Abstract
A class-C floral homeotic gene of Petunia, pMADS3, is specifically expressed in the stamen and carpels of developing flowers. We had previously reported the ect-pMADS3 phenomenon in which introduction of a part of the pMADS3 genomic sequence, including intron 2, induces ectopic expression of endogenous pMADS3. Unlike transcriptional or posttranscriptional gene silencing triggered by the introduction of homologous sequences, this observation is unique in that the gene expression is up-regulated. In this study, we demonstrated that the ect-pMADS3 phenomenon is due to transcriptional activation based on RNA-directed DNA methylation (RdDM) occurring in a particular CG in a putative cis-element in pMADS3 intron 2. The CG methylation was maintained over generations, along with pMADS3 ectopic expression, even in the absence of RNA triggers. These results demonstrate a previously undescribed transcriptional regulatory mechanism that could lead to the generation of a transcriptionally active epiallele, thereby contributing to plant evolution. Our results also reveal a putative negative cis-element for organ-specific transcriptional regulation of class-C floral homeotic genes, which could be difficult to identify by other approaches.
Keywords: epiallele, flower, MADS-box, petunia, RdDM
DNA methylation and histone modifications have been implicated in many biological processes including transcriptional gene silencing (TGS), genome imprinting, and paramutation in plants and animals. In many instances, RNA molecules play a crucial role as a trigger to induce a series of reactions leading to the modulation of gene expression mediated by DNA methylation and/or histone modifications. Small double-stranded RNAs (dsRNAs) of 21–24 bp trigger RNA-directed DNA methylation (RdDM) of homologous DNA sequences, leading to TGS in plants (1, 2). Induction of TGS by small RNAs has also been reported in human cells (3, 4). Involvement of dsRNAs due to RNA-dependent RNA polymerase (RdRP) and possibly small RNAs has been shown in paramutation of the b1 locus in maize (5, 6). These epigenetic regulations reported so far have been the down-regulation of transcription. Recently, however, 2 research groups have reported that the addition of small dsRNAs homologous to transcriptional regulatory sequences of certain endogenous genes up-regulates the transcription of target genes in human cultured cells (7, 8). This finding is striking because it has shown the existence of a previously undescribed type of small dsRNA-mediated regulation, a regulation opposite to that of TGS. However, only limited numbers of observations have been reported and their underlying mechanisms are still unclear.
We had previously reported that the introduction of a part of the genomic sequence of pMADS3, a class-C floral homeotic gene, induces ectopic up-regulation of endogenous pMADS3 in the flowering plant Petunia hybrida (9). This gene is a petunia ortholog of AGAMOUS (AG), an Arabidopsis class-C floral homeotic gene involved in the specification of stamens and carpels (10). AG contains a long second intron (intron 2), which contains a regulatory sequence that is sufficient for stamen- and carpel-specific expression (11–13). Several regulatory factors that positively or negatively control AG expression through interaction with intron 2 have been identified (12, 14–17). In our previous research, we introduced a chimeric gene, pMADS3:GUS, in which the genomic sequence of pMADS3 including its upstream and intragenic sequence up to intron 2 was fused with the coding sequence of the β-glucuronidase gene (GUS), into petunia lines. This transgene induced an unexpected homeotic conversion of floral organs due to disturbed expression of endogenous pMADS3. In some transgenic lines, pMADS3 expression was silenced, accompanied by conversion of stamens into carpels. In other lines, endogenous pMADS3 was ectopically up-regulated in petals, sepals, and leaves resulting in floral homeotic changes and curled leaves, which were unpredictable outcomes. In pMADS3:GUS plants, the 2 homeotic phenotypes—due to silencing and ectopic expression of endogenous pMADS3—interconverted somatically during development and were inherited independently of the transgenes. Aberrant RNAs, including sense and antisense sequences of pMADS3 intron 2, were detected in these transformants. On the basis of these findings, we speculated that an epigenetic regulatory mechanism, presumably involving trigger RNAs and DNA methylation, underlies this unique phenomenon (the ect-pMADS3 phenomenon).
In this study, we investigated the molecular mechanism underlying the ect-pMADS3 phenomenon. We showed that the ect-pMADS3 phenotype was reproduced by expressing inverted repeat (IR) sequences for subregions of pMADS3 intron 2 in transformant petunia lines. Then, we demonstrated that DNA methylation at specific CGs within the region targeted by IR in intron 2 is correlated with the ectopic up-regulation of pMADS3, leading to the conclusion that RdDM in particular regulatory sequences can induce transcriptional up-regulation. This acquired trait—due to transgene introduction—was inherited by the T1 generation independent of transgenes, suggesting a mechanism to generate a transcriptionally active epiallele. In addition, our findings fortuitously suggested a previously uncharacterized negative cis-element for spatiotemporal expression of class-C floral homeotic genes.
Results
Induction of pMADS3 Ectopic Expression by Expressing IR Sequences for pMADS3 Intron 2.
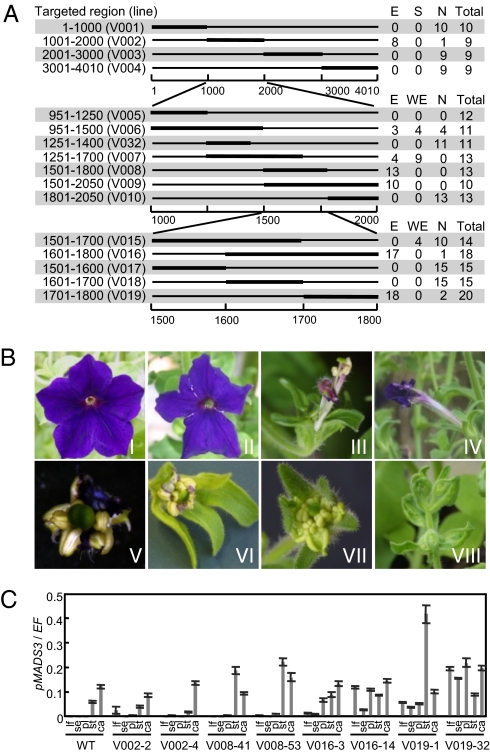
Our previous studies had showed that transgene-dependent expression of pMADS3 transcripts, including sense and antisense sequences of pMADS3 intron 2, was strongly correlated with the ect-pMADS3 phenotype in pMADS3:GUS transformant petunia lines. On the basis of this observation, we suggested the involvement of dsRNAs in this phenomenon (9), similar to the previous report of the silencing of chalcone synthase in petunia lines (18, 19). It has been shown that the expression of transcripts coding for IR sequences can result in the production of small dsRNAs through an RNA processing (RNAi) mechanism, and the small dsRNAs guide DNA methylation to homologous DNA sequences (i.e., RdDM), eventually leading to TGS (2). To test the hypothesis that an RdDM-like mechanism is involved in the ect-pMADS3 phenomenon, we expressed IR sequences for a subregion of pMADS3 intron 2 to see whether expressed RNAs induce the ect-pMADS3 phenotype. We first targeted 1-kb subregions of pMADS3 intron 2 (4 kb) by expressing corresponding IR sequences. Transformants of V002 lines expressing IR sequences corresponding to nucleotides (nts) 1001–2000 (with the first nucleotide of intron 2 as nt 1) frequently showed homeotic changes with staminoid structures developed at the margins of petal limbs and along the midveins (ect-pMADS3 flower phenotype) (Fig. 1 A and B). Quantitative PCR analyses showed that endogenous pMADS3 was expressed ectopically in petals in these plants (Fig. 1C). No homeotic change was observed in other lines expressing IR sequences corresponding to nts 1–1000, 2001–3000, and 3001–4010 (Fig. 1A). These results indicate that the 1001–2000 region includes sequences responsible for the ect-pMADS3 phenotype. Then, we targeted smaller subregions of the 1001–2000 region by IR sequences (lines V005–V010). Among the transformants, those of V008 lines, in which nts 1501–1800 were targeted, frequently showed the ect-pMADS3 phenotype (Fig. 1A). Further dissection of the 1501–1800 region showed that transformants expressing IR sequences that include the 1701–1800 (V008, V009, V016, and V019) region displayed strong ect-pMADS3 phenotypes at high frequencies (Fig. 1A). The ect-pMADS3 phenotype was not observed in transformants of V018 and V010 lines in which neighboring 1601–1700 and 1801–2050 regions, respectively, were targeted. Interestingly, the ect-pMADS3 phenotype became stronger with shorter target sequences. In V016 and V019 plants, in which the 1601–1800 and 1701–1800 regions were targeted, respectively, high levels of pMADS3 expression were also induced in sepals and leaves in addition to petals, accompanied by strong homeotic changes, including staminoid petals and curled leaves (Fig. 1 B and C). These results indicate that the 1701–1800 region in pMADS3 intron 2 is critical for the induction of pMADS3 ectopic expression.
Fig. 1.
Targeting of pMADS3 intron 2 subregions by inverted repeat (IR) sequences. (A) Diagrams for target regions of IR expression in pMADS3 intron 2. Scales represent nucleotide numbers with the first nucleotide of pMADS3 intron 2 as nt 1. To the right are the numbers of transformant lines showing ect-pMADS3 (E), weak ect-pMADS3 (WE), pMADS3 silencing (S), and normal (N) phenotypes (9). (B) Flower phenotypes of transformant plants expressing IRs. (Bi) A flower of wild-type petunia (V26); (Bii) a typical flower of V002 plants showing the ect-pMADS3 phenotype with antheroid sectors along midveins and reduced petal limbs; flowers of V016 (Biii) and V019 (Biv) plants showing ect-pMADS3 phenotypes with antheroid tissues on petal limbs; (Bv) a flower of V019 plants showing strong ect-pMADS3 phenotype with antheroid petals (some flowers showed strong phenotypes as shown but others in the same plants showed milder ones); (Bvi) a typical flower of V034 plants showing strong ect-pMADS3 phenotype with petal-to-anther conversion; (Bvii) extreme flower phenotype of 71–14 × M (T1) showing complete conversion of petals to anthers; and (Bviii) curled cauline leaves of the V019 line. (C) Levels of pMADS3 transcripts in leaves (lf), sepals (se), petals (pl), stamens (st), and carpels (ca) of transgenic lines. Expression levels of pMADS3 relative to those of the elongation factor gene (EF) were measured by RT-qPCR. Mean ± SE (n = 3).
DNA Methylation in pMADS3 Intron 2 Is Correlated with pMADS3 Ectopic Expression.
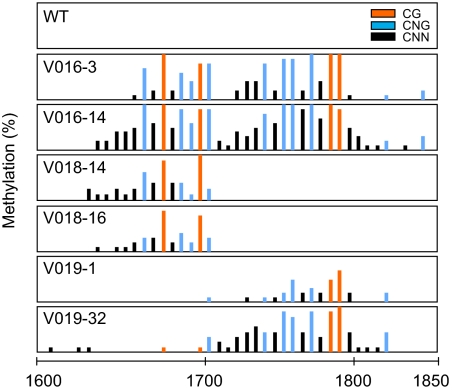
It is known that small dsRNAs resulting from expressed IR sequences can trigger RdDM to homologous DNA sequences in plants (2). We detected small dsRNAs including the sequences corresponding to expressed IR sequences in V019 plants showing the ect-pMADS3 phenotype (supporting information (SI) Fig. S1). Therefore, we analyzed the DNA methylation status in the 1701–1800 region with bisulfite sequencing by using genomic DNA from petals. In wild-type petunia plants, no cytosine methylation was detected; in contrast, cytosines in all of the CG, CNG, and CNN contexts were found to be methylated in IR-targeted sequences of the transformants (Fig. 2). These results indicate that the expressed IR sequences induced de novo RdDM. In general, cytosines in symmetric sites (CG and CNG) were more heavily methylated than those in asymmetric sites (CNN) (Fig. 2). In V016 and V019 plants showing strong ectopic expression of pMADS3, cytosines in the 1701–1800 region were heavily methylated. In contrast, in lines that did not show the ect-pMADS3 phenotype (e.g., V010 and V018), cytosine methylation was detected in IR-targeted sequences but not in the 1701–1800 region. Thus, DNA methylation at the 1701–1800 region of pMADS3 intron 2 is strongly correlated with pMADS3 ectopic expression.
Fig. 2.
DNA methylation patterns in pMADS3 intron 2 of inverted repeat (IR)-expressed transgenic plants. V016 and V019 plants show the ect-MADS3 phenotype, whereas V018 plants show normal phenotypes. At least 9 clones are sequenced for each bisulfite-treated sample. Bars represent proportions of methylated clones at each site; red, blue, and black bars represent CG, CNG, and CNN sites, respectively. The scale represents nucleotide numbers in pMADS3 intron 2 with the first nucleotide of intron 2 as nt 1.
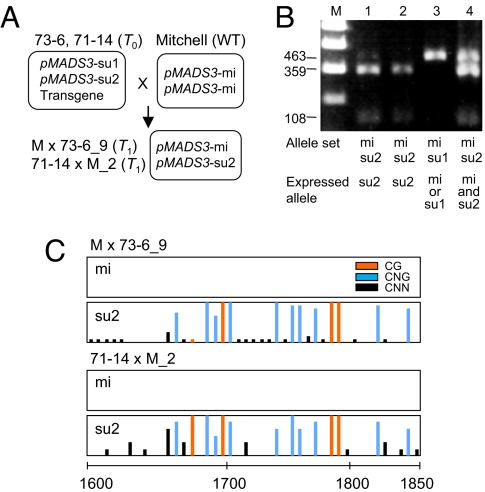
These results prompted us to examine whether DNA methylation in the same sequence region is also involved in the ect-pMADS3 phenotype of pMADS3:GUS lines, in which we had originally observed this phenotype. In some pMADS3:GUS lines, the ect-pMADS3 phenotype was maintained in the T1 generation even after the transgene had segregated away (9). The presence of transgenes hampers DNA methylation analysis because they are usually indistinguishable from their corresponding endogenous gene in DNA sequences. However, the lack of a pMADS3 transgene enabled DNA methylation analysis of these T1 plants. Moreover, pMADS3:GUS plants have a genetic background of a F1 hybrid (Surfinia); therefore, slight differences in the DNA sequences between 2 pMADS3 alleles (pMADS3-su1 and pMADS3-su2) allowed us to analyze the expression and DNA methylation patterns of the individual pMADS3 alleles (9). We crossed T0 transformants of 2 pMADS3:GUS lines (73–6 and 71–14), having pMADS3-su1 and pMADS3-su2 alleles in addition to the pMADS3:GUS transgene, with a pure variety of P. hybrida cv. Mitchell having a pMADS3-mi allele (Fig. 3A). From among the T1 offspring obtained, 2 plants (M × 73-6_9 and 71-14 × M_2) having pMADS3-su2 and pMADS3-mi alleles but not the transgenes were selected and analyzed for DNA methylation. Both plants inherited pMADS3-su2 and pMADS3-mi alleles from their parent pMADS3:GUS and Mitchell plants, respectively, but only the pMADS3-su2 allele was preferentially expressed in the petals of both plants (Fig. 3B). Bisulfite sequence analysis revealed high levels of CG and/or CNG methylation and negligible levels of CNN methylation in the 1701–1800 region of the pMADS3-su2 allele (Fig. 3C). In contrast, no cytosine methylation in any sequence context was detected in the pMADS3-mi allele (Fig. 3C). These results further support the correlation between the CG and/or CNG methylation in this region and the ect-pMADS3 phenotype, and also indicate that the effects of DNA methylation in pMADS3 intron 2 act only in cis because only the methylated allele was up-regulated. We also detected small dsRNAs corresponding to the 1701–1800 sequence in the T0 transformants of pMADS3:GUS lines (Fig. S1), indicating that basically common mechanisms are involved in the induction of ect-pMADS3 phenotypes of pMADS3:GUS and IR-expressed lines.
Fig. 3.
DNA methylation patterns in outcrossed pMADS3:GUS plants. (A) Inheritance of individual pMADS3 alleles in the T1 generation. Transgenes segregated out in both M × 73-6_9 and 71-14 × M_2 plants showing the ect-pMADS3 phenotype. (B) Allele-specific expression of endogenous pMADS3 in the petals of pMADS3:GUS T1 plants. The transformants analyzed were those without transgenes showing the ect-pMADS3 phenotype. The allele-specific expression was analyzed as described previously (9). M: marker; lane 1: 71-14 × M_2; lane 2: M × 73-6_9; lane 3: M × 73-6_7; lane 4: 71-14 × M_11. (C) DNA methylation patterns in pMADS3-mi and pMADS3-su2 alleles of M × 73-6_9 and 71-14 × M_2 plants. Bars represent proportions of methylated clones at each site; red, blue, and black bars represent CG, CNG, and CNN sites, respectively. The scale represents nucleotide numbers in pMADS3 intron 2 with the first nucleotide of intron 2 as nt 1.
Recent studies have highlighted the roles of chromatin remodeling via histone modification in transcriptional regulation. In both plants and mammals, DNA methylation is often associated with histone modification (H3K9me) (20, 21). In Arabidopsis, H3K27 trimethylation by CURLY LEAF (CLF), a polycomb-group protein, has been implicated in the transcriptional repression of AG (22). Therefore, we examined whether histone modification is involved in the ect-pMADS3 phenomenon in addition to DNA methylation. In the transformant line (V016) showing pMADS3 ectopic expression, H3K4me3 and H3K27me3 were detected in the 1601–1800 IR-targeted region but at similar levels to that in the wild type (WT) (Fig. S2). Acetyl-H3, H3K9me2, H3K27me1, and H3K36me2, which are often associated with transcriptional regulation, were not detected in either WT or V016 plants (Fig. S2). These results suggest that pMADS3 ectopic expression is a direct consequence of DNA methylation rather than that mediated by histone modification.
Candidate cis-Element Responsible for RdDM-Induced Up-Regulation of pMADS3.
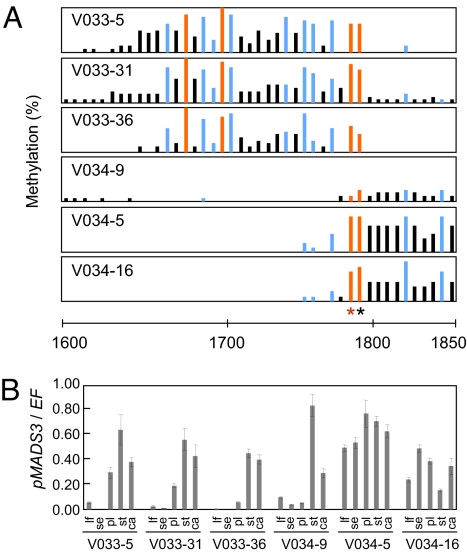
The 1701–1800 region responsible for pMADS3 ectopic expression contains 2 CG and 5 CNG sites. To identify the specific cytosine(s) responsible for the ect-pMADS3 phenotype, we further dissected the 1701–1800 sequence by expressing IRs for its subregions. This region contains a 53-bp sequence (nts 1712–1764) including 2 CCAATCA boxes that are highly conserved in several plant species, as revealed by shadowing and footprinting analyses (23) (Fig. S3). We focused on these sequences and expressed IR sequences containing the 2 CCAATCA boxes (nts 1601–1767; V033) and the region downstream of them (nts 1768–1900; V034). In both V033 and V034 plants, we observed ectopic expression of endogenous pMADS3 in petals (Fig. 4B). In all of the V033 plants examined, we detected methylation at all of the CG and CNG sites in the IR-targeted region (nts 1601–1767) (Fig. 4A). In addition, 2 CG sites at nts 1768 and 1771 immediately downstream of the targeted region were also methylated in these plants. These methylations are presumably due to a mechanism related to transitive silencing in which RNA silencing spreads outside target regions in both 3′ and 5′ directions in plants (24). In V034 plants, cytosines in the IR-targeted region including the 2 CG sites at nts 1768 and 1771 were methylated. In the V034–9 plant, only the 2 CG sites at nts 1768 and 1771 were methylated within the 1701–1800 region (Fig. 4A), besides a CNN methylation (nt 1766) that is unlikely to be responsible for the phenotype (see previous discussion). This plant still showed pMADS3 ectopic expression, although weakly (Fig. 4B). Taken together, the 2 CG sites at nts 1768 and 1771 were the only sites that were commonly methylated in all of the transformants showing the ect-pMADS3 phenotype. These results strongly suggest that methylation of the CGs at nts 1768 and/or 1771 is responsible for pMADS3 ectopic expression.
Fig. 4.
DNA methylation patterns and pMADS3 expression in V033 and V034 plants. (A) DNA methylation patterns in the 1600–1850 region. Bars represent proportions of methylated clones at each site; red, blue, and black bars represent CG, CNG, and CNN sites, respectively. The scale represents nucleotide numbers in pMADS3 intron 2 with the first nucleotide of intron 2 as nt 1. Asterisks indicate CG sites at nts 1768 (red) and 1771 (black). (B) pMADS3 mRNA levels in leaves (lf), sepals (se), petals (pl), stamens (st), and carpels (ca) of transgenic lines. Expression levels of pMADS3 relative to those of the elongation factor gene (EF) were measured by RT-qPCR. Mean ± SE (n = 3).
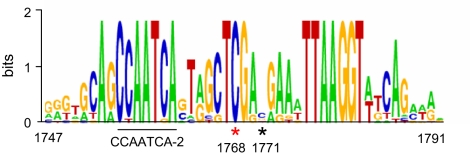
We aligned the sequences corresponding to the 1701–1800 region of pMADS3 intron 2 in 15 AG orthologs of 13 plant species belonging to 9 different families (Table S1). This alignment allowed us to find a highly conserved region immediately downstream of the 2 CCAATCA boxes (Fig. 5 and Fig. S3). Notably, the CG at nt 1768, 1 of the 2 CGs (nts 1768 and 1771) correlated with pMADS3 ectopic expression, is within this region and was perfectly conserved in all of the plant species examined. The sequence surrounding this CG, TAGCTCGA, was also highly conserved among the plant species, suggesting that this sequence motif is a cis-element that plays a role in transcriptional regulation of pMADS3. In an attempt to functionally characterize the TAGCTCGA motif as a cis-element, we fused the entire 4-kb sequence of pMADS3 intron 2 upstream of the CaMV 35S minimal promoter connected to the GUS reporter gene, as reported for Arabidopsis AG (12). Unfortunately, however, this construct gave no GUS activity in transgenic petunia lines for unknown reasons; therefore, we were unable to characterize the in vivo activity of this motif as a cis-element.
Fig. 5.
Sequence logo for conserved motifs in the intron 2 of pMADS3 homologs. The sequence logo (created by weblogo.berkeley.edu) was generated from 15 pMADS3 homologs of 13 plant species belonging to 9 different families (see Table S1). Asterisks indicate CG sites at nts 1768 (red) and 1771 (black). Numbers at the bottom indicate the positions in pMADS3 intron 2.
Discussion
On the basis of the unexpected expression patterns of endogenous pMADS3 incidentally observed in pMADS3:GUS transgenic petunia plants, we characterized the molecular mechanism underlying this epigenetic phenomenon. The accumulation of aberrant transcripts including sense and antisense sequences for pMADS3 intron 2 in the transformants (9) led us to speculate that the aberrant transcripts may have triggered the disturbance of endogenous pMADS3 expression. To prove this hypothesis and further investigate the underlying molecular mechanisms, we expressed IR sequences for pMADS3 intron 2, demonstrating that the expression of the IR sequences did induce the ect-pMADS3 phenotype. Small dsRNAs for pMADS3 intron 2 were detected in both pMADS3:GUS and IR-targeted transformants showing pMADS3 ectopic expression. Moreover, we found that DNA methylation of a particular CG was strongly correlated with pMADS3 ectopic expression in both transformants. On the basis of these observations, we propose the following mechanism for the ect-pMADS3 phenomenon occurred in pMADS3:GUS transformants: transgene-derived aberrant transcripts including pMADS3 intron 2 sequences were converted into dsRNAs by a reaction such as RdRP reaction and dsRNAs were processed into small dsRNAs, which triggered RdDM in their homologous DNA sequences in endogenous pMADS3, leading to the induction of its ectopic expression. We have not provided more direct evidence for the involvement of RdDM by using RNAi pathway mutants, such as the Dicer mutant; however, this model seems to be quite reasonable considering generally accepted outcomes resulting from the expression of IR constructs into plants.
Several reports have shown that DNA methylation in transcriptional regulatory regions is associated with gene silencing (TGS) in plants and mammals (25, 26). Deng et al. (27) provided evidence that DNA methylation within, or near sequences of, a positive cis-element (enhancer) interferes with the binding of a cognate transcription factor to this cis-element, which in turn causes TGS. The pMADS3 silencing observed in some pMADS3:GUS lines is presumably due to this mechanism (9, 28). A mechanism analogous to this can account for the up-regulation of pMADS3 in the ect-pMADS3 phenomenon. If DNA methylation occurs in a negative cis-element (silencer), instead of a positive cis-element (enhancer), the binding of its cognate transcriptional repressor to this cis-element would be interfered with, leading to derepression of transcription. So far, such regulation has been reported for Igf2, an imprinted gene in mice. CG methylation in a negative regulatory sequence of Igf2 derepressed this gene by interfering with the binding of a transcriptional repressor GCF2 to the sequence (29, 30).
Detailed dissection of pMADS3 intron 2 by IR expression followed by DNA methylation analysis revealed a specific CG, the methylation of which is strongly correlated with the up-regulation of pMADS3 transcription. This CG is located within the TAGCTCGA motif that is highly conserved among AG orthologs in various plant species. Taken together, this sequence motif is most likely a negative cis-element involved in spatiotemporal transcriptional regulation of pMADS3, and CG methylation within this motif presumably interferes with the binding of its cognate transcriptional repressor, leading to transcriptional derepression of pMADS3. Spatiotemporal regulation of the transcription of class-C floral homeotic genes has been extensively investigated as a model system to study floral-organ specification. Our finding of a putative negative cis-element for pMADS3 transcription may shed light on the regulation of class-C genes. In Arabidopsis, although positive regulation of AG had been well defined, characterization of its negative regulation, which is crucial for precise patterning of AG expression, has rather lagged behind. Negative regulators of AG expression, such as AP2, LEUNIG, SEUSS, and BELLRINGER, have been identified and all of them act through AG intron 2 (15–17). However, cis-elements for AG repression, which are the target sites for those putative transcriptional repressors, have been elusive (12, 13). Recently, binding sites for BELLRINGER were identified in AG intron 2 (17). These sites were, however, absent in pMADS3 intron 2 in petunia, questioning the generality of this regulation (17). Complexes of SEUSS and SEPALLATA3 bind to the 3′ region of AG intron 2, thereby repressing AG expression (31), although the precise binding sites for this complex have not been identified (32). The TAGCTCGA motif in pMADS3 intron 2 was located in the region corresponding to that including the binding site of the SEUSS-SEPALLATA3 complex in AG intron 2. In addition, other sequence motifs highly conserved among plant species, such as the CCAATCA boxes, were located adjacent to the TAGCTCGA motif (Fig. S4). This finding led us to speculate that a transcriptional repressor complex (or complexes), such as the SEPALLATA3-SEUSS complex, may interact with this region, with each component binding to a different sequence motif. Unfortunately, our attempts to verify cis-element activity of the TAGCTCGA motif by using a GUS reporter system were unsuccessful. Such experiments, however, may be possible with Arabidopsis AG, because AG intron 2 reproduces the spatiotemporal pattern of AG in Arabidopsis when placed upstream of the reporter gene (12). We also attempted to detect sequence-specific DNA-binding activity in nuclear extracts from petunia flowers by gel-shift assays, but TACTCGA-specific binding activity was undetectable, possibly due to technical reasons.
In RNA-induced gene activation (RNAa) in human cultured cells, small dsRNAs homologous to promoter sequences induce transcriptional activation when added to cultured cells (7, 8). Apparently, RNAa resembles the ect-pMADS3 phenomenon in that small RNAs activate transcription. In the case of RNAa, however, significant changes were not detected in DNA methylation in the sequences targeted by small dsRNAs, but changes were detected in histone modification patterns (7). These observations obviously distinguish RNAa from the ect-pMADS3 phenomenon, in which DNA methylation but not histone modifications is involved. Thus, the 2 phenomena are unlikely to be closely related mechanistically.
Naturally occurring epimutation has been shown to have significant effects on plant growth and development. For example, an epiallele of Lcyc gene in Linaria vulgaris is silenced due to DNA methylation, resulting in the alteration of floral symmetry (33). DNA methylation in promoter sequences silenced the LeSPL-CNR gene in the tomato, resulting in repressed ripening of tomato fruits (34). Paramutation of the b1 locus in maize is another example of a naturally occurring epiallele (35). It has previously been observed in tobacco that TGS of a marker gene was inherited independent of RNA triggers, representing an artificially generated epiallele (36). The heritability of these epigenetic traits is usually based on DNA methylation of maintenance methylation sites. Presumably, RNA triggers are also involved in the generation of some, if not all, naturally occurring epialleles, as is the case of the b1 paramutation (6). The ect-pMADS3 phenotype is due to DNA methylation of maintenance methylation site(s) and is heritable over generations. Although it is an artificially induced one, the same could occur in nature because RNA triggers can be generated by RdRP or by hybridization of antisense RNA transcribed from many genomic regions (37) with their corresponding sense RNA. Thus, an active epiallele for a gene that is otherwise silent can be generated in nature by a mechanism similar to that involved in the ect-pMADS3 phenomenon, suggesting a potentially broader contribution of epimutations in the formation of natural variation and plant evolution. We fortuitously noticed the ect-pMADS3 phenotype because it is easily visible, which would otherwise have been missed because of its unpredictability. This situation is reminiscent of the finding of cosuppression, which was initially recognized as unexpected color changes in petunia flowers (38, 39).
At present, we have no information regarding the generality of ect-pMADS3-like regulation. Requirements for its occurrence are the presence of maintenance methylation sites in negative cis-elements of transcription and the generation of RNA triggers for them, which do not seem to be rare.
Materials and Methods
Plant Materials.
Wild-type petunia, P. hybrida cv. V26 (pure line), P. hybrida cv. Mitchell (pure line), P. hybrida cv. Surfinia Purple Mini (F1 hybrid), and transgenic petunia plants were grown under standard greenhouse conditions in a commercial potting medium.
Generation of IR Constructs and Plant Transformation.
Fragments of pMADS3 intron 2 corresponding to the regions shown in Fig. 1A were amplified by PCR with the primer sets listed in Table S2. The PCR fragments were cloned in sense and antisense orientation downstream of the CaMV 35S promoter and upstream of the octopin synthase terminator in a cloning vector, with a fragment of GUS (372 bp) inserted between the sense and antisense sequences as a spacer. These synthetic genes were cloned into the pBINPLUS (40) binary transformation vector and the resulting chimeric constructs were introduced into P. hybrida cv. V26 by means of Agrobacterium tumefaciens (GV3101)-mediated transformation (41). After regeneration on a selective medium, transformed petunia lines were transferred to a glasshouse.
Quantitative Real-time RT-PCR.
Total RNA was isolated by using Trizol reagents (Invitrogen) and treated with cloned DNase I (takara Bio). Synthesis of cDNA was carried out with Oligo-dT primers by using SuperScript II reverse transcriptase (Invitrogen). PCRs were performed by using SYBR Premix ExTaq (takara Bio) with a Thermal Cycler Dice real time PCR system (Takara). To normalize the data, transcript levels of target genes relative to those of the elongation factor gene as an internal control were calculated. The primers for pMADS3 were MD-c614F-RT (5′-CAAAATCCGAGCCAAAAAGA-3′) and MD-c761R-RT (5′-CCCAGGCATCAAGTTCATCT-3′), and those for EF were EF-F1-RT (5′-ACCACTGGTGGTTTTGAAGC-3′) and EF-R1-RT (5′-GGGTGGTAGCATCCATCTTG-3′).
Analysis of DNA Methylation.
Genomic DNA was extracted with DNeasy plant mini kit (Qiagen). One microgram of DNA was digested by EcoRI, treated with Proteinase K, and cleaned up by using Wizard DNA Clean-Up systems (Promega). Bisulfite treatment was performed by using BisulFast DNA modification kit (Toyobo) according to the manufacturer's instruction. PCR reactions were performed as described by Mishiba et al. (42). Primers for the PCR reactions were MI-1482F-Bs (5′-GGYTGYTGAAAATGGAYGGTTG-3′) and MI-1894R-Bs (5′-CAATCTRTCTACTCAAACATRACATTRA-3′). The amplified PCR fragments were gel-purified and cloned into pGEM-T easy vector (Promega), and then 9–12 independent clones were sequenced.
Supplementary Material
Acknowledgments.
We thank Sumiko Tatsumi and Tomiko Yasuhara for technical assistance. This work was supported by a Grant-in-Aid for Scientific Research (to H.T.) and a Research Fellowship for Young Scientists (to K.S.) from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809294106/DCSupplemental.
References
- 1.Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 3.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 4.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 6.Chandler VL. Paramutation: From maize to mice. Cell. 2007;128:641–645. doi: 10.1016/j.cell.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Li LC, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janowski BA, et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 9.Kapoor M, et al. Transgene-triggered, epigenetically regulated ectopic expression of a flower homeotic gene pMADS3 in Petunia. Plant J. 2005;43:649–661. doi: 10.1111/j.1365-313X.2005.02481.x. [DOI] [PubMed] [Google Scholar]
- 10.Tsuchimoto S, van der Krol AR, Chua NH. Ectopic expression of pMADS3 in transgenic petunia phenocopies the petunia blind mutant. Plant Cell. 1993;5:843–853. doi: 10.1105/tpc.5.8.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busch MA, Bomblies K, Weigel D. Activation of a floral homeotic gene in Arabidopsis. Science. 1999;285:585–587. doi: 10.1126/science.285.5427.585. [DOI] [PubMed] [Google Scholar]
- 13.Deyholos MK, Sieburth LE. Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell. 2000;12:1799–1810. doi: 10.1105/tpc.12.10.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohmann JU, et al. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, Meyerowitz EM. LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development. 1995;121:975–991. doi: 10.1242/dev.121.4.975. [DOI] [PubMed] [Google Scholar]
- 16.Franks RG, Wang C, Levin JZ, Liu Z. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development. 2002;129:253–263. doi: 10.1242/dev.129.1.253. [DOI] [PubMed] [Google Scholar]
- 17.Bao X, Franks RG, Levin JZ, Liu Z. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell. 2004;16:1478–1489. doi: 10.1105/tpc.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzlaff M, O'Dell M, Hellens R, Flavell RB. Developmentally and transgene regulated nuclear processing of primary transcripts of chalcone synthase A in petunia. Plant J. 2000;23:63–72. doi: 10.1046/j.1365-313x.2000.00793.x. [DOI] [PubMed] [Google Scholar]
- 19.O'Dell M, Metzlaff M, Flavell RB. Post-transcriptional gene silencing of chalcone synthase in transgenic petunias, cytosine methylation and epigenetic variation. Plant J. 1999;18:33–42. [Google Scholar]
- 20.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 21.Johnson LM, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubert D, et al. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong RL, Hamaguchi L, Busch MA, Weigel D. Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell. 2003;15:1296–1309. doi: 10.1105/tpc.009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen BO, Albrechtsen M. Evidence implying only unprimed RdRP activity during transitive gene silencing in plants. Plant Mol Biol. 2005;58:575–583. doi: 10.1007/s11103-005-7307-4. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita Y, et al. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 2007;49:38–45. doi: 10.1111/j.1365-313X.2006.02936.x. [DOI] [PubMed] [Google Scholar]
- 26.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 27.Deng G, Chen A, Pong E, Kim YS. Methylation in hMLH1 promoter interferes with its binding to transcription factor CBF and inhibits gene expression. Oncogene. 2001;20:7120–7127. doi: 10.1038/sj.onc.1204891. [DOI] [PubMed] [Google Scholar]
- 28.Kapoor M, et al. Role of petunia pMADS3 in determination of floral organ and meristem identity, as revealed by its loss of function. Plant J. 2002;32:115–127. doi: 10.1046/j.1365-313x.2002.01402.x. [DOI] [PubMed] [Google Scholar]
- 29.Eden S, et al. An upstream repressor element plays a role in Igf2 imprinting. EMBO J. 2001;20:3518–3525. doi: 10.1093/emboj/20.13.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murrell A, et al. An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO Rep. 2001;2:1101–1106. doi: 10.1093/embo-reports/kve248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z. Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA. 2004;101:11494–11499. doi: 10.1073/pnas.0403055101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sridhar VV, Surendrarao A, Liu Z. APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development. 2006;133:3159–3166. doi: 10.1242/dev.02498. [DOI] [PubMed] [Google Scholar]
- 33.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 34.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 35.Stam M, Belele C, Dorweiler JE, Chandler VL. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 2002;16:1906–1918. doi: 10.1101/gad.1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones L, Ratcliff F, Baulcombe DC. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol. 2001;11:747–757. doi: 10.1016/s0960-9822(01)00226-3. [DOI] [PubMed] [Google Scholar]
- 37.Stolc V, et al. Identification of transcribed sequences in Arabidopsis thaliana by using high-resolution genome tiling arrays. Proc Natl Acad Sci USA. 2005;102:4453–4458. doi: 10.1073/pnas.0408203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Engelen FA, et al. pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Res. 1995;4:288–290. doi: 10.1007/BF01969123. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA. Chalcone synthase cosuppression phenotypes in petunia flowers: Comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol Biol. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- 42.Mishiba K, et al. Consistent transcriptional silencing of 35S-driven transgenes in gentian. Plant J. 2005;44:541–556. doi: 10.1111/j.1365-313X.2005.02556.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.