and McGowan, 2004). We therefore focused on AID/Apobec enzymes and Mbd4 for our studies.
Involvement of AID/Apobec enzymes in demethylation was assessed by knockdown experiments using antisense morpholino-modified oligonucleotides (hereafter referred as morpholinos). We note that AID knockdown was only partial, whereas Apobec2a/b knockdowns were efficient (Figures S3A–S3C), with Apobec knockdowns verified by immunoblot analysis using antisera we raised against zebrafish Apobec2a or 2b. Knockdown of all three AID/Apobec members (but not each separately) attenuated demethylation of injected M-DNA and demethylation of the bulk genome caused by M-DNA injection at 13 hpf (Figure 1G and 1H, respectively). To test whether AID/Apobec enzymes affect the methylation status of the genome during normal development, in the absence of M-DNA injection, we measured global methylation levels in embryos injected with morpholinos against all three deaminase members (AID, Apobec2a and Apobec2b; termed AAAmo). Notably, at 24 hpf, AAAmo embryos harbor ~12% more methylation than wild-type embryos (WT, 8.55+/−0.04%, AAAmo = 9.61+/−0.28%). Together, these results suggest that AID/Apobec enzymes normally reduce steady-state methylation levels.
Coexpression of AID/Apobec and Mbd4 Causes Widespread DNA Demethylation
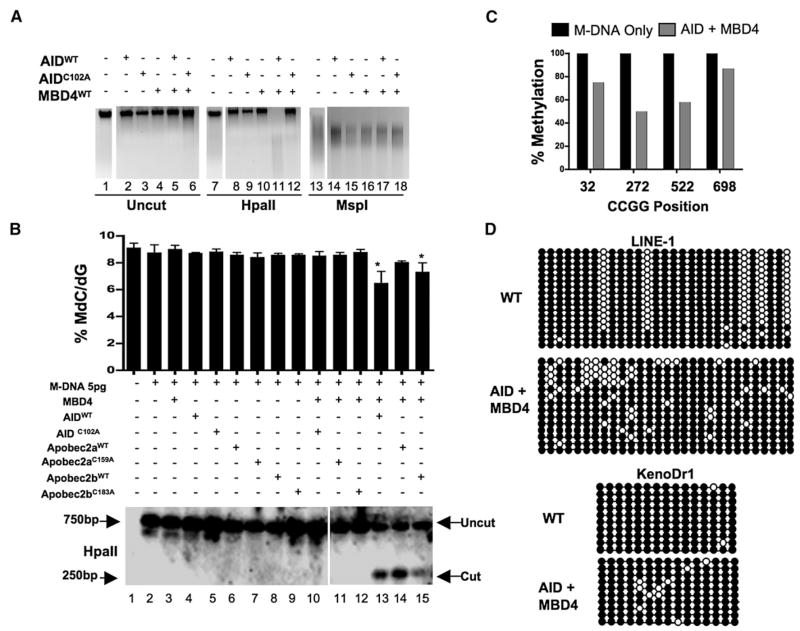
Notably, AID or Apobec2a/b overexpression (by RNA injection at the single-cell stage) had little or no impact (at 13 hpf) on HpaII cleavage of the bulk genome or the injected M-DNA fragment, or on 5-meC levels assessed by mass spectrometry (Figures 2A and 2B). This suggests that overexpressed AID/Apobec enzymes were either not targeted, or not activated, or both. Furthermore, the predicted product of successful deamination (a G:T mismatch) is not cleavable by HpaII; restoration of cleavage requires thymine removal (possibly by base excision) and cytosine reincorporation by repair or replication. We therefore tested whether coupling the deaminase with a glycosylase would provide demethylation. However, the current zebrafish genome-build remains incomplete, and lacks the 5′ end of zMbd4. Therefore, our overexpression studies instead employed full-length human Mbd4. The relevance of this approach is supported by the common observation that human orthologs effectively complement their zebrafish orthologs in knockdown/complementation studies (Lan et al., 2007; Rai et al., 2006) and expression of hMbd4 effectively complemented zMbd4 knockdown for phenotypic defects (Figure S4, discussed further below). As with AID/Apobec enzymes, overexpression of hMbd4 alone had little or no affect on methylation status (Figures 2A and 2B).
Figure 2. Overexpression of a Deaminase/Glycosylase Pair Elicits DNA Demethylation.
(A–C) Methylation status assessed by HpaII digestion of total genomic DNA (A), LC-MS quantitation ([B] upper panel), HpaII digestion of M-DNA (Southern analysis) ([B] lower panel), and bisulphite sequencing of M-DNA (C). Lanes 1, 7, and 13 in (A) and lane 1 in (B) correspond to wild-type sample. For (B), M-DNA was injected at 5 pg, below the threshold level for eliciting demethylation on its own (See Figures 1C and 1D). For (C), twenty clones were subjected to bisulphite sequencing, and the methylation status of each HpaII/MspI (CCGG) site reported as a percentage of total sites tested.
(D) Repeat elements from DNA isolated from embryos (13 hpf) injected at the single-cell stage with RNA encoding wild-type AID, along with MBD4 wild-type mRNA.
For each experiment, one representative of at least three biological repeats is shown except in LC-MS measurement where graph is prepared from values of two biological replicates. Asterisks (*) depict statistical significance (p < 0.05). Error bars are ± one standard deviation.
Remarkably, coexpression of AID or Apobec2a/b along with hMbd4 provided clear DNA demethylation at 13 hpf as assessed by several methods and at multiple loci: HpaII cleavage of genomic DNA (Figure 2A, lane 11), mass spectrometric analysis of the bulk genome (Figure 2B, upper panel, lanes 13–15), HpaII cleavage of M-DNA (coinjected at 5 pg (subthreshold), Figure 2B, lower panel, lanes 13–15), and the bisulphite sequencing of M-DNA (Figure 2C), where demethylation was pronounced. AID involvement was not entirely unexpected given its known ability to deaminate 5me-C in single-stranded DNA in vitro; however the efficacy of Apobec2a/b (especially Apobec2b) was somewhat unexpected, as substrates have heretofore been elusive. Furthermore, demethylation required the catalytic activity of each AID/Apobec enzyme (Figures 2A and 2B), and mutation of the catalytic residue did not affect enzyme abundance (Figures S5A–S5C). Notably, the combination of AID/Apobec and hMbd4 overexpression, but not catalytic mutants, caused lethality by 24 hpf, consistent with observations in mice and zebrafish that even moderate changes in DNA methylation are highly detrimental or lethal (Ooi and Bestor, 2008; Rai et al., 2006). Finally, we emphasize that AID or Apobec expression alone did not prevent MspI cleavage of M-DNA or the bulk genome. Here, deamination of 5-meC (if it occurred) would prevent MspI cleavage due to creation of G:T mismatches, but was not observed. These results suggest that deamination activity by AID/Apobec may not occur unless Mbd4 (and possibly other factors) are present and/or activated, explored below.
Our overexpression experiments showed reductions in bulk methylation which we wanted to validate by the definitive bisulphite sequencing method. To this end, we examined the methylation status of two repetitive elements. Here, LINE-1 elements showed a clear difference in methylation pattern but little change in total methylation levels (Figure 2D, top panels), whereas KenoDr1 showed moderate demethylation (Figure 2D, bottom panels). As these loci are highly and constitutively methylated, we did not expect them to be normal targets of the demethylase system in somatic cells, but may have been targeted to these loci by overexpression and the presence of an MBD on Mbd4. Gene/promoter targets of the system are characterized later, following phenotypic analysis of morphants, and characterization of the demethylation mechanism.
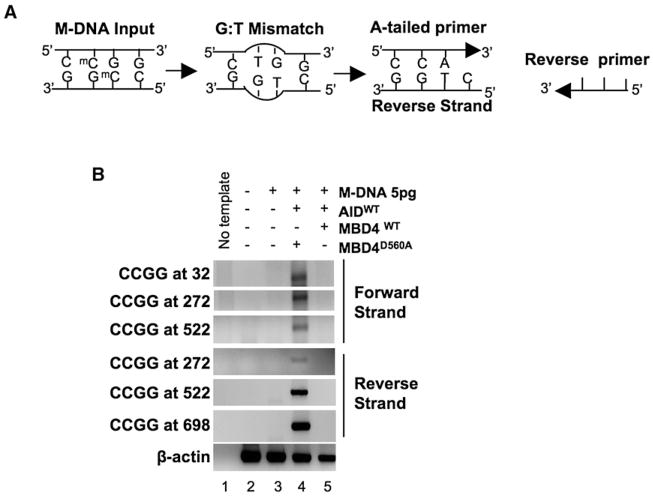
Evidence for Demethylation via a G:T intermediate
To test whether the demethylation reaction following AID and MBD4 coexpression proceeded through a G:T intermediate, two assays were performed on M-DNA (5 pg, coinjected with AID and Mbd4) reisolated from 13 hpf embryos: (1) PCR analysis of all four HpaII/MspI sites on M-DNA for the presence of a G:T base pair. The technique uses a ‘forward’ primer with a 3′-terminal adenosine complementary to the thymine base derived from the deamination of 5-meC at the initial G:5meC base pair in M-DNA (Figure 3A). The “reverse” primer is perfectly complimentary to a downstream region of M-DNA. Here, a PCR product will be generated only when a G:T intermediate is formed. (2) Isolation and sequencing of the M-DNA fragment for the frequency of 5-meC > T transitions (48 clones). Here, we reasoned that following deamination by AID, the thymine base of the mutagenic G:T intermediate is likely rapidly removed by Mbd4 glycosylase activity. To stabilize the putative G:T intermediate, and prevent rapid thymine removal, we coexpressed a catalytically inactive hMbd4 derivative (D560A) along with wild-type AID. Using our PCR strategy (Figure 3A), we clearly detected the diagnostic PCR product of the G:T intermediate (Figure 3B, lane 4) at all four initial HpaII sites (see Figure 1A), and on both DNA strands. In addition, sequencing of the recovered M-DNA fragment revealed a small number (2/48) of C > T transitions at the internal cytosine of the HpaII/MspI site, but no other mutations. Importantly, when wild-type AID and wild-type hMbd4 were coinjected neither assay yielded evidence for the proposed G:T intermediate (Figure 3B, lane 5). Furthermore, the catalytically inactive Mbd4 derivative (D560A) was expressed at levels equivalent to active hMbd4 (Figure S5D). Together, these results provide evidence that the G:T intermediate is created by AID deaminase activity and removed by Mbd4 glycosylase activity, and also suggest the possible physical coupling of the deaminase and the glycosylase to help ensure rapid thymine removal, addressed further below.
Figure 3. A PCR Strategy to Detect a G:T Intermediate.
(A) Schematic of the PCR reaction for thymine (CmeCGG > CTGG) detection at M-DNA HpaII/MspI sites using an A-tailed primer (only 3 of the ~22 bases shown) with an adenosine at the 3′ end.
(B) Detection of a G:T mismatch on M-DNA by PCR. M-DNA, AID mRNA, and RNA encoding either wild-type or catalytically inactive hMbd4 (D560A) was injected at the single-cell stage and assessed at 13 hpf.
Gadd45 Proteins Promote DNA Demethylation in Zebrafish Embryos
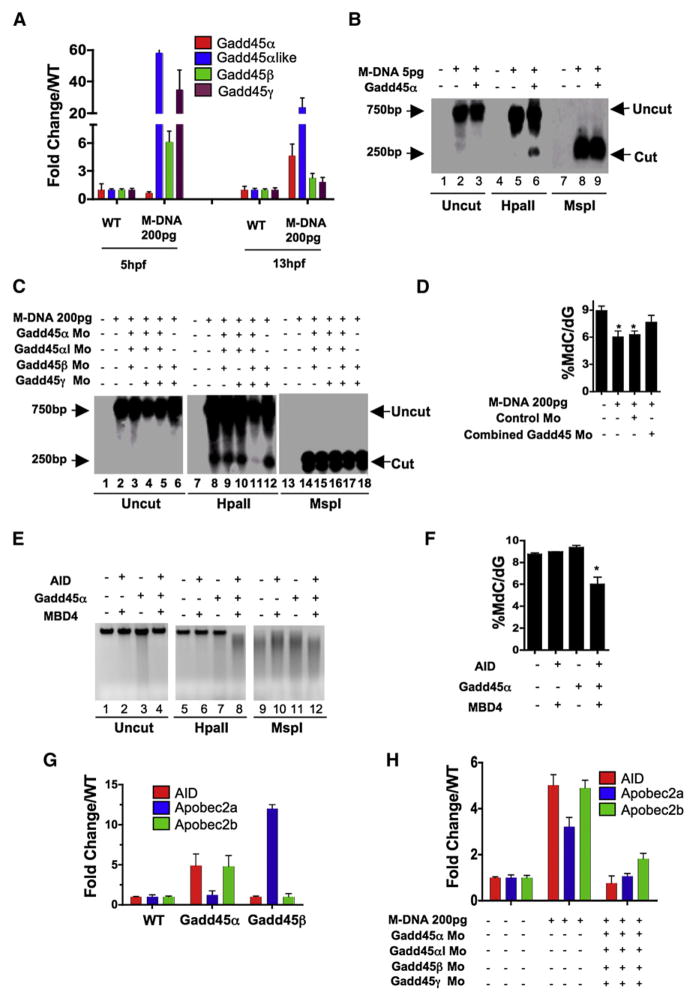
We next addressed the regulation of the DNA demethylation activity and the coupling of deamination and thymine base excision by a glycosylase. Above, we showed that M-DNA demethylation occurs in a window of development and requires a threshold of DNA, raising the possibility that DNA damage signaling pathways activate this demethylation system. Notably, we found that Gadd45α (and Gadd45α-like), a gene activated by DNA damage (Hollander and Fornace, 2002) and implicated previously in DNA demethylation (Barreto et al., 2007), was upregulated prior to and during the demethylation window elicited by M-DNA injection (200 pg, which is sufficient to elicit demethylation; Figure 4A). However, zebrafish contain six Gadd45-family members (α/β/γ and α/β/γ-like, all highly similar to their human counterparts) making a comprehensive analysis of the entire family untenable. Therefore, we focused on the family member best associated with DNA damage signaling, Gadd45α, and extended our work to other family members when appropriate. Zebrafish Gadd45α overexpression elicited moderate demethylation of the M-DNA fragment (injected at 5 pg, below the threshold level for eliciting demethylation on its own, Figure 4B), and of the bulk genome (causing a ~15% (or 9%) reduction by MS analysis: WT, 9.31 ± 0.17%; Gadd45α, 7.91 ± 0.2% (15% reduction); Gadd45αlike, 8.5 ± 0.27% (9% reduction)). Our results are consistent with the demethylation of the bulk genome by Gadd45α in human cells (Barreto et al., 2007), though of lower magnitude. Notably, knockdown of four of the six Gadd45 isoforms, but not separate knockdowns (morpholino efficacy, Figure S6), greatly attenuated M-DNA demethylation or the genome-wide demethylation caused by M-DNA injection at 200 pg (Figures 4C and 4D), suggesting the involvement of Gadd45 members, and their partially redundant roles in M-DNA demethylation.
Figure 4. Gadd45 Proteins Promote Demethylation and Selectively Upregulate Deaminases.
(A) Gadd45 family members are upregulated by M-DNA, assessed by RT-PCR.
(B) Gadd45α induces moderate demethylation of M-DNA as detected by HpaII digestion and subsequent Southern blotting. M-DNA is injected at 5 pg which does not induce demethylation on its own. Lanes 1, 4, and 7 correspond to wild-type sample.
(C and D) Lowering the levels of Gadd45 family members via morpholino injection attenuates de-methylation. Demethylation of 5-methylcytosine as assessed by HpaII digestion and Southern blotting (C), or LC-MS quantitation of 5-meC (D) in total genomic DNA isolated from 13 hpf old embryos injected at the single-cell stage with M-DNA alone (200 pgs) or along with morpholinos as shown. Lanes 1, 7 and 13 correspond to wild-type sample. Combined Gadd45 Mo refers to the combination of morpholinos to all four Gadd45 family members tested (α,α-like, β, and γ; 2 pg each).
(E and F) Synergy among AID, hMbd4, and Gadd45 for demethylation. Methylation status of total genomic DNA from embryos (13 hpf) was assessed by HpaII susceptibility (E) or by LC-MS quantitation of global 5-methylcytosine levels (F) injected at single-cell stage with mRNAs encoding factors as indicated. Lanes 1, 5, and 9 correspond to wild-type sample. Note: AID, MBD4, and Gadd45α were injected at subthreshold amounts (at 25 pgs each), levels which are not sufficient to induce demethylation alone.
(G and H) Quantitative RT-PCR for deaminase family members (assessed at 13 hpf) injected at single-cell stage with mRNA encoding Gadd45α or Gadd45β (F) and M-DNA (200 pg) alone or with morpholinos as shown (G). For each experiment, one representative of at least three biological repeats is shown except in LC-MS measurement where graph is prepared from values of two biological replicates.
Asterisks (*) depict statistical significance (p < 0.05). Error bars: are ± one standard deviation.
Synergy Among Gadd45α, AID, and Mbd4
Our data raised the possibility that Gadd45α might cooperate with AID/Apobec and Mbd4. As an initial test for cooperativity, we titrated AID, hMbd4 and Gadd45α injections to derive levels that individually (or as an AID/hMbd4 combination) would not cause demethylation. Interestingly, injection of all three at these subthreshold levels elicited DNA demethylation of M-DNA and also the bulk genome, assessed by both HpaII cutting (Figure 4E) and mass spectrometric analysis (Figure 4F).
Gadd45 Proteins Upregulate Specific AID/Apobec Proteins
We next tested whether Gadd45α overexpression would upregulate AID/Apobec expression, which would reflect cooperativity at the transcriptional level. Interestingly, overexpression of Gadd45α greatly enhanced AID and Apobec2b expression at 13 hpf (Figure 4G). In counter distinction, overexpression of Gadd45α strongly stimulated only Apobec2a expression (Figure 4G). These observations strongly suggest specific transcriptional relationships between Gadd45- and AID/Apobec-family members that may help coordinate the functional interactions among Gadd45 members and particular AID/Apobec enzymes, which remain to be further developed. Moreover we note that, the upregulation of all three AID/Apobec members at 13 hpf was greatly attenuated by knocking down four Gadd45 family members (Figure 4H). We then explored whether Gadd45 might further influence demethylation by promoting the coupling of deaminase and glycosylase function.
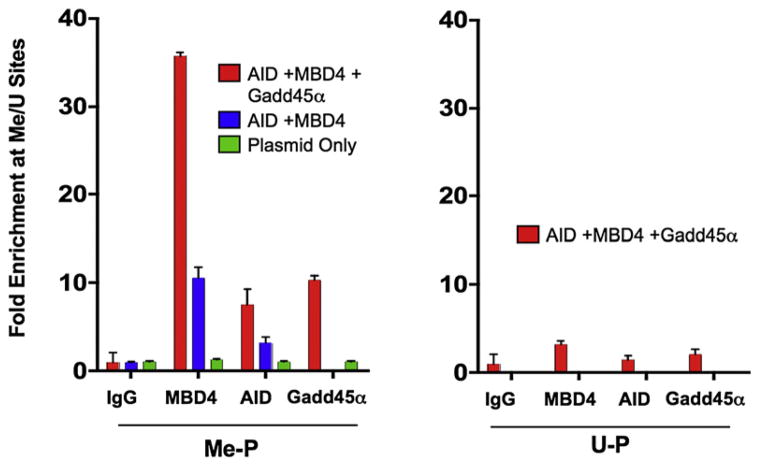
AID and MBD4 Occupy Methylated DNA Loci that Undergo Demethylation
We next tested whether AID and Mbd4 interact directly with a methylated DNA substrate in vivo, and whether this interaction was influenced by Gadd45α. Here, we utilized a plasmid that has a region dense with HpaII sites and regions lacking HpaII sites, and methylated the plasmid in vitro with HpaII methylase. This provides a methylated region, and an unmethylated region on the same plasmid which can be compared for factor occupancy. This plasmid was also used for in vivo demethylation in Figure S1. The plasmid was injected into single-cell embryos along with DNA constructs encoding epitope-tagged derivatives of AID or hMbd4, and DNA binding was tested by chromatin immunoprecipitation (ChIP) in zebrafish embryos at 12 hpf. In the absence of Gadd45, binding of hMbd4 and AID was clearly detectable on the methylated region of the methylated plasmid (Me-P) compared to the unmethylated region (Figure 5, blue bars). Interestingly, coexpression of Gadd45 enhanced the interaction of hMbd4 and AID proteins with the methylated region on Me-P (Figure 5 left panel, red bars), but had little affect on the unmethylated plasmid (U-P, Figure 5, right panel, red bars). This suggests that Gadd45 activity or abundance can affect AID or hMbd4 targeting.
Figure 5. Gadd45α Promotes Recruitment of AID and MBD4 to Methylated Regions of a Plasmid In Vivo.
Enrichment of AID, MBD4, and Gadd45α on pCMV-Luc, which contains both methylated (Me) and unmethylated (U) regions. ChIP experiments with extracts from embryos (12 hpf) injected at the single-cell stage with V5-tagged AID, HA-tagged hMbd4, His-tagged Gadd45α and in vitro-methylated (by HpaII methylase) pCMV-Luc (Me-P). Y–axis values represent the ratio of enrichment on a DNA segment containing in vitro methylated CmeCGG sites to enrichment on a site (also on pCMV-Luc) containing no CCGG elements. Me-P and U-P on axis depict methylated and unmethylated plasmid, respectively.
Graph shows one representative experiment of three biological repeats. Error bars: are ± one standard deviation.
Gadd45α Promotes Deaminase/Glycosylase Interactions in Human Cells
Next, we tested for physical interactions between AID or Apobec enzymes and Mbd4, and whether this was influenced by Gadd45α induction. Here, zebrafish embryos proved intractable for this assay due to low endogenous levels of AID/Apobec and Gadd45 and difficulties in deriving extracts from early embryos. Therefore, we utilized extracts derived from transfected human RKO cells to test for interactions by coimmunoprecipitation. Zebrafish AID or Apobec2a enzymes displayed a weak but detectable interaction with hMbd4, whereas a more robust interaction between hMbd4 and Apobec2b was detected (Figures S7A, S7C, and S7E). Interestingly, coexpression of Gadd45α moderately enhanced the interaction of AID and Apobec2a enzymes with Mbd4 (Figures S7B, S7D, and S7F). Furthermore, Gadd45 coprecipitated well with hMbd4, AID, Apobec2a, and Apobec2b (Figures S7G–S7J), raising the possibility that Gadd45 helps bridge the enzymes. Thus, in this heterologous system, physical interactions can occur among these proteins, though the modest IP efficiencies argue against a highly stable ternary complex.
AID, zMbd4, and Gadd45α Morphants Display Hypermethylation at neurod2
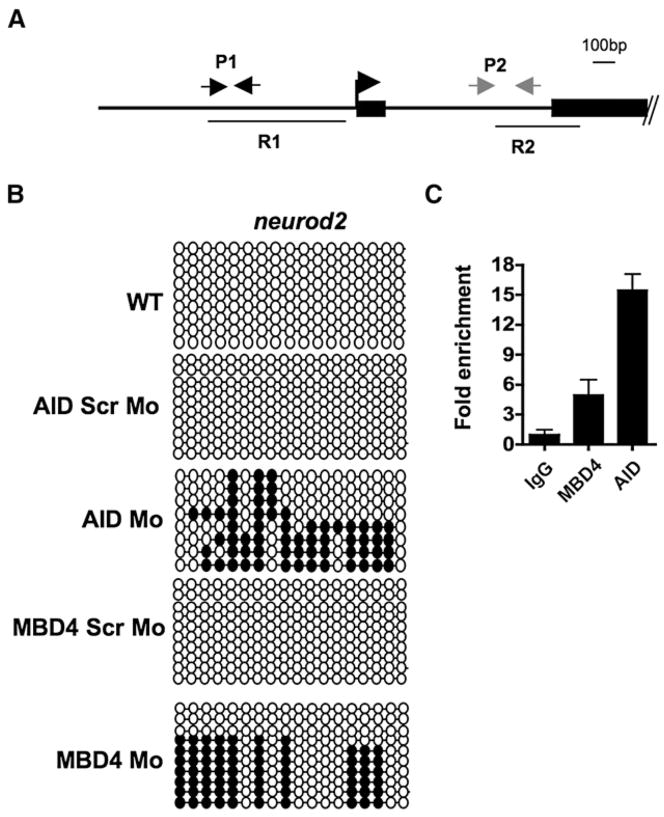
To begin to address whether AID/Apobec enzymes, Gadd45, or zebrafish Mbd4 have a role in the control of DNA methylation during normal zebrafish development, we knocked down (by morpholino injection) a subset of the enzyme family members and examined their impact on development. Interestingly, either AID, Gadd45α, or Mbd4 knockdown caused the loss of neurons at 24 hpf, shown by the absence of pro-neuronal markers such as neurogenin-1 or sox-2 (Figure S4 and Table SI). Specificity was demonstrated by rescue of neuronal markers by coinjecting (along with the morpholino) a spliced RNA refractory to the morpholino (Figure S4). As neurons form in mice lacking Mbd4 (or AID), zebrafish may rely more on these proteins for neurogenesis than do mice, or alternatively, neurogenesis in zebrafish may be more sensitive to misregulation of DNA methylation levels. We note that we have not evaluated the impact on other organ systems. We then tested for differences in methylation status at neurod2 (Figure 6A) and at sox2, two transcription factors involved in neurogenesis, in AID or zMbd4 morphants compared to control morphants at 80% epiboly; this is the latest time point in development where AID and MBD4 morphant embryos are indistinguishable from wild-type embryos (or control morphants), providing an appropriate examination point for methylation differences that might impact future phenotypes. Notably, we observed a pronounced increase in CpG methylation near the neurod2 transcription start site (Figure 6B), but not 1.6 kb downstream, nor at the sox2 promoter (data not shown). To address whether the demethylation near the neurod2 TSS was direct, we tested for physical association of tagged exogenous AID and hMbd4 at the TSS of neurod2 by ChIP, which revealed occupancy relative to the downstream control locus (Figure 6C). Although these results do not prove a direct demethylation of the neurod2 promoter by these enzymes during normal neurogenesis, they do show occupancy of a gene involved in the morphant phenotype (neurogenesis) by the candidate enzymes, and provide a correlation between the loss of demethylase candidates and an increase in neurod2 promoter methylation.
Figure 6. AID and hMbd4 Occupy the neurod2 Promoter and Affect the Methylation Status at 80% Epiboly.
(A) Schematic of the neurod2 promoter and start site region. R1 and R2 show regions of bisulfite sequencing (Results shown for only R1; R2 remains unmethylated and unaffected). P1 and P2 depict the amplicons used for ChIP determinations.
(B) Bisulphite sequencing of the R1 region of the neurod2 promoter in the WT (uninjected) animals or animals injected with morpholinos (all 2 pg) as shown. Scr Mo; a control morpholino where the base composition is maintained, but the order scrambled.
(C) Enrichment of AID and hMbd4 at neurod2 (P1 versus P2). ChIP experiments with extracts from embryos at 80% epiboly, which were initially injected at the single-cell stage with V5-tagged AID and HA-tagged hMbd4. Graph shows one representative biological experiment (two biological repeats), with the average of three technical replicates shown. Error bars are ± one standard deviation.
To reveal additional candidate gene targets for this demethylation system, we performed a pilot genomic Me-DIP experiment on AID morphants at 24 hpf. This dataset provided candidate affected and unaffected loci, a small portion of which were examined further for methylation differences by qPCR and bisulphite sequencing. We tested these candidates at 80% epiboly, the latest time where AID morphants and control morphants are indistinguishable. These approaches confirmed an additional transcription factor important for neuorgenesis (Sox1a), as well as additional transcription factors and housekeeping genes (Figures S8 and S9). Notably, loci initially identified as highly impacted by the AID morpholino were also highly affected by the Mbd4 morpholino, but not by a control morpholino. Taken together, we provide initial evidence for a demethylase system that functions at multiple genes during development.
DISCUSSION
Evidence is accumulating for an active DNA demethylation process in vertebrates, including studies on the activation of the interleukin-2 locus (Bruniquel and Schwartz, 2003), active demethylation following fertilization (Mayer et al., 2000; Oswald et al., 2000; Reik, 2007), and recent evidence for cyclical DNA demethylation at ERα targets (Kangaspeska et al., 2008; Metivier et al., 2008). However, candidate enzymes for active demethylation have remained elusive or controversial.
Our identification of demethylase candidates was enabled by a versatile zebrafish embryo assay system, where demethylation could be elicited and monitored, and in which candidates could be evaluated by overexpression or knockdown. Our work provides multiple lines of evidence that demethylation involves a coordinated system involving at least three factors: an AID/Apobec deaminase, an Mbd4-related G:T glycosylase, and a Gadd45 family member (Figure 7). First, the demethylation of an injected DNA fragment (or plasmid), along with 20%–40% of the 5meC nucleotides in the zebrafish genome, was temporally correlated with the upregulation of Gadd45 and AID/Apobec members (Figure 1). Importantly, morpholino knockdown studies showed a reliance on these factors for demethylation. Although zMbd4 was not upregulated by M-DNA injection, there are high levels of the RNA encoding Mbd4 in the early embryo (Figure S2) and Mbd4 protein might be activated following M-DNA injection in a posttranscriptional manner. Evidence that these enzymes conduct demethylation in the normal developing embryo was provided by examining the AAAmo (which lowers AID and eliminates Apobec members), which caused ~12% increase in bulk methylation levels, a result comparable to the observation in Arabidopsis mutants lacking all three major 5-meC glycosylases, which likewise show only a modest upregulation of bulk 5-meC levels (Lister et al., 2008; Penterman et al., 2007). Furthermore, we localized AID and hMbd4 to methylated regions on a plasmid (Figure 5), and also to a gene (neurod2) that is related to a phenotype (loss of neurons) observed in AID and Mbd4 morphants (Figure 6), supporting roles in vivo at genes. We expanded the list of gene targets by examining candidates revealed in a pilot genomic Me-DIP study in AID morphants, which revealed transcription factors and housekeeping genes that relied on AID, and on Mbd4, for their demethylated status (at 80% epiboly), though the full scope of this system at different times in development remains to be examined in future studies.
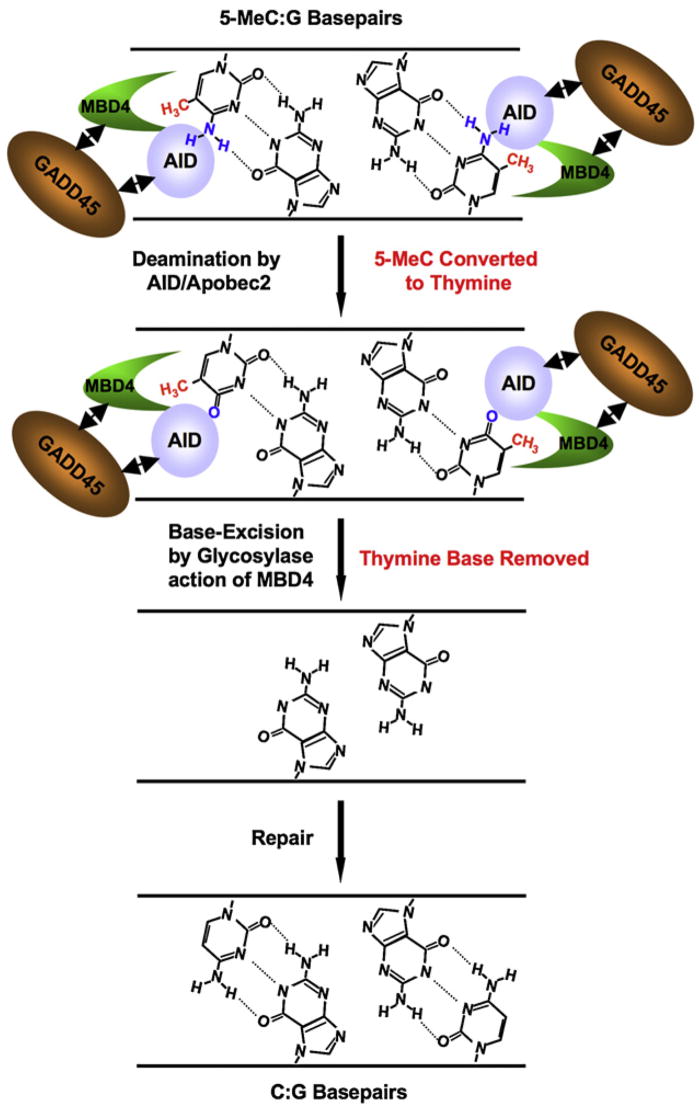
Figure 7. Model for 5-meC Demethylation.
Demethylation may occur through a two-step coupled enzymatic process, promoted by Gadd45. The first enzymatic step involves deamination of 5-meC by AID (amine group removed, in blue), generating a thymine product and a G:T mismatch. The second step involves thymine base removal by Mdb4, generating an abasic site. As the transient G:T intermediate is not detected in cells with active Mbd4, but is with catalytically inactive Mbd4, the thymine is likely rapidly removed, suggesting a coupling between deaminase and glycosylase activity. Gadd45 may promote functional or physical interactions between AID and Mbd4 at the site of demethylation. Mbd4 may couple with a lyase to help promote base replacement through base excision repair (neither shown nor addressed). Targeting of AID/Mbd4 may be promoted by recognition of the 5-meCpG (methyl group in red), or through other mechanisms (data not shown).
Importantly, overexpression of AID/Apobec along with hMbd4, but not either alone, caused significant demethylation of the bulk genome (Figure 2). The magnitude of this reduction (20%–40%) is pronounced, as studies in zebrafish and mice strongly suggest that perturbations beyond these levels are not compatible with viability (Ooi and Bestor, 2008; Rai et al., 2006). Notably, AID/Apobec expression alone did not prevent MspI cleavage, suggesting that AID/Apobec activity is promoted by Mbd4 and/or another cofactor (such as Gadd45) at 5-meC sites. Here, physical association of AID/Apobec with Mbd4 may help prevent the persistence of mutagenic G:T intermediates, as Mbd4 could rapidly remove the thymine (Figure S7). Indeed, our ability to detect a G:T intermediate (using a PCR priming strategy) only in hMbd4 catalytic mutants strongly supports the reaction mechanism proposed, while also underscoring the importance for proper regulation (Figure 3). Notably, coexpression of a catalytically inactive version of hMbd4 (along with AID) elicited a small number of 5-meC > T transitions in the injected M-DNA fragment, but not other mutations. By analogy, misregulation of demethylation might underlie the preponderance of C > T transitions in certain genes linked to cancer in mammals. We note that although mice lacking Mbd4 are viable, they show a higher frequency of mutations at CpG sites (Millar et al., 2002; Wong et al., 2002).
Two impressive recent studies clearly showed cyclical demethylation (~2 hr cycles of methylation and demethylation) at ERα targets (Kangaspeska et al., 2008; Metivier et al., 2008), and one study (Metivier et al., 2008) suggested a related stepwise deamination/glycosylase mechanism for demethylation that utilized different enzymes and deamination chemistry than our proposed mechanism. One counterintuitive aspect of their work was the proposed use of DNMT3 both to methylate cytosine and to deaminate 5-meC, relying on an inefficient deaminase activity (displayed in vitro) that will deaminate 5-meC only under conditions of low S-adenosylmethionine (SAM). Although plausible, it is unclear how SAM levels are held low in vivo, or how the low turnover number accounts for the demethylation kinetics. In counter distinction, the mechanism proposed here (Figure 7) employs a dedicated deaminase family (AID/Apobec).
New roles are suggested for Gadd45 family members in regulating DNA demethylation. First, as observed in human cells, Gadd45α (and α-like) overexpression promotes moderate DNA demethylation (Figure 4). Furthermore, Gadd45 family members appeared important in, but also redundant for, promoting DNA demethylation elicited by M-DNA injection. Notably, particular Gadd45 members upregulated the transcription of specific AID/Apobec enzymes, suggesting possible partnerships. Importantly, we observe physical interactions between Gadd45α and both AID/Apobec and Mbd4 (Figure S7), raising the possibility that Gadd45 might couple these two enzymes physically and/or activate them functionally, either alone or through known interactions with kinases downstream of Gadd45. Taken together, our results argue for a regulated demethylase system that involves enzyme and regulator families functioning in a coordinated manner.
Several interesting questions remain regarding the regulation of the demethylation system revealed here. First, it remains to be determined whether this demethylase system is utilized for the bulk demethylation of the paternal genome (and likely a portion of the oocyte genome) following fertilization. Second, it is not clear how these enzymes are targeted to particular loci in the genome, or the role of the MBD domain in Mbd4. Third, our studies suggest 5-meC as one possible substrate for Apobec2 class enzymes in vivo (Figure 2); thus far, in vitro approaches by others have not yielded substrates. Fourth, it remains curious why M-DNA (or methylated plasmid) induced demethylation is first observed ~8 hpf, peaks ~13 hpf, and then diminishes ~28 hpf (Figures 1 and S1). However, as we measure steady state levels of methylation, the remethylation of M-DNA around 28 hpf may reflect more robust DNA methylation activity rather than the true loss of demethylation activity. Fifth, it is of high interest to understand which repair pathways conduct cytosine replacement following AID and Mbd4 action. Furthermore, other glycosylases such as TDG should be examined for their ability to substitute for Mbd4 function, and for redundancy with Mbd4 in certain genetic assays; mouse Mbd4 knockouts are viable, but have accelerated rates of CpG mutations (Millar et al., 2002; Wong et al., 2002). Finally, we note that our studies do not diminish the possibility for other active mechanisms for 5-meC removal.
Taken together, we have utilized the developing zebrafish embryo as an assay system to uncover a regulated DNA demethylase system involving the coupled action of deaminase/glycosylase pairs promoted by Gadd45 family members. Further work will address more precisely when in development, and where in the genome, the demethylation system identified here is normally utilized, and how the misregulation of this system might contribute to cancer.
EXPERIMENTAL PROCEDURES
Genomic DNA Preparation, Restriction Digestion, and Southern Hybridization
Embryos were harvested at the designated time points and the genomic DNA was harvested using Puregene DNA isolation kit (QIAGEN/Gentra) according to the manufacturer’s instructions. Total genomic DNA was digested with HpaII (100 units) or Msp1 (100 units) for 16 hr at 37°C. Uncut, HpaII cut, and MspI cut DNA were then separated on a 1% agarose gel. For Southern blotting, the gel was first incubated in denaturing solution (0.4 M NaOH and 1M NaCl) twice for 15 mins, transferred to nylon membrane (Amersham), dried, crosslinked, and prehybridized in a buffer containing 6× SSC, 5× Denhardt’s solution, 0.5% SDS, and 100 mg/ml salmon sperm DNA. Hybridization was carried out in the same pre-hyb solution with a probe (M-DNA) prepared using Rediprime kit (Amersham). Following hybridization, the membrane was washed twice in buffer 1 (1× SSC, 0.1% SDS) and buffer 2 (0.5× SSC, 0.1% SDS), and exposed to a phosphoimager (Amersham).
Morpholino, Plasmid, and mRNA Injections
Wild-type zebrafish (Tuebingen strain) were maintained in a 14 hr:10 hr light:dark cycle in a Z-Mod at 28.5°C. Embryos were injected at single-cell stage and then grown in a 28.5°C incubator. Morpholinos were obtained from Gene-tools LLC Ltd. Morpholino sequences are provided in the Supplemental Data. Plasmid information for making mRNA and transfections is also provided in the Supplemental Data.
Chromatin Immunoprecipitation
After injections with plasmids as indicated, embryos were harvested at 12 hpf and crosslinked in 2.2% paraformaldehyde. Crosslinking was stopped using 0.125 M glycine, and after washing in PBS nuclei were isolated by breaking embryos in cell lysis buffer (10 mM Tris-Cl, [pH 8.0], 10mM NaCl, 0.5% NP-40, and protease inhibitors). Nuclei were then precipitated and broken in nuclei lysis buffer (50 mM Tris-Cl, [pH 8.0], 10 mM EDTA, 1% SDS, protease inhibitors, and phosphatase inhibitors). Extracts were then frozen at −80°C. Extract was then sonicated to produce DNA fragments between 300–600 bp. After sonication and dilution in IP dilution buffer (16.7 mM Tris-Cl, [pH 8.0], 167 mM NaCl, 1.2mM EDTA, 1.1% Triton X-100, and 0.01% SDS, protease inhibitors) extracts were precleared using sheep anti-rabbit dynabeads (Invitrogen) and then incubated with the respective antibodies overnight. Immunocomplexes were collected by sheep anti-rabbit dynabeads which were then washed twice each in dialysis buffer (50 mM Tris-Cl, [pH 8.0], 2 mM EDTA, 0.2% Sarkosyl) and wash buffer (100 mM Tris-Cl, [pH 9.0], 500mM LiCl, 1% NP-40, and 1% deoxycholic acid). Finally, DNA was eluted off the beads in elution buffer (50 mM NaHCO3 and 1% SDS) and eluate incubated in 0.3 M NaCl and 100 ng RNaseA at 50°C overnight. DNA was then purified using PCR purification kit (QIAGEN).
RT-PCR
Embryos were harvested at the designated time points and total RNA was isolated using Trizol reagent (Invitrogen) according to manufacturer’s instructions. Complimentary DNA library was prepared from 2ug of total RNA using Superscript III reverse transcriptase enzyme (Invitrogen) according to the manufacturer’s instructions.
LC-MS
LC-MS analysis of genomic 5mdC levels was performed as described previously (Song et al., 2005). Briefly, purified genomic DNA (500 ng) was denatured and hydrolysed through sequential digestion by S1 nuclease (Fermentas), venom phosphodiesterase I (Sigma), and alkaline phosphatase (Fermentas). A volume equivalent to 80 ng of the original DNA sample was then subjected to HPLC (Agilent; model G1322A) first with a guard column (providing background reduction) followed by an Atlantis DC18 silica column (Waters, # 186001301). MS determinations were performed using an Applied Biosystems MDS Sciex API 3000 triple quadrupole mass spectrometer coupled to the LC system through a TurboIonSproay ion source interface (Song et al., 2005).
Bisulfite Sequencing
Total genomic DNA was isolated from wild-type or injected embryos using Puregene DNA isolation kit (Gentra/QIAGEN). Genomic DNA (2 αg) was heat denatured in the presence of NaOH and bisulfite converted using 3 M sodium metabisulfite, (pH 5.0) (Sigma) and 0.5mM hydroquinone (Sigma) overnight. The reaction mixture was then desalted using a DNeasy spin column (QIAGEN) and desulphonated in 0.3 M NaOH. Finally, DNA was recovered by ethanol precipitation.
Statistical Analysis
P values were calculated using an unpaired t test.
Supplementary Material
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures, nine figures, and two tables and can be found with this article online at http://www.cell.com/supplemental/S0092-8674(08)01517-1.
Acknowledgments
We thank Jacqui Wittmeyer for generating antibodies for Apobec2a/b, Richard Dorsky for probes, Sung Wan Kim for plasmids, M. Ben Major for Gadd45 morpholino efficacy determinations, Bob Schackmann for oligos and Helaman Escobar for sequencing. We thank Brenda Bass, Don Ayer, and Barbara Graves for comments on the manuscript. Support has been received from: Huntsman Cancer Institute (supplies, genomics and support of K.R., I.H., and D.A.J.), the National Cancer Institute (for D.A.J, S.R.J., and A.R.K.), CA24014 and CA16056 (for core facilities), and the Howard Hughes Medical Institute (supplies, genomics, and support of B.R.C.).
References
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- Collas P. Modulation of plasmid DNA methylation and expression in zebrafish embryos. Nucleic Acids Res. 1998;26:4454–4461. doi: 10.1093/nar/26.19.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic Cytosine Methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Hollander MC, Fornace AJ., Jr Genomic instability, centrosome amplification, cell cycle checkpoints and Gadd45a. Oncogene. 2002;21:6228–6233. doi: 10.1038/sj.onc.1205774. [DOI] [PubMed] [Google Scholar]
- Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Mhanni AA, McGowan RA. Global changes in genomic methylation levels during early development of the zebrafish embryo. Dev Genes Evol. 2004;214:412–417. doi: 10.1007/s00427-004-0418-0. [DOI] [PubMed] [Google Scholar]
- Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet . 2005;14:R47–R58. doi: 10.1093/hmg/ddi114. Spec No 1. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Ozer A, Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat Chem Biol. 2007;3:144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, Cairns BR, Jones DA. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol. 2006;26:7077–7085. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Sedgwick B. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol. 2004;5:148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- Song L, James SR, Kazim L, Karpf AR. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77:504–510. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
- Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, Fazzari M, Jin B, Brown AM, Lipkin M, Edelmann W. Mbd4 inactivation increases Cright-arrowT transition mutations and promotes gastrointestinal tumor formation. Proc Natl Acad Sci USA. 2002;99:14937–14942. doi: 10.1073/pnas.232579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Angliker H, Schwarz S, Thiry S, Siegmann M, Jost JP. 5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence. Nucleic Acids Res. 2000;28:4157–4165. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures, nine figures, and two tables and can be found with this article online at http://www.cell.com/supplemental/S0092-8674(08)01517-1.