Abstract
The circadian clock regulates the daily rhythms in the physiology and behavior of many organisms, including mice and humans. These cyclical changes at molecular and macroscopic levels affect the organism's response to environmental stimuli such as light and food intake and the toxicity and efficacy of chemo- and radiotherapeutic agents. In this work, we investigated the circadian behavior of the nucleotide excision repair capacity in the mouse cerebrum to gain some insight into the optimal circadian time for favorable therapeutic response with minimal side effects in cancer treatment with chemotherapeutic drugs that produce bulky adducts in DNA. We find that nucleotide excision repair activity in the mouse cortex is highest in the afternoon/evening hours and is at its lowest in the night/early morning hours. The circadian oscillation of the repair capacity is caused at least in part by the circadian oscillation of the xeroderma pigmentosum A DNA damage recognition protein.
Keywords: cancer treatment, chronotherapy, circadian clock, xeroderma pigmentosum A
Circadian rhythm is the daily oscillation in the biochemical, behavioral, and physiological functions of organisms (1, 2). Circadian clock disruption by environmental factors and behavioral patterns has been implicated as a contributing factor in carcinogenesis (2–4). Similarly, a limited number of studies have indicated that the efficacy of chemo- and radiotherapy of cancer and the side effects of these treatments are markedly influenced by the circadian time of administration of these agents (4, 5). Although attempts have been made to put these findings to practice in preventive and clinical medicine, the empirical nature of the observations and the lack of mechanistic explanations for the findings have been serious obstacles to making use of these findings in cancer prevention and treatment.
Important progress in the past decade has provided a reasonably detailed model for the mammalian circadian clock. In mice and humans, the clock is present in essentially every cell and is generated by an autoregulatory transcription-translation feedback loop (TTFL) (1, 2): Clock and Bmal1 transcription factors bind to the E-box promoter elements of the Cryptochrome (Cry)1 and 2 and Period (Per)1 and 2 genes and activate their transcription. The Cry and Per proteins make oligomeric complexes that, after a time delay, inhibit the Clock·Bmal1 complex and hence the transcription of the Cry and Per genes. The time delay between the synthesis of Cry and Per and their action as repressors generates an oscillatory pattern of expression of Crys and Pers and other clock controlled genes that are regulated by Clock·Bmal1 but are not part of the TTFL. It is this oscillation of gene expression that affects cellular and organismic function to give rise to the circadian rhythm at a macroscopic level. These peripheral circadian oscillations are synchronized with one another by the master circadian clock that is located above the optic chiasma, in the suprachiasmatic nuclei (SCN). The SCN coordinates the peripheral clocks in organs such as liver, kidney, and heart to give rise to the circadian rhythm at the organism level.
Because of such a pervasive influence of the circadian clock on mammalian physiology, it is expected that any response of the organism to internal and external agents that cause perturbation of tissue and organismic homeostasis would be gated by the clock. One such response is the cellular response to DNA-damaging agents. This response includes DNA repair, DNA damage checkpoints, and apoptosis (6). In this work we have investigated the effect of the circadian clock on nucleotide excision repair in mice. Excision repair is a multicomponent system that removes virtually all DNA base lesions, and it is the sole system in mice and humans for the repair of bulky lesions such as cyclobutane pyrimidine dimers, (6-4) photoproducts, and cisplatin-d(GpG), and cisplatin-d(GpXpG) intrastrand diadducts (7). Because of the significance of these adducts in UV-induced carcinogenesis and in the treatment of a variety of cancers by cisplatin, respectively, we decided to investigate the effect of the circadian clock on nucleotide excision repair in mice.
Results
Effect of Circadian Time on Excision Repair Activity in Mouse Cerebrum.
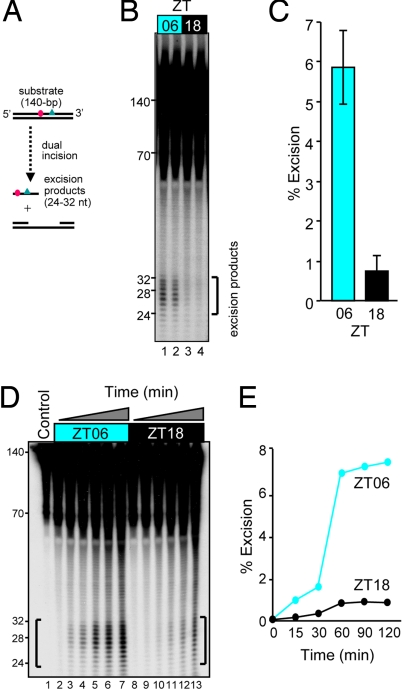
To determine whether the circadian time affects nucleotide excision repair activity in mice, we harvested several organs at ZT06 and ZT18 (ZT0 is time of light-on under a 12-h light:12-h dark lighting regimen) and tested them for nucleotide excision repair activity. We used the highly sensitive and specific “excision assay” (7, 8) to measure repair (Fig. 1A). We found that nonspecific nucleases in liver and kidney degraded the substrate and seriously interfered with the assay (data not shown). In contrast, cerebrum had an acceptable level of nonspecific nucleases, and the excision assays with cerebrum cell-free extracts yielded consistently good quality data that were amenable to quantitative analysis. We performed the excision assay with brains harvested at ZT06 and ZT18 (Fig. 1B). We found that the extracts made at ZT06 were ≈6-fold more active than those made at ZT18 (Fig. 1C). To ensure that the difference in the levels of excision observed represented real differences in the excision rates, we performed a kinetic assay with the two types of extracts. The results (Fig. 1, D and E) show that the ZT06 extract excises the (6-4) photoproduct at a 6- to 7-fold faster rate than the ZT18 extract, suggesting that nucleotide excision repair in the mouse brain may be regulated by the circadian clock.
Fig. 1.
Day and night nucleotide excision repair activity in the mouse brain. (A) Diagram of the excision assay used to measure repair capacity. A 140-bp duplex containing a centrally located (6-4) photoproduct (triangle) adjacent to a 32P label (circle) is incubated with CFE, which removes the damage by dual incisions in the form of 24- to 32-nt-long oligomers. (B) Excision activities in cerebral CFEs of two mice killed at ZT06 (1300) and two killed at ZT18 (0100). Each lane represents brain extract from one mouse. (C) Quantitative analysis of the excision activity as a function of two circadian (ZT) times. The averages and standard deviations were calculated from three independent excision assays for each mouse (n = 6 for both ZT06 and ZT18). (D) Kinetics of excision activity of CFE, from ZT06 and ZT18 brains. Kinetic assays were conducted with two of the brain extracts used in B. Ten femtomoles of substrates were loaded as a control (lane 1). (E) Quantitative analysis of data in D.
Circadian Oscillation of Excision Repair in the Mouse Cerebrum.
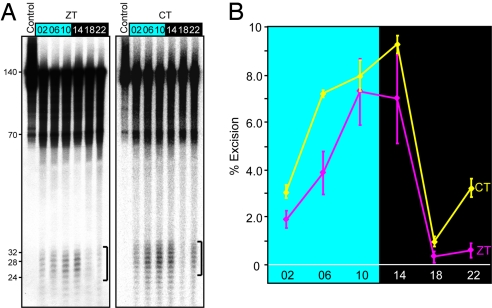
Next, we measured the excision repair activity in the cerebrum over the entire course of a circadian cycle to ascertain whether the excision activity exhibits a bona fide circadian rhythm and, if so, to locate the zenith and nadir of the activity. The results (Fig. 2A Left) reveal that excision activity does indeed show a circadian pattern with an activity maximum at ZT10–14 that is ≈5- to 10-fold higher than the minimum value at ZT18–22 (Fig. 2B). This oscillatory behavior, however, was obtained under conditions of 12-h light:12-h dark (LD) cycle and, although unlikely, it could have been a response at the tissue level to the LD cycles rather than to an intrinsic oscillatory regulatory mechanism. Hence, to test for a circadian rhythm in the conventional sense, the cerebral excision repair activity was measured in mice under constant conditions (in dark, DD) for the duration of the experiment, and the results are presented as a function of circadian time (CT, where CT0 = subjective dawn and CT12 = subjective dusk). The results (Fig. 2A Right), in agreement with those obtained under LD cycles, show a circadian pattern of oscillation of the excision repair activity. Therefore, we conclude that excision repair activity in mice brain, and quite likely in other organs, oscillates with a circadian periodicity.
Fig. 2.
Circadian oscillation of nucleotide excision repair activity in the mouse brain. (A) Excision assays with mice cerebrum extracts prepared from brains harvested at the indicated ZT and CT times. Ten femtomoles of substrates were loaded as a control. (B) Quantitative analysis of the excision activity as a function of ZT or CT. For ZT, each data point represents the average of excision activity from the cerebrums of three mice, and the bars represent SE. For CT, the data points represent the average of duplicate experiments conducted with extracts from a single mouse for each data point. Bars indicate SD.
Effect of the Circadian Clock on Excision Protein Levels.
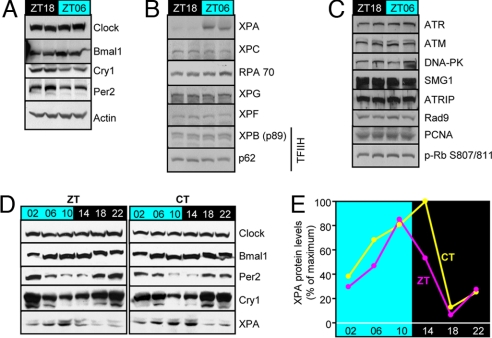
To find out how the circadian clock controls the rhythmic activity of nucleotide excision repair, we determined the levels of the core clock proteins (Fig. 3A), core excision repair proteins (Fig. 3B), and some of the key checkpoint and cell cycle proteins (Fig. 3C) that may indirectly affect repair activity at two circadian times. As expected, Clock does not exhibit circadian oscillation (9), Bmal1 is high at ZT06 and low at ZT18. Cry1 and Per2 are antiphase with Bmal1, again in agreement with previous reports and the known properties of these proteins (10). When the six core excision repair factors were analyzed, only xeroderma pigmentosum A (XPA), one of the three DNA damage recognition proteins, exhibited a circadian time-dependent change in expression with a markedly higher level at ZT06 compared with ZT18 (Fig. 3B). None of the DNA damage checkpoint proteins tested exhibited circadian variability between these two phases (Fig. 3C). Next, we analyzed the levels of Per2, Cry1, Bmal1, and XPA over the course of a circadian period at 4-h intervals: XPA levels are in phase with Bmal1 and antiphase with Cry1 and Per2 levels (Fig. 3D), consistent with Fig. 3A and with the notion that Xpa is a clock-controlled gene positively regulated by Clock·Bmal1 and negatively regulated by Cry and Per as also evidenced by the ≈3-fold elevated level of XPA protein in Cry1−/−Cry2−/− mouse fibroblasts relative to wild-type control (data not shown). Quantitative analysis of the data in Fig. 3D reveals a robust circadian oscillation for XPA with ≈10-fold difference between the zenith and nadir values (Fig. 3E). In support of our finding, a comprehensive analysis of clock-controlled genes by microarray profiling has revealed that Xpa expression in the mouse liver exhibits a high-amplitude circadian pattern (11) and moreover our immunoblot analysis revealed that oscillation of XPA in the liver is in phase with that in the brain (data not shown).
Fig. 3.
Effect of the circadian clock on day and night expression patterns of clock, excision repair, and checkpoint proteins. (A–C) Extracts from brains harvested at ZT18 (night) and ZT06 (day) were analyzed by immunoblotting. (A) Clock proteins. Clock shows no oscillation, whereas Cry1 and Per2 oscillate antiphase with Bmal1. (B) Nucleotide excision repair proteins. XPA is highly expressed during the day and down-regulated at night. The other excision repair proteins do not exhibit a measurable variation between ZT18 and ZT06. (C) Checkpoint proteins. None of the checkpoint/cell cycle proteins tested exhibits a measurable difference between day (ZT06) and night (ZT18) values. (D) Circadian oscillation of XPA. Brains were harvested from mice under either LD or DD conditions, and the levels of XPA over a circadian period were analyzed by immunoblotting along with the four core clock proteins as a reference. Note the zenith values for XPA at afternoon/evening hours under both ZT and CT conditions are in phase with Bmal1 and antiphase with Cry1 and Per2 as expected for a Clock·Bmal1-controlled protein. Clock that does not oscillate is used as a loading control for a nonoscillating protein. (E) Quantitative analysis of XPA oscillation. Each data point represents XPA value from the cerebrum of one mouse, and the values are expressed relative to that of XPA at CT14, which was the highest.
Restoration of Excision Activity by Supplementing the Circadian Nadir Extract with Recombinant XPA.
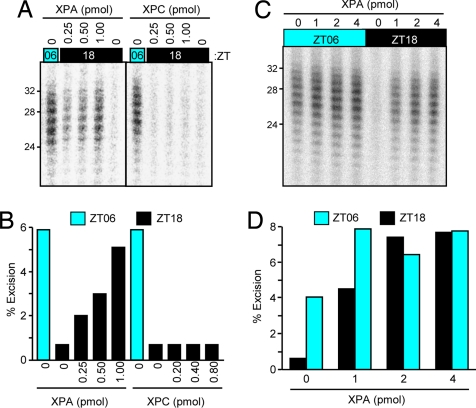
To determine whether the reduced excision repair in the ZT18 brain extracts was indeed caused by the low level of XPA at this circadian time, we supplemented the extract with recombinant XPA protein and tested for excision activity. As seen in Fig. 4 A and B, addition of XPA restored the excision activity to nearly ZT06 levels, indicating that the main cause of circadian oscillation of excision repair activity in mouse cerebrum is the circadian oscillation of the XPA protein level. In contrast to XPA, addition of another key damage recognition protein, XPC, to the ZT06 extract had no measurable effect on the excision repair activity (Fig. 4 A and B). To ascertain that the restoration of excision repair capacity to the ZT18 extract by addition of XPA was caused by the restoration of the low level of XPA protein in the extract from this circadian time and not to overall stimulation of excision by XPA added to all extracts, we supplemented both the ZT06 and ZT18 extracts with increasing concentration of XPA and measured excision activity. Fig. 4 C and D shows that although XPA has a minor stimulatory effect on ZT06 extract, its effect on the ZT18 extract is such that extracts from the two circadian times supplemented with 2–4 pmol of XPA have indistinguishable excision activities, supporting the conclusion that the main cause of the oscillatory behavior of excision repair is the XPA protein level.
Fig. 4.
Complementation of excision repair-defective nighttime brain extract with XPA protein. (A) Specificity of XPA complementation. To the ZT06 brain extract XPA or XPC was added at the indicated amounts in the reaction mixtures of the excision assays. Only the part of the gels containing the excision products is shown. The assay with the ZT06 extract is included as positive control. (B) Quantification of data shown in A. (C) Comparison of the effect of XPA on night- and daytime cerebrum extracts excision activities. ZT06 and ZT18 extracts were supplemented with the indicated amounts of XPA, and the excision assays were carried out for 60 min. (D) Quantitative analysis of data in C.
Discussion
Exicision Repair and Brain Physiology.
Neurons rely almost exclusively on oxidative phosphorylation for energy metabolism, and as a consequence their genome is under constant threat of damage by reactive oxygen species that are generated as side products of oxidative phosphorylation. Although many DNA lesions caused by oxidative damage are repaired by base excision repair, it has been demonstrated that nucleotide excision repair removes the two major oxidative DNA lesions, 8-oxoguanine and thymine glycol, as efficiently as the cyclobutane thymine dimer, which is considered the classic substrate for this repair system (12). Hence, it is likely that nucleotide excision repair plays an important role in maintaining neuronal integrity and function under physiological conditions. In addition to its physiological role, nucleotide excision repair plays an important role in the cellular response to xenobiotic and chemotherapeutic agents, and therefore its regulation, circadian and otherwise, must be taken into account in preventive and therapeutic approaches to cancer management.
Exicision Repair and Chronotherapy.
In this work we find that excision repair activity in the mouse brain exhibits circadian oscillation with zenith at ZT10–14 and nadir at ZT18–22. Interestingly, a previous study on the effect of circadian clock on genotoxicity of cyclophosphamide in mice also found that the animals exhibited maximum resistance at ZT10–14 and maximum sensitivity to the lethal effects of the drug at ZT18–22 (13). Although the base monoadducts caused by cyclophosphamide and similar alkylating agents are substrates for nucleotide excision repair (14), it is not known whether excision repair is the main repair mechanism for these adducts. In addition, this class of drugs makes DNA interstrand cross-links that are presumed to be major contributors to the cytotoxic and chemotherapeutic effects of these drugs. However, in mammals excision repair is not the main pathway for cross-link repair (15), casting further doubt about a causal relationship between the circadian oscillations of excision repair and the sensitivity to cyclophosphamide. Clearly, further work is required to determine whether there is a causal relationship or a simple coincidence between the circadian times of maximum excision repair activity and maximum resistance to the lethal effects of cyclophosphamide that was ascribed to the depletion of B cells in the bone marrow and peripheral blood (13).
Nevertheless, the findings presented in this report provide a plausible molecular explanation for the empirical observation that circadian time of delivery of chemotherapeutic drugs such as cisplatin, whose major DNA lesions are cisplatin-d(GpG) and cisplatin-d(GpXpG) diadducts (6, 7), may be a significant contributing factor to the efficacy of the drug and the severity of its side effects (4, 5). The potential practical applications of our findings are 2-fold. First, if the circadian behavior of the tumor can be determined, the drug would have maximum efficacy when delivered at a time corresponding to the CT18–22 of cerebrum circadian time (16). Second, a significant fraction of cancer patients who undergo chemotherapy report cognitive impairment after the treatment (“postchemotherapy cognitive impairment syndrome” or “chemobrain”) (17). Although the cause and even the existence of such a syndrome is a matter of some debate, it is conceivable that the syndrome, at least in part, is caused by DNA damage to neurons that impair cognitive functions. The extent of the damage caused by certain drugs such as cisplatin may be minimized by administering the drug at the time of maximum excision repair activity in the brain (ZT10–14), provided that this administration scheme does not reduce the efficacy of the drug on the tumor cells, which may or may not be in phase with the circadian clock in the brain. It is hoped that a systematic approach that incorporates the circadian clock as a parameter in designing chemotherapeutic regimens would enhance the therapeutic index of cancer chemotherapy by cisplatin and other drugs that produce base damage repairable by nucleotide excision repair.
Excision Repair and Preventive Medicine.
Finally, it must be noted that even though in this article we have analyzed only the circadian oscillation of excision repair in the brain, we have found that the XPA protein, which appears to be responsible for the oscillatory behavior of the repair activity, oscillates in the liver with amplitude and phase quite similar to those of the brain (data not shown). Hence, we believe that the results obtained with the mouse cerebrum would be applicable to most other tissues as well. Therefore, we suggest that our findings must be taken into account to minimize the harmful effects of DNA damage by recreational activities such as sunbathing and occupational activities such as working with genotoxic chemicals. Further work on ascertaining that human excision repair exhibits circadian oscillation similar to that in mice and determination of the phases of maximum and minimum repair capacities are necessary steps to expand our findings to clinical applications and taking preventive measures to reduce cancer incidence by solar UV, which is the major environmental carcinogen.
Materials and Methods
Harvesting Mice Cerebrums.
C57BL/6J male mice (8–10 weeks old) were obtained from the Jackson Laboratory and were maintained on a LD 12:12 schedule for at least for 2 weeks before killing. For the DD experiments, mice that were maintained under LD were kept under constant darkness for an additional day. ZT0 is the time of light-on (0700), and ZT12 is the time of light-off (1900). For animals under DD, 0700 was taken to be CT0 (35 h in DD), and 1900 was considered CT12 (47 h in DD) even though because of the 23.7-h period of mice these values are off of actual CT times by ≈1%. The mice were handled in accordance with the guidelines of National Institutes of Health and the University of North Carolina School of Medicine. At the indicated times the mice were killed by carbon dioxide exposure, and their brains were removed and washed extensively with cold PBS. The cerebrum was separated from the cerebellum, and the spinal cord and was flash frozen in dry ice/ethanol and stored at −80 °C until use. All surgical procedures were carried out under red light.
Preparation of Cell-Free Extract (CFE) from Cerebrum.
Frozen mouse cerebrum was pulverized by using porcelain mortar and pestle under liquid nitrogen. CFE was prepared as described in ref. 18 with some modifications. Briefly, the homogenized tissue was resuspended in 4 packed cell volume (PCV) (1 mL for one cerebrum homogenate) of hypotonic lysis buffer [10 mM Tris·HCl (pH 8.0), 1 mM EDTA, 5 mM DTT, and protease inhibitor mixture (Roche)]. Cells were cracked by three cycles of freeze/thaw in liquid nitrogen and were then incubated on ice for 20 min. Four PCV of buffer containing 50 mM Tris·HCl (pH 8.0), 25% sucrose (wt/vol), 50% glycerol (vol/vol), 10 mM MgCl2, 2 mM DTT, and protease inhibitor mixture were added. Then, 1 PCV of saturated (NH4)2SO4 (pH 7.0) was added slowly over a period of 30 min at 4 °C with mechanical mixing. Insoluble material was removed by centrifugation at 30,000 × g at 4 °C for 1 h. To the supernatant an equal volume of 50% saturated (NH4)2SO4 (pH 7.0) was added and mixed mechanically for 30 min at 4 °C. The precipitated proteins were collected by centrifugation at 12,000 × g for 20 min at 4 °C and resuspended in 100 μL of excision buffer [25 mM Hepes (pH 7.9), 100 mM KCl, 12 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 12% glycerol (vol/vol), and protease inhibitor mixture] and transferred to slide-a-lyzer minidialysis units 10,000 MWCO (Pierce) and dialyzed for 16 h. After dialysis the insoluble material was removed by centrifugation, and the protein in the supernatant was further concentrated by using a centrifugal filter device with 10-kDa exclusion (Millipore) until the concentration was 20–30 mg/mL by Bradford assay.
Excision Repair Assay.
Nucleotide excision repair activity with the brain extracts was carried out with cell-free extracts supplemented with 35 nM replication protein A (RPA) for optimal activity as reported in ref. 7. Briefly, 10 fmol of 140-bp duplex with a (6-4) photoproduct in the center and 32P-label at the 5th phosphodiester bond 5′ to the damage was incubated with 70 μg of CFE protein in 25 μL of excision buffer at 30 °C for 1 h unless indicated otherwise. We used this substrate rather than cisplatin-d(GpXpG) diadduct because of its ready availability. The (6-4) photoproduct-containing oligomer was from the synthetic organic chemistry core (University of Texas Medical Branch). Purified recombinant XPA and XPC proteins (7) were added to the reaction mixtures at the indicated amounts in complementation experiments. Note that because only a fraction of recombinant XPA is active, the amount added to the ZT18 extract to reach activity comparable with that of ZT06 extract is more than the difference in the levels of XPA between the two extracts. Quantification of excision was performed by using ImageQuant 5.2 software (Molecular Dynamics).
Immunoblotting.
Conventional immunoblotting procedures were used to determine the levels of a select number of proteins involved in the circadian clock, nucleotide excision repair, and cell cycle. The following antibodies were obtained from commercial sources: Clock, actin, PCNA, ATR, RPA70, XPA, XPC, XPG, XPF, XPB, and the p62 subunit of TFIIH (Santa Cruz Biotechnology), Per2 (BD Transduction Laboratories), p-Rb S807/811 (Cell Signaling Technology), ATM (Novus Biologicals), DNA-PK (Neomarkers), SMG1 (Bethyl Laboratories), and ATRIP (Zymed). Bmal1 antibodies (19) were a kind gift from Choogon Lee (Florida State University), and Cry1 antibodies were made in our laboratory (20).
Acknowledgments.
We thank Drs. Tadayoshi Bessho (University of Nebraska) and John Hogenesch (University of Pennsylvania) for helpful comments. This work was supported by National Institutes of Health Grants GM31082 and GM32833.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 2481.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 3.Stevens RG. Circadian disruption and breast cancer: From melatonin to clock genes. Epidemiology. 2005;16:254–258. doi: 10.1097/01.ede.0000152525.21924.54. [DOI] [PubMed] [Google Scholar]
- 4.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi M, Wood PA, Hrushesky WJ. Circadian chemotherapy for gynecological and genitourinary cancers. Chronobiol Int. 2002;19:237–251. doi: 10.1081/cbi-120002600. [DOI] [PubMed] [Google Scholar]
- 6.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 7.Reardon JT, Sancar A. Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems. Methods Enzymol. 2006;408:189–213. doi: 10.1016/S0076-6879(06)08012-8. [DOI] [PubMed] [Google Scholar]
- 8.Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panda S, Hogenesch JB. It's all in the timing: Many clocks, many outputs. J Biol Rhythms. 2004;19:374–387. doi: 10.1177/0748730404269008. [DOI] [PubMed] [Google Scholar]
- 11.Hughes M, et al. High-resolution time course analysis of gene expression from pituitary. Cold Spring Harbor Symp Quant Biol. 2007;72:381–386. doi: 10.1101/sqb.2007.72.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: Possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbacheva VY, et al. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci USA. 2005;102:3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant DF, Bessho T, Reardon JT. Nucleotide excision repair of melphalan monoadducts. Cancer Res. 1998;58:5196–5200. [PubMed] [Google Scholar]
- 15.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 16.Kondratov RV, Antoch MP. Circadian proteins in the regulation of cell cycle and genotoxic stress responses. Trends Cell Biol. 2007;17:311–317. doi: 10.1016/j.tcb.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Hede K. Chemobrain is real but may need new name. J Natl Cancer Inst. 2008;100:162–163. 169. doi: 10.1093/jnci/djn007. [DOI] [PubMed] [Google Scholar]
- 18.Smeaton MB, Miller PS, Ketner G, Hanakahi LA. Small-scale extracts for the study of nucleotide excision repair and nonhomologous end joining. Nucleic Acids Res. 2007;35:e152. doi: 10.1093/nar/gkm974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 20.Partch CL, Shields KF, Thompson CL, Selby CP, Sancar A. Posttranslational regulation of the mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc Natl Acad Sci USA. 2006;103:10467–10472. doi: 10.1073/pnas.0604138103. [DOI] [PMC free article] [PubMed] [Google Scholar]