Abstract
Sorting of eukaryotic membrane and secretory proteins depends on recognition of ribosome-bound nascent chain signal sequences by the signal recognition particle (SRP). The current model suggests that the SRP cycle is initiated when a signal sequence emerges from the ribosomal tunnel and binds to SRP. Then elongation is slowed until the SRP-bound ribosome–nascent chain complex (RNC) is targeted to the SRP receptor in the endoplasmic reticulum (ER) membrane. The RNC is then transferred to the translocon, SRP is released, and translation resumes. Because RNCs do not target to the translocon efficiently if nascent chains become too long, the window for SRP to identify its substrates is short. We now show that a transmembrane signal–anchor sequence (SA) significantly enhances binding of SRP to RNCs even before the SA emerges from the ribosomal tunnel. In this mode, SRP does not contact the SA directly but is in close proximity to the portion of the nascent polypeptide that has already left the ribosomal tunnel. Early recruitment of SRP provides a mechanism to expand the window for substrate identification. We suggest that the dynamics of the SRP–ribosome interaction is affected not only by the direct binding of SRP to an exposed signal sequence but also by properties of the translating ribosome that are triggered from within the tunnel.
Keywords: chaperones, site-specific cross-linking, yeast, nascent polypeptide-associated complex, Ssb1/2
A prerequisite to maintaining the integrity of a eukaryotic cell is the efficient targeting of proteins to their correct subcellular localization and to the extracellular space. The targeting information for proteins initially sorted to the endoplasmic reticulum (ER) is contained in so-called signal sequences. These are segments of 15–50 aa comprising a central hydrophobic core, which is flanked by an N-terminal, positively charged and a C-terminal, hydrophilic region (1). For cotranslational delivery of signal sequence-containing proteins to the ER, signal recognition particle (SRP) is the essential targeting factor. In yeast and mammalian cells SRP is an oligomeric complex of 6 proteins and an RNA that directly interacts with both ribosomes and the signal sequences of ribosome-bound nascent chains. Recent structural and functional investigations have unraveled the mechanistic principles of SRP-dependent protein targeting [for recent reviews on structure and function of eukaryotic SRP (2–4)]. In eukaryotic cells SRP has 2 primary roles. The first is a transient translation arrest that is required to maximize the efficiency of protein translocation into the ER (5–7). The second is the targeting of ribosome–nascent chain complexes (RNCs) to the SRP receptor in the ER membrane. Subsequent transfer of the signal sequence to the translocon in the ER membrane is controlled by GTPases contained in SRP and SRP receptor (2–4, 8).
SRP binds more tightly to translating than to nontranslating ribosomes (9, 10). This effect can be detected even before nascent chains have emerged from the tunnel and is thus related to the general translational status of the ribosome rather than to direct interaction between SRP and the nascent chain. Previous studies have addressed the question of whether or not specific amino acid sequences of segments inside the tunnel can further the affinity of SRP for RNCs (9, 10). Because signal sequences would be prime candidates for such effects, this possibility was tested in the eukaryotic system by using RNCs carrying preprolactin, a secreted protein with a cleavable signal sequence. However, when nascent preprolactin was too short to exit the tunnel, the affinity of SRP for RNCs was not enhanced above a control (9). In the bacterial system, which contains a simpler version of SRP consisting of only 1 protein and an RNA molecule (2), the affinity of SRP for RNCs carrying nascent chains too short to exit the tunnel is also higher than to nontranslating ribosomes. However, no significant influence of the amino acid sequence of such short nascent chains was detected (10) (see also Discussion).
When a signal sequence has fully emerged from the tunnel, it interacts with the Srp54 subunit of ribosome-bound SRP (11, 12). Concomitantly the affinity of SRP for the RNC greatly increases and even harsh experimental conditions do not release SRP from the RNC (9, 13). In this high-affinity mode, SRP slows down translation to keep the nascent polypeptide competent for ER targeting (6, 7). Yet not every signal sequence targets its client protein to the ER via the cotranslational, SRP-dependent pathway. In yeast a significant number of proteins are targeted into the ER posttranslationally and independent of SRP (14–17). Proteins that reach the ER on a posttranslational route are often secreted proteins with N-terminal, cleavable signal sequences. Prime substrates for the SRP-dependent route are integral membrane proteins that are inherently aggregation prone in an aqueous environment and require cotranslational insertion into the membrane (18). Such integral membrane proteins in many cases contain a noncleavable transmembrane signal sequence, the so-called signal-anchor sequence (SA) (1, 16). SAs have a dual function, first providing the signal for SRP-dependent targeting and second serving as membrane anchors. One of the best-characterized proteins in terms of SRP-dependent targeting is yeast Dap2 (16, 19, 20). This type II membrane protein contains a short, but strongly hydrophobic, SA localized between amino acids 30 and 45 (21) (Fig. 1 A and B). Here, we have addressed when during translation eukaryotic SRP starts to discriminate between a cytosolic protein like Pgk1 and a membrane protein like Dap2. We find that SRP is able to recognize Dap2 as its substrate early during its biogenesis when the SA is still far inside the ribosomal tunnel. The results support a model in which strongly hydrophobic signal sequences, such as transmembrane domains, that are located inside the ribosomal tunnel can allosterically alter interactions of RNCs with downstream partners such as SRP.
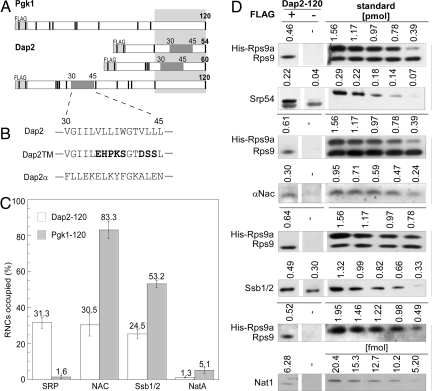
Fig. 1.
The exposed SA of Dap2 attracts SRP and disfavors binding of other ribosome-associated protein biogenesis factors to RNCs. (A) Schematic representation of nascent Pgk1 of 120 aa, and Dap2 of 54, 60, and 120 aa. The type II membrane protein Dap2 contains a SA localized between amino acids 30 and 45. The SA is indicated in dark gray; the portion of the nascent chains covered by the ribosomal tunnel is indicated in light gray. Nascent chains contained an N-terminal FLAG tag that was used for the purification of RNCs. The positions of lysines, important for chemical cross-linking, are marked as black bars. (B) SA of Dap2 and of the mutant versions, Dap2TM, and Dap2α. (C) Occupation of Dap2-120 or Pgk1–120 RNCs with RPBs in percent of RNCs. Error bars indicate the standard error of the mean (SEM). (D) Immunoblots for the quantification of Srp54, αNAC, Ssb1/2, and Nat1 on Dap2-120 RNCs exemplifies the data in C. RNCs carrying FLAG-tagged nascent Dap2 were isolated by native immunoprecipitation using αFLAG-coated beads. Aliquots of the material recovered and purified standard proteins were analyzed by immunoblotting. Signals obtained from the same exposure of a single gel are boxed. In case of the standard, numbers above the lanes give the amount of the respective purified protein loaded to the gel in picomoles or femtomoles, respectively. The standard protein His-Rps9a was supplemented with total yeast extract to ensure quantitative recovery during the procedure. Standard curves (Fig. S2) were used to determine the amount of RNCs and attached factors pulled down via the FLAG-tagged nascent chains. The background derived from an identically treated sample containing RNCs carrying the same, but nontagged, nascent chain was subtracted. The amount of His-Rps9a in the samples was divided by the factor 0.7 corresponding to Rps9a/b deviation from the mean value of ribosomal proteins previously determined (ref. 22 and Fig. S2).
Results
A Nascent Chain That Contains an Exposed SA Attracts SRP and Disfavors Binding of Other Ribosome-Associated Protein Biogenesis Factors.
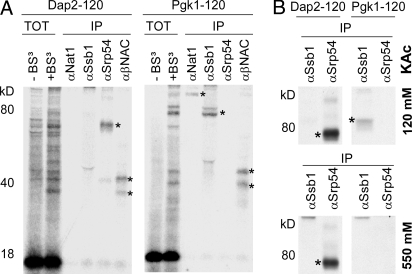
To quantify SRP recruitment to Dap2 RNCs with a fully exposed SA, a 120-aa nascent chain was generated (Fig. 1A). The N-terminal 120 aa of the cytosolic enzyme Pgk1, which does not possess a signal sequence, served as a control (Fig. 1A). Translation reactions were performed in a yeast translation extract, and subsequently, RNCs that carried a FLAG-tagged nascent chain were isolated [for details on the experimental setup, refer to supporting information (SI) Figs. S1–S5]. Approximately 30% of the Dap2-120 RNCs had SRP bound to them, whereas < 2% of the Pgk1-120 RNCs were isolated in complex with SRP (Fig. 1 C and D and Fig. S2). In contrast, the Hsp70 homolog Ssb1/2, nascent polypeptide-associated complex (NAC), and the Nat1 subunit of Nα-acetyltransferase NatA each bound substantially less well to Dap2-120 RNCs than to Pgk1-120 RNCs (Fig. 1 C and D). Like SRP, Ssb1/2, NAC, and NatA are ribosome-associated protein biogenesis factors (RPBs) that can interact with ribosomes as well as with nascent chains (22, 23). Parallel cross-linking experiments using a homobifunctional, amino-reactive cross-linker (BS3) revealed that nascent Dap2-120 was adjacent to Srp54, the signal sequence-binding subunit of SRP, and also NAC, whereas Pgk1-120 was close to Ssb1/2, NAC, and NatA, but not to SRP (Fig. 2A). SRP binding to Dap2-120 RNCs was highly salt resistant due to a direct interaction between SRP and the signal sequence (Fig. 2B; and see Fig. 4A) (13).
Fig. 2.
Interaction of RPBs with nascent chains. (A) 35S-labeled RNCs carrying untagged nascent chains were isolated by centrifugation through a sucrose cushion containing 120 mM potassium acetate (pH 7.4) and subsequently incubated either in the absence (TOT − BS3) or presence (TOT + BS3) of the homobifunctional cross-linker BS3. Aliquots containing 4 times as much material as the TOT + BS3 samples were subjected to immunoprecipitation (IP) under denaturing conditions with antibodies directed against Ssb1, Srp54, α/βNAC, or Nat1. Samples were run on Tris-Tricine gels and subsequently analyzed by autoradiography. (B) Interaction of SRP with RNCs after treatment with low salt or high salt. The experiment was performed as described in A with the exception that RNCs carrying FLAG-tagged nascent chains were isolated although sucrose cushions containing either 120 mM or 550 mM potassium acetate. Ssb1/2 served as a control because it no longer forms a cross-link after treatment of RNCs with 550 mM potassium acetate. Asterisks indicate the relevant cross-links.
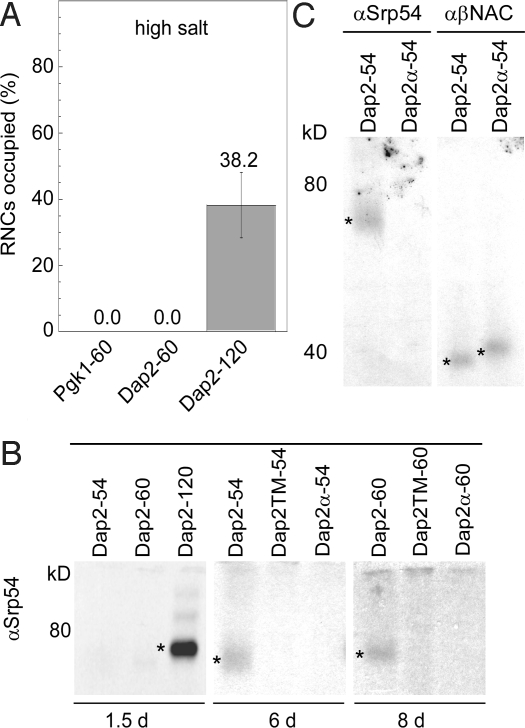
Fig. 4.
SRP is close to nascent chains with a Dap2 SA localized within the tunnel but requires SA exposure for salt-resistant binding. (A) SRP occupancy of RNCs carrying 60-residue Pgk1, or 60-, and 120-residue nascent Dap2 after isolation at high salt concentration (550 mM potassium acetate). Error bars indicate the SEM. Analysis and quantification was as in Fig. 1D. (B) Interaction of SRP with FLAG-tagged 54-, 60-, and 120-residue nascent Dap2, Dap2TM, or Dap2α. (C) Interaction of SRP and NAC with FLAG-tagged nascent Dap2-54 or Dap2α-54. Cross-link products are labeled with an asterisk. Cross-linking with BS3 and isolation of cross-link products was as in Fig. 2A.
Amino Acid Substitutions Within a SA Abolish the Interaction of a Nascent Chain with SRP and Promote the Interaction with the Ribosome-Bound Chaperone Ssb.
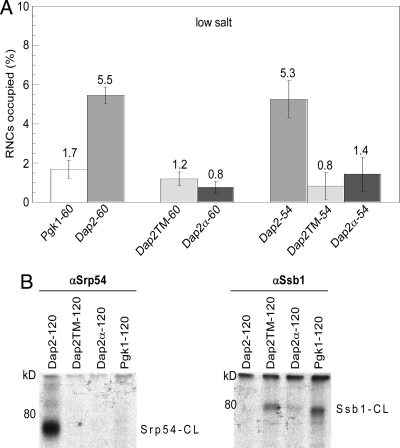
Next we generated RNCs with nascent Dap2 and Pgk1 polypeptides of 60 aa (Dap2-60 and Pgk1-60). At this length, the Dap2 SA is still fully inside the ribosomal tunnel (Fig. 1A). However, when compared to Pgk1-60, SRP was enriched by a factor of three on Dap2-60 RNCs (Fig. 3A). To exclude preferential SRP recruitment due to differences between the exposed, N-terminal portions of Pgk1 and Dap2 we designed 2 mutant versions of Dap2. The first mutant, designated Dap2TM, contained 8 amino acid exchanges within the SA that were intended to disrupt both its α-helical and hydrophobic character. In the second mutant, designated Dap2α, the complete 15-aa signal–anchor sequence was replaced by a 15-aa α-helical segment derived from Pgk1 (Fig. 1B). Cross-linking with BS3 confirmed that Dap2TM-120 and Dap2α-120 RNCs had lost the ability to directly interact with SRP (Fig. 3B). We also tested cross-linking of Dap2TM and Dap2α to the ribosome-bound chaperone Ssb1/2 that did not form cross-links to Dap2-120 (Fig. 2A), however, can form cross-links to a variety of nascent chains (22, 24, 25). Indeed, a cross-link between Ssb1/2 and Dap2TM-120 or Dap2α-120 was detected (Fig. 3B). It is interesting to recall that a significant amount of Ssb was bound also to ribosomes carrying Dap2-120 (Dap2-120 RNC occupation with Ssb: 24.5%, Fig. 1C). Thus, an exposed SA negatively affects the ability of Ssb to form a cross-link to an otherwise identical nascent chain (Fig. 3B). Disruptive mutations in the SA did not only abolish interaction of the nascent chain with SRP but promoted its interaction with Ssb1/2.
Fig. 3.
The SA of Dap2 within the ribosomal tunnel enhances SRP association with ribosomes. (A) SRP occupancy of RNCs carrying different lengths of nascent Dap2, Dap2TM, Dap2α, or Pgk1 at low salt concentration (120 mM potassium acetate). The number of amino acids in a nascent chain, exclusive of the FLAG-tag, follows the hyphen. Error bars indicate the SEM. Analysis and quantification was as in Fig. 1D and Fig. S2. (B) Dap2TM and Dap2α have lost the ability to interact with SRP and have gained the ability to interact with Ssb1. Cross-linking with BS3 was performed as described in Fig. 2A on RNCs carrying FLAG-tagged 120-residue nascent Dap2, Dap2TM, Dap2α, and Pgk1 after isolation through sucrose cushions containing 120 mM potassium acetate. Immunoprecipitations were performed with αSrp54 or αSsb1 as indicated. Srp54-CL: cross-link between the nascent chain and Srp54; Ssb1-CL: cross-link between the nascent chain and Ssb1/2.
A Nascent Chain's SA Can Attract SRP to RNCs Before the SA Arrives at the Exit of the Ribosomal Tunnel.
Dap2TM and Dap2α had lost the characteristics of a SRP substrate. We thus tested the recruitment of SRP to short nascent Dap2TM and Dap2α of 60 aa (Dap2TM-60 and Dap2α-60). These nascent chains differ from wild-type Dap2-60 exclusively in amino acids covered by the ribosomal tunnel (Fig. 1A). Nevertheless, Dap2α-60 and Dap2TM-60 recruited significantly less SRP than Dap2-60. Importantly, the amount of SRP bound to Dap2TM-60 and Dap2α-60 RNCs was similar to that bound to RNCs carrying Pgk1-60 (Fig. 3A). The data are consistent with a model in which the SA presence within the tunnel stimulated SRP recruitment. We then addressed the question whether the recruitment of SRP to RNCs carrying short nascent Dap2 was confined to a specific positioning of the signal–anchor within the tunnel. To that end, the SA was moved 6 aa toward the peptidyl transferase center by shortening the nascent chains. The fraction of Dap2-54 RNCs occupied by SRP (5.3%) was essentially the same as of Dap2-60 RNCs (5.5%) (Fig. 3A and Fig. S2). In contrast, Dap2TM-54 RNCs (0.8%) and Dap2α-54 RNCs (1.4%) recruited significantly less SRP (Fig. 3A and Fig. S2). These data show that the SA was able to stimulate SRP binding to an RNC from different locations within the ribosomal tunnel, albeit less efficiently than when the SA was exposed (Dap2-120 RNC occupation with SRP: 31.3%, Fig. 1C).
A Nascent Chain's SA Inside the Ribosomal Tunnel Can Induce a Low-Efficiency Cross-Link Between SRP and Exposed Portions of the Nascent Chain.
In the case of the bacterial system, it has been suggested that SRP may dip into the ribosomal tunnel (26). If this were true for yeast SRP, it might interact directly with the SA even though the SA has not arrived at the tunnel exit. In the case of such a scenario, we would expect the association of SRP with Dap2-60 RNCs to be salt resistant, because direct contact of SRP with a signal sequence induces salt resistance of RNC–SRP complexes (13). In addition, SRP should be in direct contact with the SA. To test these possibilities, we compared the salt resistance of SRP binding to Pgk1-60, Dap2-60, and Dap2-120 RNCs. After high-salt treatment, the amount of RNC-bound SRP was below the detection limit for both Pgk1-60 and Dap2-60 RNCs, whereas binding to Dap2-120 RNCs was entirely salt resistant (Figs. 2B and 4A). Consistently, cross-linking with BS3 revealed that SRP did not interact with Dap2-54 and Dap2-60 nascent chains in the same way as with Dap-120 nascent chains because the extent of cross-linking was much higher with the latter (Fig. 4B, 1.5 days exposure). However, upon longer exposure, SRP cross-linking could be observed even to Dap2-54 and Dap2-60 nascent chains in which the SA was positioned well inside the ribosomal tunnel. The interaction of SRP with nascent Dap2-54 and Dap2-60 was poorly efficient (Fig. 4B, 6- and 8-day exposures). However, the efficiency of the cross-link between nascent Dap2-54 and SRP resembled the efficiency of the cross-link between Dap2-54 and NAC (Fig. 4C). In contrast to SRP, NAC formed cross-links not only with wild-type Dap2-54 but also with Dap2α-54 (Fig. 4C). We conclude that nascent Dap2-54 and Dap2-60 contain exposed lysines amenable for cross-linking and that a cross-link to SRP was formed only when a SA was present inside the ribosomal tunnel.
SRP Does Not Directly Contact a SA That Is Still Inside the Ribosomal Tunnel.
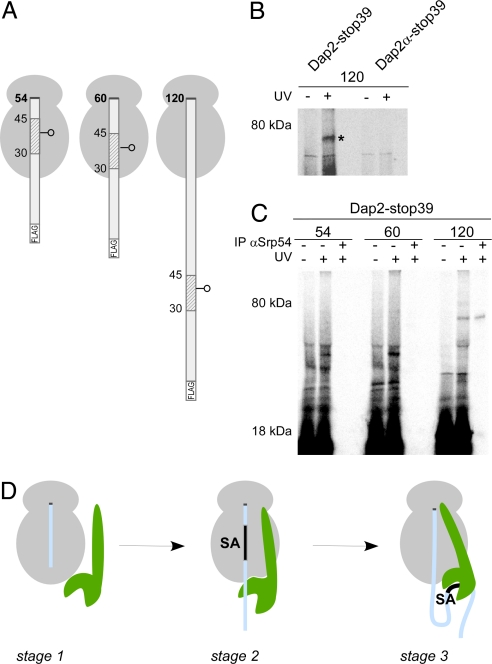
To determine whether SRP cross-linking to the short nascent chains occurred via lysines near the SA or rather via portions of the nascent chain that had already emerged from the tunnel, we used a photocross-linking approach. Using an amber suppressor tRNA, an uncharged photoreactive lysine analog was incorporated at position 39 of Dap2 in the middle of the SA to assess its proximity to SRP (Dap2-stop39). In parallel, the photoreactive residue was placed at position 39 of Dap2α (Dap2α-stop39) (Fig. 5A). When RNCs with 120-residue nascent chains were photolyzed, a prominent cross-link was generated in the case of Dap2-stop39, but not Dap2α-stop39 (Fig. 5B). The product of photocross-linking was efficiently immunoprecipitated with antibodies recognizing Srp54 (Fig. 5C). Thus SRP was in direct contact with a residue in the exposed SA of Dap2, but not with a residue at the same position of a mutant version of Dap2 that lacked the SA. When RNCs containing 54-, 60-, and 120-residue nascent chains of Dap2-stop39 were prepared and photolyzed, covalent photoadducts between SRP and the nascent chain were observed only after the SA had emerged from the tunnel (Fig. 5C). The absence of Srp54 photocross-linking to a SA in RNCs with short nascent chains shows that SRP is not adjacent to a SA in these complexes and disfavors the possibility that SRP dips into the ribosomal tunnel.
Fig. 5.
A Dap2 SA inside the ribosomal tunnel is not adjacent to SRP. (A) RNCs with 54-, 60-, and 120-residue Dap2 nascent chains with a single photo probe in the middle of the SA sequence are depicted. (B) 35S-methionine-labeled RNCs with εANB-Lys probes at position 39 of 120-residue length FLAG-Dap2 or FLAG-Dap2α were photolyzed (+UV). As a control, a parallel sample was not illuminated with UV light (−UV). RNCs were then isolated via the FLAG tag and analyzed by autoradiography. The cross-link between Dap2-120 and Srp54 is labeled with an asterisk. (C) 35S-methionine-labeled RNCs with εANB-Lys probes at position 39 of nascent FLAG-Dap2 were photolyzed (+UV) or kept in the dark (−UV) and were then isolated via the FLAG tag. A sample corresponding to 2 times the amount of the material shown in the +UV lane was subjected to immunoprecipitation under denaturing conditions using αSrp54 coupled to protein A Sepharose beads (IP αSrp54). (D) Three-stage model of SRP binding to RNCs. Stage 1: SRP (green) binds weakly to translating ribosomes (gray). This mode of interaction is independent of the length and type of nascent chain (light blue), but SRP affinity is higher than to nontranslating ribosomes. Stage 2: Upon synthesis of a SA, the interaction with the ribosome is strengthened and SRP localizes close to the exposed portion of the nascent polypeptide. The interaction is salt sensitive, suggesting that it is mainly electrostatic in nature. Stage 3: When the SA emerges from the tunnel, SRP binds to it with high affinity. The interaction is now salt resistant, thereby suggesting that hydrophobic interactions contribute significantly. In this stage, translation is arrested and the RNC–SRP complex is targeted to SRP receptor in the ER membrane. For details and references, see Discussion.
The combined data therefore indicate that SRP binding to ribosomes becomes highly stable and salt resistant when the SA emerges from the ribosomal tunnel and has unimpeded access to the SRP. The low amount of chemical cross-link to SRP detected when the SA was located within the tunnel presumably originates from positioning of SRP in the proximity of already exposed portions of the nascent chain. Importantly, no such cross-linking was observed with nascent chains that lacked a SA within the tunnel (Fig. 4B). Thus, a SA inside the tunnel can stimulate SRP binding to an RNC and position Srp54 close enough to any exposed nascent chain to cross-link to it.
Discussion
Research done during the last decade has revealed the complexity of the SRP cycle (2–4). In the early phase of the cycle, before SRP docks to SRP receptor, current models distinguish between 2 modes of interaction. SRP binds to RNCs independent of the nature of the nascent chain in a low-affinity, salt-sensitive mode (9, 13) (Fig. 5D, stage 1). SRP binds to an RNC tightly and in a salt-resistant manner upon contact with a signal sequence (13) (Fig. 5D, stage 3). Based on our data, we put forward a model that adds a distinct mode of SRP–RNC interaction to the cycle (Fig. 5D, stage 2). Stage 2 is induced from stage 1 when a SA (or possibly another strong cotranslational targeting sequence) enters the lumen of the ribosomal tunnel. This causes an increase in the affinity of the ribosome for SRP and positions it close to the exposed part of the nascent chain. When the SA finally exits the tunnel and is accessed directly by SRP, SRP binding switches to the high-affinity stage 3 (Fig. 5D).
This model provides a mechanism by which SRP can, early in biogenesis, select RNCs with substrates that are destined for cotranslational translocation. Such early recruitment of SRP might be beneficial for efficient targeting in a living cell. The large ribosomal subunit covers more than 30 aa in an extended conformation (27–29). Thus, full exposure of the Dap2 SA requires the synthesis of at least 75 aa (Fig. 1A). Segments of nascent chains inside the ribosomal tunnel have previously been shown to fold stably far inside the tunnel, probably by adopting α-helical conformation (29, 30). As the propensity of the Dap2 SA to form an α-helix is high, this may further increase the length of nascent chain required to fully expose the SA. In vitro, SRP binding to RNCs exposing a signal sequence declines when the nascent chain reaches a length of ≈110–140 aa (9, 11, 31). In vivo, membrane integration of Dap2, an 818-aa protein, is complete at a nascent chain length of ≈200 aa (20). Considering that in living yeast the elongation rate exceeds 9 aa per second (32), SRP is left with only a few seconds to recognize and interact with Dap2. Consistently, in vivo experiments performed in mammalian cells revealed that the window for SRP to recognize and target substrates to the cotranslational pathway is on the order of seconds (33).
Further complicating the process, proteins that enter the posttranslational route to the ER possess signal sequences that can interact with SRP during translation (16, 34). Because a yeast cell contains only 1–2 molecules of SRP per 100 ribosomes (22), early recognition of transmembrane proteins has to be promoted. It is attractive to speculate that in vivo a preference of SRP for its prime substrates is facilitated by recruitment to RNCs translating hydrophobic SAs even before these emerge at the tunnel exit. Although the mechanism is not understood, it may resemble ribosomal recognition of a nascent transmembrane domain that was found to affect the operational mode of the ribosome-bound translocon in the ER membrane (30, 35, 36). Consistent with such a model, it is the hydrophobicity of a signal sequence that determines whether or not a protein enters the co- or posttranslational pathway in vivo (16). Our observations differ from recent findings on the interaction of prokaryotic SRP with ribosomes. In prokaryotes, a SA inside the ribosomal tunnel does not enhance the affinity of SRP for ribosomes (10). Also, specific interactions between nascent transmembrane segments and components of the prokaryotic ribosomal tunnel have not been detected (26). Thus, the prokaryotic ribosome may not be affected allosterically by a SA or other transmembrane domain inside the exit tunnel. Alternatively, the function of bacterial SRP, which is much simpler and, importantly, also lacks the elongation-arrest domain of eukaryotic SRP, may not require to sense a signal sequence as early as its eukaryotic counterpart. Based on our data, we propose that the eukaryotic ribosome has evolved to directly recognize a hydrophobic SA, transmit the information to SRP, and hence influence the targeting route to the ER early on.
Materials and Methods
DNA Manipulations.
Plasmids encoding yeast Dap2 (dipeptidyl aminopeptidase B), or yeast Pgk1 (3-phosphoglycerate kinase) are described in ref. 22. Dap2TM and Dap2α mutant versions were generated by replacing a NheI/BstZ17-I fragment within Dap2 in pSPUTK-Dap2-E (2)K (22) and pSPUTK-FLAG-Dap2 (22) with PCR products containing the mutations resulting in the amino acid sequences as depicted in Fig. 1B. Plasmids are designated pSPUTK-FLAG-Dap2TM, pSPUTK-Dap2TM, pSPUTK-FLAG-Dap2α, and pSPUTK-Dap2α. N-terminally FLAG-tagged nascent polypeptides were used for quantitative pulldown experiments as well as for cross-linking experiments (Fig. S2). Chemical cross-linking to untagged and tagged nascent chains gave the identical set of cross-links (data not shown). For photocross-linking experiments, a TAG amber stop codon was substituted for a TGG coding for Trp at position 39 in Dap2 and a TTC coding for Phe in the corresponding position in Dap2α using a PCR fragment and the BsrGI and BstZ17I sites in pSPUTK-FLAG-Dap2 and pSPUTK-FLAG-Dap2α (Fig. 1B). The resulting plasmids are pSPUTK-FLAG-Dap2-stop39 and pSPUTK-FLAG-Dap2α-stop39.
In Vitro Transcription and Translation.
The following plasmids were used as a template to generate PCR products for SP6 polymerase based transcription: pSPUTK-Dap2-E (2)K, pSPUTK-FLAG-Dap2, pSP64-Pgk1-S (2)K, pSPUTK-FLAG-E-Pgk1; pSPUTK-FLAG-Dap2TM, pSPUTK-FLAG-Dap2α, pSPUTK-Dap2TM, pSPUTK-Dap2α, pSPUTK-FLAG-Dap2-stop39, and pSPUTK-FLAG-Dap2α-stop39. All transcripts lacked terminal stop codons. As a result translation products remained bound to ribosomes as peptidyl-tRNAs. Yeast translation extracts were prepared as previously described (37) from strains JK9-3dα or MH272-3fα. Translation reactions were performed in a yeast translation system in the presence of 35S-methionine (37). For quantitative pulldown experiments, radiolabeled amino acids were omitted during the translation reaction and subsequent quantification was via immunoblotting.
Chemical Cross-Linking.
Cross-linking was performed with the homobifunctional cross-linker bis-(sulfosuccinimidyl)-suberate (BS3, spacer length 1.14 nm; Pierce). RNCs were isolated via sedimentation through sucrose cushions as previously described (22). After sedimentation the ribosomal pellet was resuspended carefully in resuspension buffer [20 mM Hepes-KOH (pH 7.4), 2 mM magnesium acetate, 100 mM potassium acetate, 600 mM sorbitol, 2 mM DTT, 1 mM PMSF, 4 milliunits/μL RNase inhibitor]. A standard reaction of 80–20 μL was supplemented with BS3 to a final concentration of 500 μM and was incubated for 30 min on ice. The reaction was stopped by the addition of glycyl-glycine (pH 7.5) to a final concentration of 15 mM. Immunoprecipitations under denaturing conditions were performed as described using protein A-Sepharose (CL-4B; GE Healthcare) precoated with antibodies against Ssb1, Srp54, α/βNAC, and Nat1 (22).
Photocross-Linking.
A standard 80-μL translation reaction for photocross-linking contained 48 pmol of εANB-Lys-tRNAamb (Fig. S4). Translations were performed in the dark for 80 min at 20 °C with truncated FLAG-Dap2-stop39 or FLAG Dap2α-stop39 mRNAs encoding for nascent chains of 54, 60, or 120 aa plus the N-terminal 8 aa derived from the FLAG tag. Samples were photolyzed in an ice/water mixture for 10 min using a 500-W mercury arc lamp. For each reaction a control was kept in the dark. RNCs were then immunoprecipitated under denaturing conditions using protein A-Sepharose precoated with IgGs directed against Srp54. Alternatively, RNCs were isolated via the FLAG tag on the nascent chain on ANTI-FLAG M2 affinity gel (αFLAG-beads; Sigma) under native conditions. To that end, translation reactions were added to 20 μL of αFLAG-beads resuspended in 500 μL of buffer P1 [20 mM Hepes-KOH (pH 7.4), 150 mM potassium acetate, 2 mM magnesium acetate, 0.1% vol/vol Triton X-100, 1 mM PMSF, and a protease inhibitor mix yielding a final concentration of 1.25 μg/mL leupeptin, 0.75 μg/mL antipain, 0.25 μg/mL chymostatin, 0.25 μg/mL elastinal, and 5 μg/mL pepstatin A], and reactions were incubated for 2 h at 4 °C on a shaker. The αFLAG beads were separated from the supernatant by centrifugation and were washed twice with 500 μL of ice-cold buffer P2 [20 mM Hepes-KOH (pH 7.4), 150 mM potassium acetate, 2 mM magnesium acetate, 300 mM NaCl, 1 mM PMSF, and protease inhibitor mix as above]. Nascent chains and photoadducts were released from the αFLAG beads by incubation in SDS/PAGE sample buffer for 10 min at 95 °C. Samples were analyzed on Tris-Tricine gels followed by autoradiography.
Quantification and Statistical Analysis of RPBs on RNCs Carrying FLAG-Tagged Nascent Chains.
Quantitative pulldown experiments and purifications of standard proteins were carried out as previously described (22). For a detailed description of the procedure, refer to Figs. S2–S4. His6-Rps9a was used as a standard for the quantification of RNCs isolated via the FLAG-tagged nascent chain. FLAG-tagged nascent chains were detected using anti-mouse FLAG M2 antibody (Stratagene). A dilution series of relevant standard proteins was loaded on each gel used for quantification. Immunoblots were developed using ECL with horseradish peroxidase-conjugated goat anti-rabbit IgG (Pierce) as the secondary antibody. Quantifications of Western blots were performed using the AIDA ImageAnalyzer (Raytest). For statistical analysis 3 or more individual experiments for each RPB were examined. Error bars in the figures indicate the SEM.
Supplementary Material
Acknowledgments.
We thank Drs. K. Pfanner, A. Chacinska, Y. Dubaquié, and M. Gautschi for discussion and critical reading of the manuscript. This work was supported by SFB 746, Forschergruppe 967, and by the Excellence Initiative of the German Federal and State Governments (EXC 294) (to S.R).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808584106/DCSupplemental.
References
- 1.Martoglio B, Dobberstein B. Signal sequences: More than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 2.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 3.Shan SO, Walter P. Co-translational protein targeting by the signal recognition particle. FEBS Lett. 2005;579:921–926. doi: 10.1016/j.febslet.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 4.Halic M, Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr Opin Struct Biol. 2005;15:116–125. doi: 10.1016/j.sbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason N, Ciufo LF, Brown JD. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000;19:4164–4174. doi: 10.1093/emboj/19.15.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shan SO, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of the signal reception particle and its receptor drive initiation of protein translocation. J Cell Biol. 2007;178:611–620. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanagan JJ, et al. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J Biol Chem. 2003;278:18628–18637. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- 10.Bornemann T, Jöckel J, Rodnina MV, Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat Struct Mol Biol. 2008;15:494–499. doi: 10.1038/nsmb.1402. [DOI] [PubMed] [Google Scholar]
- 11.Kurzchalia TV, et al. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- 12.Krieg UC, Walter P, Johnson AE. Photocross-linking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc Natl Acad Sci USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powers T, Walter P. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr Biol. 1996;6:331–338. doi: 10.1016/s0960-9822(02)00484-0. [DOI] [PubMed] [Google Scholar]
- 14.Chirico WJ, Waters MG, Blobel G. 70 k heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- 15.Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- 16.Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 18.Kleizen B, van Vlijmen T, de Jonge HR, Braakman I. Folding of CFTR is predominantly cotranslational. Mol Cell. 2005;20:277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Halic M, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 20.Cheng Z, Gilmore R. Slow translocon gating causes cytosolic exposure of transmembrane and lumenal domains during membrane protein integration. Nat Struct Mol Biol. 2006;13:930–936. doi: 10.1038/nsmb1146. [DOI] [PubMed] [Google Scholar]
- 21.Roberts CJ, Pohlig G, Rothman JH, Stevens TH. Structure, biosynthesis, and localization of dipeptidyl aminopeptidase B, an integral membrane glycoprotein of the yeast vacuole. J Cell Biol. 1989;108:1363–1373. doi: 10.1083/jcb.108.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raue U, Oellerer S, Rospert S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J Biol Chem. 2007;282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- 23.Rospert S, Gautschi M, Rakwalska M, Raue U. In: Protein Folding Handbook. Buchner J, Kiefhaber T, editors. Vol. II. 1. Weinheim, Germany: Wiley-VCH; 2005. pp. 429–458. [Google Scholar]
- 24.Pfund C, et al. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome–nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautschi M, et al. The yeast Nα-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol Cell Biol. 2003;23:7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houben EN, Zarivach R, Oudega B, Luirink J. Early encounters of a nascent membrane protein: specificity and timing of contacts inside and outside the ribosome. J Cell Biol. 2005;170:27–35. doi: 10.1083/jcb.200503035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 28.Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005;44:8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- 30.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 31.Siegel V, Walter P. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J. 1988;7:1769–1775. doi: 10.1002/j.1460-2075.1988.tb03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonven B, Gullov K. Peptide chain elongation rate and ribosomal activity in Saccharomyces cerevisiae as a function of the growth rate. Mol Gen Genet. 1979;170:225–230. doi: 10.1007/BF00337800. [DOI] [PubMed] [Google Scholar]
- 33.Goder V, Crottet P, Spiess M. In vivo kinetics of protein targeting to the endoplasmic reticulum determined by site-specific phosphorylation. EMBO J. 2000;19:6704–6712. doi: 10.1093/emboj/19.24.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plath K, Rapoport TA. Spontaneous release of cytosolic proteins from posttranslational substrates before their transport into the endoplasmic reticulum. J Cell Biol. 2000;151:167–178. doi: 10.1083/jcb.151.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao S, Lin J, Do H, Johnson AE. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- 36.Daniel CJ, Conti B, Johnson AE, Skach WR. Control of translocation through the Sec61 translocon by nascent polypeptide structure within the ribosome. J Biol Chem. 2008;283:20864–20873. doi: 10.1074/jbc.M803517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia PD, Hansen W, Walter P. In vitro protein translocation across microsomal membranes of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:675–682. doi: 10.1016/0076-6879(91)94049-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.